Introduction
Pancreatobiliary tract cancer is a term used to
describe malignant carcinoma in pancreatic, gallbladder, and
extrahepatic bile ducts. Pancreatobiliary tract cancer is a highly
fatal cancer (1–4). Detection of pancreatobiliary tract
cancer is difficult because it lacks typical clinical symptoms and
because of its anatomical location, especially in the early stage.
Biomarker discovery is therefore important for identification of
increased risk, early diagnosis, and prediction of response to
therapy and prognosis of pancreatobiliary tract cancer (5,6). The
serum carbohydrate antigen 19-9 (CA19-9) level is one clinically
useful biomarker related to pancreatobiliary tract cancer (6–8).
However, there is also still a limit to detect pancreatobiliary
tract cancer using the existing methods (9). Therefore, further studies are needed
to develop biomarkers for pancreatobiliary tract cancer.
Recently, microRNAs (miRNAs) have been reported as
potential biomarkers for various types of cancers including
pancreatobiliary tract cancer (10–15).
They are single-stranded non-coding RNAs of ~21–23 nucleotides in
length that can affect gene expression at the post-transcriptional
level by binding to the three prime untranslated region (3′-UTR) of
target messenger RNAs (16). It has
been noted that miRNAs could be detected in body fluids (17). Several studies have suggested that
miRNAs circulating in blood are useful as biomarkers for the
diagnosis of pancreatic cancer (10–14,18).
Recent studies have also focused on salivary miRNAs
as biomarkers for various diseases (19). Saliva collection is simple and
non-invasive. Saliva contains proteins, nucleic acids, and hormones
originating from both local and systemic sources. Notably, salivary
miRNAs have been shown to be highly stable because exosomes or
protein complexes protect them (20). A few studies have suggested that
salivary miRNAs are useful as biomarkers for the diagnosis of
pancreatic cancer (21,22). However, in these studies, exosomes
were not extracted from saliva. Most miRNAs in human saliva likely
exist in exosomes (23,24). For diagnosis of pancreatobiliary
tract cancer, miRNAs in salivary exosomes may be superior to those
in saliva.
To the best of our knowledge, there is no literature
regarding the relationship between miRNAs in salivary exosomes and
pancreatobiliary tract cancer. On the other hand, one report is
available regarding the relationship between miRNAs in blood
exosomes and pancreatic cancer (18). In that study, four miRNAs (miR-1246,
miR-3976, miR-4306, and miR-4644) in serum exosomes were identified
as biomarkers for pancreatic cancer. miRNA expression profiles in
saliva could be similar to those in serum (25). Therefore, in the present study, we
hypothesized that miR-1246, miR-3976, miR-4306, and miR-4644 in
salivary exosomes might be useful biomarkers for pancreatobiliary
tract cancer. Thus, the aim of this study was to examine whether
these four miRNAs in salivary exosomes could be useful biomarkers
for pancreatobiliary tract cancer.
Materials and methods
Study population
The study design was a case-control study. It was
not possible to estimate the sample size preliminarily, because
there was no prior information on which to base a sample size
(26).
Twelve patients (6 males and 6 females) with
pancreatobiliary tract cancers were referred to the Department of
Preventive Dentistry, Okayama University Hospital for saliva
collection in the morning before the onset of cancer therapy from
July 2013 to July 2014.
As a control group, 13 healthy participants were
recruited at the Department of Preventive Dentistry, Okayama
University Hospital from August 2014 to December 2014 for saliva
collection in the morning. Inclusion criteria for healthy control
participants were >50 years of age and no history of any cancer.
To avoid the effect of systemic conditions on circulating miRNA
expressions, the exclusion criteria for healthy control
participants were as follows: diabetes (27); pulmonary diseases (28); cardiovascular diseases (29); kidney diseases (30); liver diseases (31); and autoimmune diseases (32) at the time of saliva collection.
According to the inclusion/exclusion criteria, the control
participants with hypertension and dyslipidemia (2 participants)
and hyperuricemia (1 participant) were included in this study. This
study was approved by the Ethics Committee, Okayama University
Graduate School of Medicine, Dentistry and Pharmaceutical Sciences
and Okayama University Hospital (no. 1506–052). Written, informed
consent was obtained from all participants.
General status examination
Measurements were performed before the onset of
cancer therapy. Medical charts were reviewed to obtain information
about cancer and the body mass index (BMI). A personal interview
was performed to gather information about smoking habits
(pack-years). Serum levels of hemoglobin A1c (HbA1c), C-reactive
protein (CRP), albumin, carcinoembryonic antigen (CEA), and CA19-9
were evaluated. Concentrations of HbA1c, CRP, and albumin were
measured by the high-performance liquid chromatography method, the
latex agglutination method, and the bromocresol green albumin
method, respectively. Concentrations of CEA and CA19-9 were
measured by electrochemiluminescence immunoassays.
Saliva collection
Unstimulated whole saliva was collected as reported
previously with minor modification (24). Briefly, saliva was collected in the
morning (7:00 a.m.–12:00 noon). During the collection period,
participants were seated straight up and instructed to refrain from
speaking or swallowing. They allowed the saliva to accumulate in
the floor of the mouth and then spit it through a funnel into a
tube kept on ice. At least 0.5 ml of unstimulated whole saliva was
collected. After collection of saliva, it was stored at 4°C for up
to 6 h, after which it was stored at −80°C until use.
Exosome isolation
Exosomes were isolated from saliva samples (0.5–1.0
ml) using Total Exosome Isolation Reagent (Invitrogen, Carlsbad,
CA, USA), in accordance with the manufacturer's instructions
(33).
RNA extraction
Total Exosome RNA and Protein Isolation Kits
(Invitrogen) were used for extraction of total RNA from exosome
samples. Total RNA was extracted in accordance with the
manufacturer's instructions. After extraction of total RNA from
salivary exosomes, their quality was confirmed using the Agilent
2100 Bioanalyzer and the Agilent RNA 6000 Pico Kit (both from
Agilent Technologies, Santa Clara, CA, USA).
Quantitative real-time PCR (RT-qPCR)
To compare miRNA expression between the control and
cancer group, TaqMan RT-qPCR assays were performed. TaqMan MicroRNA
Assays (life Technologies, Carlsbad, CA, USA) were used for the
RT-qPCR analyses performed on the Mx3000P Real-Time QPCR System
(Agilent Technologies) according to the manufacturer's instructions
(34,35). Briefly, reverse transcription (RT)
enzymes including specific RT primers for each miRNA and total RNA
sample were mixed in a tube. RT run conditions were then set. The
plate was incubated at 16°C for 30 min, 42°C for 30 min, and 85°C
for 5 min, and then held at 4°C. Products of the RT reaction were
stored at −20°C until use. RT-qPCR was performed in triplicate
using TaqMan Universal Master Mix II, no UNG (Applied
Biosystems/life Technologies, Carlsbad, CA, USA). After holding at
95°C for 10 min, 50 thermal cycles (95°C for 15 sec and 60°C for 60
sec) were run. Threshold cycle (Ct) values were determined using
the background-based threshold (cycle-range, 5–8) calculated by the
MxPro Mx3000P v4.10 software (Stratagene/Agilent Technologies,
Edinburgh, UK). Data with a raw Ct>40 for each miRNA were
treated as Ct=40 for the subsequent statistical test (36). U6 snRNA was considered suitable as
an internal control (22). The
relative expression rates of each miRNA were calculated using U6
snRNA expression.
Discrimination power of candidate miRNA
biomarkers for pancreatobiliary tract cancer
To evaluate the discrimination power of candidate
miRNA biomarkers for pancreatobiliary tract cancer, receiver
operating characteristic (ROC) curves (37) were constructed. The ROC curves were
obtained by plots of the sensitivity (true-positive rate) and
1-specificity (false-positive rate) of the diagnostic test at
various cut-off values. The ROC curves graphically show the
relationship between the true- and the false-positive rate
according to the various cut-off values. The area under the curve
(AUC) was estimated to assess the predictive power. To decide the
optimal threshold value, the Youden index (sensitivity +
specificity − 1) was used (38).
Statistical analyses
Characteristics of the control and cancer group are
represented as continuous variables [age, BMI, smoking habit
(pack-years), HbA1c, CRP, albumin, CEA, and CA19-9] and categorical
variables (gender, cancer site, and cancer stage). The Mann-Whitney
U test, the Chi-square test, and Fisher's exact test were used to
assess significant differences in clinical variables between the
two groups, as appropriate. The Mann-Whitney U test was used to
compare the relative expression ratios of each miRNA of the control
and cancer group. To calculate the P-value for the AUC, a
non-parametric test for AUC=0.5 was performed. Two-sided P<0.05
values were considered to represent significant differences. To
assess the correlations of variables in the cancer group,
Spearman's rank correlation coefficient and its P-value were
calculated for age (years), HbA1c (NGSP) (%), CRP (mg/dl), albumin
(g/dl), CEA (ng/ml), CA19-9 (U/ml), relative expression ratios of
the miRNAs, and smoking (pack-years) as continuous variables, and
cancer stage (I–III, IVa, and IVb) as a categorical variable. These
statistical analyses were performed using SPSS software version 20
(SPSS, Inc., Chicago, Il, USA).
Results
Characteristics of the participants
Table I summarizes
the characteristics of all participants. There were no significant
differences in age and gender between the two groups. However, a
significant difference in the percent of current smokers was
observed (P=0.015). Pancreatic cancer was predominant in the
patient group (75.0%), and 83.3% of patients were diagnosed as
having the most advanced (IVa or IVb) stage. In addition, 58.3 and
66.7% of patients showed values greater than the reference values
of CEA (10 ng/ml) and CA19-9 (100 U/ml), respectively.
 | Table ICharacteristics of the control and
cancer group. |
Table I
Characteristics of the control and
cancer group.
|
Characteristics | Control
(n=13) | Cancer
(n=12) | P-valuea |
|---|
| Age (years) | | | |
| Median
(range) | 66 (53–83) | 65 (45–84) | 0.728 |
| Gender n (%) | | | |
| Male | 6 (46.2) | 6 (50.0) | 0.848 |
| Female | 7 (53.8) | 6 (50.0) | |
| Smoking, n (%) | | | |
| Current | 0 (0.0) | 5 (41.7) | 0.015 |
| Cancer site, n
(%) | | | |
| Bile duct
cancer | | 2 (16.7) | |
| Gallbladder
cancer | | 1 (8.3) | |
| Pancreatic
cancer | | 9 (75.0) | |
| T categoryb, n (%) | | | |
| T1 | | 0 (0.0) | |
| T2 | | 1 (8.3) | |
| T3 | | 2 (16.7) | |
| T4 | | 7 (58.3) | |
| Unknown | | 2 (16.7) | |
| N categoryb, n (%) | | | |
| N0 | | 6 (50.0) | |
| N1 | | 1 (8.3) | |
| N2 | | 1 (8.3) | |
| N3 | | 1 (8.3) | |
| Unknown | | 3 (25.0) | |
| M categoryb, n (%) | | | |
| M0 | | 6 (50.0) | |
| M1 | | 5 (41.7) | |
| Unknown | | 1 (8.3) | |
| Cancer
stageb, n (%) | | | |
| I | | 0 (0.0) | |
| II | | 1 (8.3) | |
| III | | 1 (8.3) | |
| IVa | | 4 (33.3) | |
| IVb | | 6 (50.0) | |
| HbA1c (NGSP)
(%) | | | |
| Median
(range) | | 5.9 (4.9–8.1) | |
| CRP (mg/dl) | | | |
| Median
(range) | | 0.42
(0.06–5.47) | |
| Albumin (g/dl) | | | |
| Median
(range) | | 3.7 (2.9–4.5) | |
| CEA (ng/ml) | | | |
| Median
(range) | | 12 (1–1,379) | |
| CA19-9 (U/ml) | | | |
| Median
(range) | | 414 (1–38,864) | |
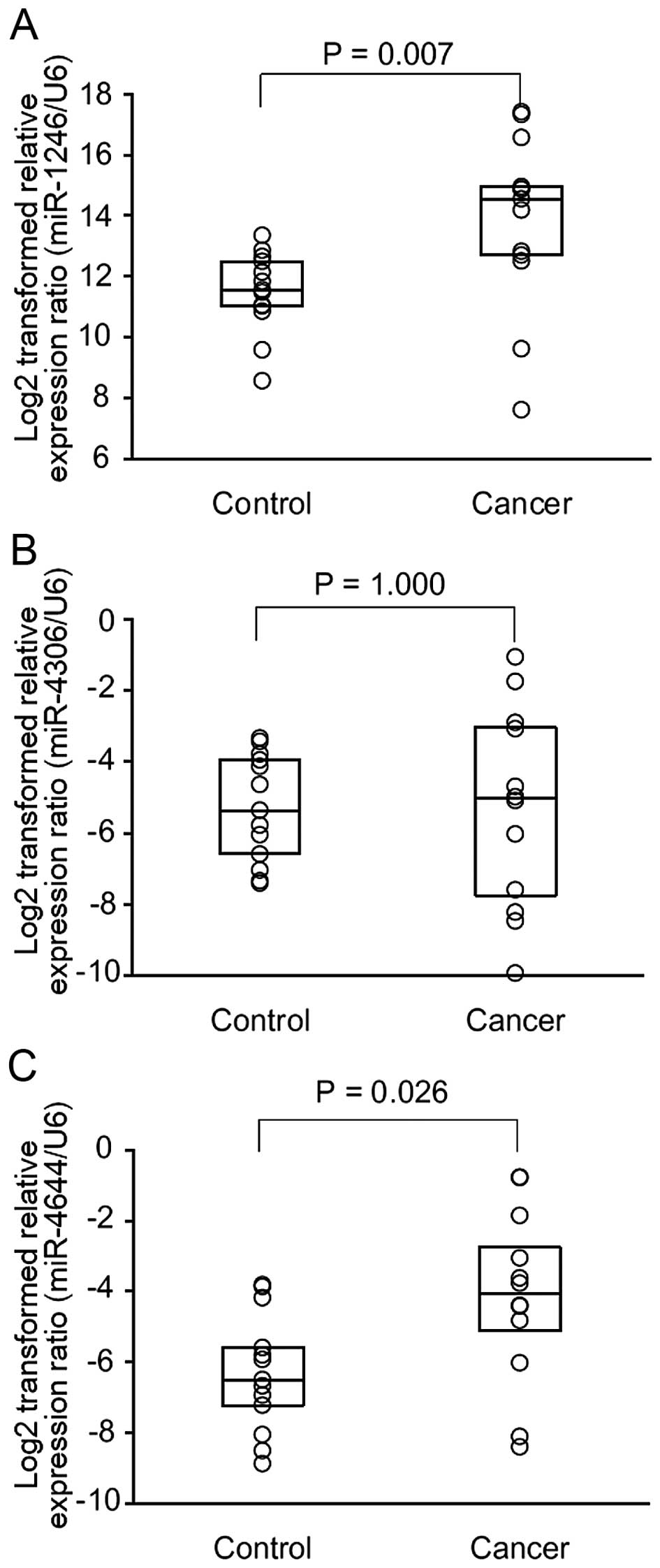
RT-qPCR validation
After the RT-qPCR assays, miR-3976 was excluded from
the following analyses because 24 of 25 (96.0%) samples had high
(>40) raw Ct-values. The number of samples having high (>40)
raw Ct-values of expression levels of miR-1246, miR-4306, miR-4644,
and U6 snRNA were 0, 2, 1 and 0, respectively. Therefore, miR-1246,
miR-4306, and miR-4644 were considered as candidate miRNAs. For the
control group, the median (25th and 75th percentile) values of
log2-transformed relative expression of miR-1246, miR-4306, and
miR-4644 were 11.6 (10.9, 12.6), −5.4 (−6.8, −3.9), and −6.5 (−7.6,
−4.9), respectively. For the cancer group, those of miR-1246,
miR-4306, and miR-4644 were 14.7 (12.6, 16.2), −5.0 (−8.1, −2.9),
and −4.1 (−5.7, −2.2), respectively. The Mann-Whitney U test showed
that the relative expression ratios of miR-1246 and miR-4644 were
significantly higher in the cancer than these ratios in the control
group (Fig. 1).
Discrimination power of miR-1246 and
miR-4644 for pancreatobiliary tract cancer
To evaluate the discrimination power of miR-1246 and
miR-4644 for pancreatobiliary tract cancer, ROC curves were
constructed (Fig. 2). For miR-1246,
the results yielded an AUC of 0.814 [95% confidence interval (CI),
0.616–1.000; cut-off, 13.77; sensitivity, 0.667; specificity,
1.000; P=0.008]. For miR-4644, the results yielded an AUC of 0.763
(95% CI, 0.564–0.961; cut-off, −5.205; sensitivity, 0.750;
specificity, 0.769; P=0.026). For the combination of miR-1246 and
miR-4644, the results yielded an increased AUC of 0.833 (95% CI,
0.630–1.000; cut-off, 8.035; sensitivity, 0.833; specificity,
0.923; P= 0.005).
 | Figure 2ROC curve analysis for the
discriminatory power of salivary exosomal miRNAs for the control
and cancer group. ROC curves for (A) miR-1246, (B) miR-4644 and (C)
combination of miR-1246 and miR-4644 are represented.
Log2-transformed relative expression ratio using U6 snRNA as
reference was used for each miRNA. P-value was calculated using
non-parametric test for AUC=0.5. Cut-off, sensitivity, specificity,
PPV, and NPV were optimized using the maximum Youden index. Black
circle represents the plot with these optimized values. ROC,
receiver operating characteristic; miRNAs, microRNAs; AUC, area
under the curve; PPV, positive predictive value; NPV, negative
predictive value. |
Correlations of variables in the cancer
group
In the cancer group, there were significant
correlations between CRP and cancer stage (r=0.822), between
albumin and HbA1c (r=0.582), between albumin and CRP (r=−0.711),
between CA19-9 and miR-1246 (r=0.818), between miR-1246 and
miR-4644 (r=0.671), between smoking and HbA1c (r=0.579), and
between smoking and albumin (r=0.647) (Table II).
 | Table IICorrelation of variables in the
cancer group. |
Table II
Correlation of variables in the
cancer group.
| Variables | Values | Age
(years) | Cancer
stage
(I–III, IVa, and IVb) | HbA1c
(NGSP) (%) | CRP
(mg/dl) | Albumin
(g/dl) | CEA
(ng/ml) | CA19-9
(U/ml) | miR-1246a | miR-4306a | miR-4644a | Smoking
(pack-years) |
|---|
| Age (years) | rb | 1.000 | | | | | | | | | | |
| P-valueb | | | | | | | | | | | |
| Cancer stage | | | | | | | | | | | | |
| (I–III, IVa, and
IVb) | r | −0.145 | 1.000 | | | | | | | | | |
| P-value | 0.652 | | | | | | | | | | |
| HbA1c (NGSP)
(%) | r | 0.483 | 0.157 | 1.000 | | | | | | | | |
| P-value | 0.111 | 0.626 | | | | | | | | | |
| CRP (mg/dl) | r | −0.311 | 0.822d | −0.209 | 1.000 | | | | | | | |
| P-value | 0.326 | 0.001 | 0.514 | | | | | | | | |
| Albumin (g/dl) | r | 0.315 | −0.192 | 0.582c | −0.711d | 1.000 | | | | | | |
| P-value | 0.318 | 0.550 | 0.047 | 0.009 | | | | | | | |
| CEA (ng/ml) | r | −0.196 | 0.527 | 0.179 | 0.214 | 0.302 | 1.000 | | | | | |
| P-value | 0.541 | 0.078 | 0.578 | 0.505 | 0.340 | | | | | | |
| CA19-9 (U/ml) | r | −0.455 | 0.321 | 0.007 | 0.231 | 0.060 | 0.098 | 1.000 | | | | |
| P-value | 0.137 | 0.310 | 0.983 | 0.470 | 0.854 | 0.762 | | | | | |
| miR-1246a | r | −0.291 | 0.122 | −0.112 | 0.116 | 0.004 | −0.091 | 0.818d | 1.000 | | | |
| P-value | 0.359 | 0.705 | 0.728 | 0.721 | 0.991 | 0.779 | 0.001 | | | | |
| miR-4306a | r | −0.123 | 0.168 | −0.098 | 0.049 | 0.134 | 0.406 | 0.336 | 0.510 | 1.000 | | |
| P-value | 0.704 | 0.602 | 0.761 | 0.880 | 0.679 | 0.191 | 0.286 | 0.090 | | | |
| miR-4644a | r | −0.077 | −0.252 | −0.249 | −0.315 | 0.250 | 0.063 | 0.406 | 0.671c | 0.790d | 1.000 | |
| P-value | 0.812 | 0.430 | 0.435 | 0.318 | 0.434 | 0.846 | 0.191 | 0.017 | 0.002 | | |
| Smoking
(pack-years) | r | 0.133 | 0.043 | 0.579c | −0.320 | 0.647c | 0.281 | 0.335 | 0.133 | 0.109 | 0.156 | 1.000 |
| P-value | 0.681 | 0.896 | 0.048 | 0.310 | 0.023 | 0.377 | 0.287 | 0.681 | 0.736 | 0.628 | |
Discussion
This study was performed to clarify whether
miR-1246, miR-3976, miR-4306, and miR-4644 levels in salivary
exosomes were useful as potential biomarkers for pancreatobiliary
tract cancer. Among these four miRNAs, two miRNAs (miR-1246 and
miR-4644) in salivary exosomes showed significantly higher
expression in pancreatobiliary tract cancer patients than that
noted in healthy control participants. In addition, in the ROC
curve analysis, the AUCs of both miR-1246 and miR-4644 were
>0.7, indicating fair discriminatory power (39). These results indicate that miR-1246
and miR-4644 in salivary exosomes could be useful biomarkers for
screening pancreatobiliary tract cancer patients. On the other
hand, in the present study, overlap of the relative expression
profiles of miR-1246 and miR-4644 was also found in the cancer and
control groups. This suggests that miR-1246 and miR-4644 might be
non-specific screening biomarkers for pancreatobiliary tract
cancer.
Compared to other body fluids including blood,
saliva may be immediately exposed to the outside environment and
confounded by a wide variety of environmental factors (19). However, some molecules in saliva are
highly stable and represent the body's health status. In this study
using salivary exosomes, the sensitivity and specificity for
detecting pancreatobiliary tract cancer were 66.7 and 100.0% for
miR-1246 (cut-off, 13.77) and 75.0 and 76.9% for miR-4644 (cut-off,
−5.205), respectively. In serum, it is reported that sensitivity
and specificity for pancreatic cancer of miR-1246 were 79.1 and
82.0%, respectively (40). It is
also known that, for pancreatic cancer, in plasma they were 82 and
73% for miR-885-5p, 82 and 82% for miR-22-3p, and 82 and 55% for
miR-642b-3p, respectively (11).
Taking these findings into account, the sensitivity for
pancreatobiliary tract cancer of miR-1246 and miR-4644 in salivary
exosomes seems to be slightly lower than other miRNAs in blood, and
their specificity seems to be similar or greater than that of other
miRNAs in blood.
In the present findings, the combination of miR-1246
and miR-4644 in salivary exosomes increased the sensitivity for
pancreatobiliary tract cancer. This is consistent with previous
results, indicating that concomitant pancreatic cancer-initiating
cell and miRNA marker expression strengthened the sensitivity for
pancreatic cancer (18). In the
present study, since drawing blood from healthy participants was
difficult ethically, we could not combine miRNAs in salivary
exosomes and serum markers to assess the predictive power. However,
serum markers would further strengthen the sensitivity for
pancreatobiliary tract cancer of miR-1246 and miR-4644 in salivary
exosomes.
Serum CA19-9 is one of the most studied and
validated serum biomarkers for pancreatobiliary tract cancer
(6–8). In the present study, the serum CA19-9
level was significantly correlated with the expression of miR-1246
in the cancer group. This observation indicates that miR-1246 in
salivary exosomes was associated with serum biomarkers for
pancreatobiliary tract cancer. On the other hand, the expression of
miR-4644 in salivary exosomes was significantly correlated with the
expression of miR-1246, but not with serum CA19-9. The relationship
between miR-4644 in salivary exosomes and the onset of
pancreatobiliary tract cancer may be indirect.
miR-1246 has been found to be aberrantly expressed
in pancreatic cancer tissue (41),
and such a condition could contribute to the increased miRNA in
blood. Due to the extensive blood supply in salivary glands, saliva
is considered to be a terminal product of blood circulation, and
molecules that are present in blood are also present in saliva.
Therefore, it is feasible that miR-1246 in salivary exosomes
originated from pancreatobiliary tract cancer tissue. However,
further studies are needed to clarify this point.
Studies have investigated the relationship between
salivary miRNA profiles and pancreatobiliary tract cancer. For
instance, a clinical study showed that miR-21, miR-23a, miR-23b,
and miR-29c were significantly upregulated in saliva of pancreatic
cancer patients compared to controls (21). Another clinical study also reported
that salivary miR-3679-5p was significantly downregulated and
miR-940 was significantly upregulated in pancreatic cancer
(22). Among the present findings,
miR-1246 and miR-4644 in salivary exosomes were confirmed to show
significant cancer-related increases. The present and previous
findings support the notion that salivary miRNAs are useful
biomarkers for pancreatobiliary tract cancer. A saliva test that is
less invasive than a blood test or endoscopy may be used as a broad
screening test to identify pancreatobiliary tract cancer patients
who would need further screening.
The limitations of this study are as follows. First,
external validity was limited because all participants were
recruited at the Okayama University Hospital. In addition, the
cancer patients were mainly diagnosed as having advanced-stage
disease when they participated in this study. Further studies
including patients with chronic pancreatitis and early stage of
pancreatobiliary tract cancer are necessary to improve the validity
of miR-1246 and miR-4644 in salivary exosomes as screening markers
of pancreatobiliary tract cancer. Furthermore, overall screening of
miRNAs in salivary exosomes using a microarray will be necessary to
discover a new biomarker in pancreatobiliary tract cancer
patients.
In conclusion, the present results demonstrated that
miR-1246 and miR-4644 in salivary exosomes could be useful
biomarkers for identification of patients with pancreatobiliary
tract cancer. We hope that clinical use of simple and non-invasive
salivary tests will contribute to the screening of pancreatobiliary
tract cancer patients and improve their survival.
Acknowledgments
This study was supported by a Grant-in-Aid for
Scientific Research (26670904) from the Ministry of Education,
Culture, Sports, Science and Technology, Tokyo, Japan.
Abbreviations:
|
AUC
|
area under the curve
|
|
BMI
|
body mass index
|
|
CA19-9
|
carbohydrate antigen 19-9
|
|
CEA
|
carcinoembryonic antigen
|
|
CRP
|
C-reactive protein
|
|
HbA1c
|
hemoglobin A1c
|
|
ROC
|
receiver operating characteristic
|
|
RT
|
reverse transcription
|
|
miRNAs
|
microRNAs
|
|
Ct
|
threshold cycle
|
References
|
1
|
Saif MW: Advancements in the management of
pancreatic cancer: 2013. JOP. 14:112–118. 2013.PubMed/NCBI
|
|
2
|
Benavides M, Antón A, Gallego J, Gómez MA,
Jiménez-Gordo A, La Casta A, Laquente B, Macarulla T,
Rodríguez-Mowbray JR and Maurel J: Biliary tract cancers: SEOM
clinical guidelines. Clin Transl Oncol. 17:982–987. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shaffer EA: Gallbladder cancer: The
basics. Gastroenterol Hepatol (N Y). 4:737–741. 2008.
|
|
4
|
Im JH, Seong J, Lee IJ, Park JS, Yoon DS,
Kim KS, Lee WJ and Park KR: Surgery alone versus surgery followed
by chemotherapy and radiotherapy in resected extrahepatic bile duct
cancer: Treatment outcome analysis of 336 patients. Cancer Res
Treat. 48:583–595. 2016. View Article : Google Scholar :
|
|
5
|
Poruk KE, Firpo MA, Adler DG and Mulvihill
SJ: Screening for pancreatic cancer: Why, how, and who? Ann Surg.
257:17–26. 2013. View Article : Google Scholar
|
|
6
|
Zeng X and Tao H: Diagnostic and
prognostic serum marker of cholangiocarcinoma (Review). Oncol Lett.
9:3–8. 2015.
|
|
7
|
Ballehaninna UK and Chamberlain RS: The
clinical utility of serum CA 19-9 in the diagnosis, prognosis and
management of pancreatic adenocarcinoma: An evidence based
appraisal. J Gastrointest Oncol. 3:105–119. 2012.PubMed/NCBI
|
|
8
|
Lin MS, Huang JX and Yu H: Elevated serum
level of carbohydrate antigen 19-9 in benign biliary stricture
diseases can reduce its value as a tumor marker. Int J Clin Exp
Med. 7:744–750. 2014.PubMed/NCBI
|
|
9
|
Fong ZV and Winter JM: Biomarkers in
pancreatic cancer: Diagnostic, prognostic, and predictive. Cancer
J. 18:530–538. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ali S, Almhanna K, Chen W, Philip PA and
Sarkar FH: Differentially expressed miRNAs in the plasma may
provide a molecular signature for aggressive pancreatic cancer. Am
J Transl Res. 3:28–47. 2010.PubMed/NCBI
|
|
11
|
Ganepola GA, Rutledge JR, Suman P,
Yiengpruksawan A and Chang DH: Novel blood-based microRNA biomarker
panel for early diagnosis of pancreatic cancer. World J
Gastrointest Oncol. 6:22–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li A, Yu J, Kim H, Wolfgang CL, Canto MI,
Hruban RH and Goggins M: MicroRNA array analysis finds elevated
serum miR-1290 accurately distinguishes patients with low-stage
pancreatic cancer from healthy and disease controls. Clin Cancer
Res. 19:3600–3610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J,
Wang X, Gong Y, Wang W and Kong X: Combination of plasma microRNAs
with serum CA19-9 for early detection of pancreatic cancer. Int J
Cancer. 131:683–691. 2012. View Article : Google Scholar
|
|
14
|
Liu R, Chen X, Du Y, Yao W, Shen L, Wang
C, Hu Z, Zhuang R, Ning G, Zhang C, et al: Serum microRNA
expression profile as a biomarker in the diagnosis and prognosis of
pancreatic cancer. Clin Chem. 58:610–618. 2012. View Article : Google Scholar
|
|
15
|
Wang M, Wen TF, He LH, Li C, Zhu WJ and
Trishul NM: A six-microRNA set as prognostic indicators for bile
duct cancer. Int J Clin Exp Med. 8:17261–17270. 2015.
|
|
16
|
Stark A, Bushati N, Jan CH, Kheradpour P,
Hodges E, Brennecke J, Bartel DP, Cohen SM and Kellis M: A single
Hox locus in Drosophila produces functional microRNAs from opposite
DNA strands. Genes Dev. 22:8–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Madhavan B, Yue S, Galli U, Rana S, Gross
W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, et al:
Combined evaluation of a panel of protein and miRNA serum-exosome
biomarkers for pancreatic cancer diagnosis increases sensitivity
and specificity. Int J Cancer. 136:2616–2627. 2015. View Article : Google Scholar
|
|
19
|
Lin X, Lo HC, Wong DT and Xiao X:
Noncoding RNAs in human saliva as potential disease biomarkers.
Front Genet. 6:1752015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Palanisamy V, Sharma S, Deshpande A, Zhou
H, Gimzewski J and Wong DT: Nanostructural and transcriptomic
analyses of human saliva derived exosomes. PloS One. 5:e85772010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Humeau M, Vignolle-Vidoni A, Sicard F,
Martins F, Bournet B, Buscail L, Torrisani J and Cordelier P:
Salivary microRNA in pancreatic cancer patients. PLoS One.
10:e01309962015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie Z, Yin X, Gong B, Nie W, Wu B, Zhang
X, Huang J, Zhang P, Zhou Z and Li Z: Salivary microRNAs show
potential as a noninvasive biomarker for detecting resectable
pancreatic cancer. Cancer Prev Res (Phila). 8:165–173. 2015.
View Article : Google Scholar
|
|
23
|
Michael A, Bajracharya SD, Yuen PS, Zhou
H, Star RA, Illei GG and Alevizos I: Exosomes from human saliva as
a source of microRNA biomarkers. Oral Dis. 16:34–38. 2010.
View Article : Google Scholar :
|
|
24
|
Gallo A and Alevizos I: Isolation of
circulating microRNA in saliva. Methods Mol Biol. 1024:183–190.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar :
|
|
26
|
Julious SA: Sample size of 12 per group
rule of thumb for a pilot study. Pharm Stat. 4:287–291. 2005.
View Article : Google Scholar
|
|
27
|
Chien HY, Lee TP, Chen CY, Chiu YH, Lin
YC, Lee LS and Li WC: Circulating microRNA as a diagnostic marker
in populations with type 2 diabetes mellitus and diabetic
complications. J Chin Med Assoc. 78:204–211. 2015. View Article : Google Scholar
|
|
28
|
Sessa R and Hata A: Role of microRNAs in
lung development and pulmonary diseases. Pulm Circ. 3:315–328.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schulte C and Zeller T: microRNA-based
diagnostics and therapy in cardiovascular disease-Summing up the
facts. Cardiovasc Diagn Ther. 5:17–36. 2015.PubMed/NCBI
|
|
30
|
Wei Q, Mi QS and Dong Z: The regulation
and function of microRNAs in kidney diseases. IUBMB Life.
65:602–614. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arrese M, Eguchi A and Feldstein AE:
Circulating microRNAs: Emerging biomarkers of liver disease. Semin
Liver Dis. 35:43–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qu Z, Li W and Fu B: MicroRNAs in
autoimmune diseases. BioMed Res Int. 2014:5278952014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Machida T, Tomofuji T, Ekuni D, Maruyama
T, Yoneda T, Kawabata Y, Mizuno H, Miyai H, Kunitomo M and Morita
M: MicroRNAs in salivary exosome as potential biomarkers of aging.
Int J Mol Sci. 16:21294–21309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schmittgen TD, Lee EJ, Jiang J, Sarkar A,
Yang L, Elton TS and Chen C: Real-time PCR quantification of
precursor and mature microRNA. Methods. 44:31–38. 2008. View Article : Google Scholar
|
|
35
|
Chen C, Tan R, Wong L, Fekete R and Halsey
J: Quantitation of microRNAs by real-time RT-qPCR. Methods Mol
Biol. 687:113–134. 2011. View Article : Google Scholar
|
|
36
|
Life Technologies: DataAssist v3.0
Software user instructions. 2011, http://cgs.hku.hk/portal/files/CGS/Genomics/Realtime-PCR/dataassist%20v3_0%20software%20user%20instructions.pdf.
|
|
37
|
Hajian-Tilaki K: Receiver Operating
Characteristic (ROC) Curve Analysis for medical diagnostic test
evaluation. Caspian J Intern Med. 4:627–635. 2013.PubMed/NCBI
|
|
38
|
Ruopp MD, Perkins NJ, Whitcomb BW and
Schisterman EF: Youden Index and optimal cut-point estimated from
observations affected by a lower limit of detection. Biom J.
50:419–430. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Swets JA: Measuring the accuracy of
diagnostic systems. Science. 240:1285–1293. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kojima M, Sudo H, Kawauchi J, Takizawa S,
Kondou S, Nobumasa H and Ochiai A: MicroRNA markers for the
diagnosis of pancreatic and biliary-tract cancers. PLoS One.
10:e01182202015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ali S, Saleh H, Sethi S, Sarkar FH and
Philip PA: MicroRNA profiling of diagnostic needle aspirates from
patients with pancreatic cancer. Br J Cancer. 107:1354–1360. 2012.
View Article : Google Scholar : PubMed/NCBI
|