Introduction
Bladder carcinoma is one of the most common
malignancies and the second leading cause of death among men in
urologic cancer (1). Approximately
75% of the patients have superficial bladder cancer, but they recur
and progress to muscle-invasive cancer after surgical intervention.
The 5-year survival rate is only ~60% for patients with
muscle-invasive bladder cancer (2,3).
Therefore, exploring and understanding the potential molecular
mechanism underlying bladder cancer is crucial for the development
of novel therapies.
MicroRNAs (miRNAs) are a class of endogenous 19–25
nucleotides small non-coding RNA molecules, which lead to target
mRNA silencing or translational repression at the
post-transcriptional level through complementary binding to the
3′-untranslated region (3′-UTR) of target genes (4). Dysregulation of miRNAs is a general
feature of many tumors, including bladder cancer (5,6).
Aberrant expression of miRNAs appear to play crucial roles in
various physiological processes of cancer (7). It has been reported that miRNAs could
serve as biomarkers for bladder cancer (8). In our preliminary investigation on the
changes of miRNA expression profiles in chemotherapy of bladder
tumor by miRNA microarray analysis, we observed differentially
expressed miRNAs after arsenic trioxide treatment in 5637 and T24
cells, particularly miR-451 showed significantly altered
expression. miR-451 is a special miRNA since it has a unique
structure and synthetic pathway, and its maturity is mediated by
Ago2. Frequent dysregulation of miR-451 was reported in several
types of cancers in recent research (9). Intriguingly, expression of miR-451 is
not consistent in different tumors. Compared with normal tissues,
miR-451 is downregulated in hypopharyngeal squamous cell carcinoma
(10), lung (11) and liver cancer (12), and osteosarcoma (13). On the contrary, the expression of
miR-451 is upregulated in glioma (14) and colorectal cancer (15). This indicated that distinction of
miR-451 expression in tumors may be determined based on the type of
tumor. However, the role of miR-451 involved in bladder cancer
development and progress is still remains largely unraveled.
Therefore, it is necessary to investigate the expression and
potential function of miR-451 in bladder cancer.
Our purpose was to analyze miR-451 differential
expression between bladder cancer and normal bladder tissues. We
used in vitro approach to study the role of miR-451 in
regulating the biological behavior of bladder cancer cells.
Furthermore, we predicted a direct downstream target gene of
miR-451 and verified their correlation to deepened our
understanding of the role of miR-451 in bladder cancer.
Materials and methods
Human tissue samples
Bladder cancer and adjacent normal tissues were
obtained from patients who received partial resection or radical
cystectomy at Department of Urological Surgery, the First
Affiliated Hospital of Harbin Medical University. None of the
patients received any preoperative therapy. After resection, fresh
tissues were immediately frozen in liquid nitrogen and then stored
at −80°C until RNA extraction. The use of the tissue samples was
approved by Institute Ethics Committee. The patients signed
informed consent forms. The characteristics of the patients are
described in Table I. Bladder
tumors were divided into low grade group (n=12) and high grade
(n=8) according to the World Health Organization pathology
classification standard (16).
 | Table IClinicopathological information of
the patients with bladder cancer. |
Table I
Clinicopathological information of
the patients with bladder cancer.
| Patient no. | Gender | Age (years) | Stage | Grade |
|---|
| 1 | Male | 56 | II | Low |
| 2 | Male | 60 | I | Low |
| 3 | Male | 62 |
III | High |
| 4 | Male | 53 |
III | High |
| 5 | Male | 61 | I | Low |
| 6 | Male | 59 |
III | High |
| 7 | Female | 63 |
III | High |
| 8 | Male | 55 | II | High |
| 9 | Male | 59 | II | Low |
| 10 | Female | 45 | I | Low |
| 11 | Male | 55 | II | High |
| 12 | Male | 50 | II | Low |
| 13 | Male | 65 | I | Low |
| 14 | Female | 68 |
III | Low |
| 15 | Female | 64 | II | Low |
| 16 | Male | 58 | II | Low |
| 17 | Male | 74 |
III | High |
| 18 | Male | 62 | II | Low |
| 19 | Female | 66 | II | Low |
| 20 | Male | 57 |
III | High |
Cell culture and transfection
Human bladder cancer cell lines T24 and 5637 were
obtained from the Cell Bank of the Chinese Academy of Science
(Shanghai, China). Cells were cultured in RPMI-1640 medium (Gibco,
Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; PAA,
USA) under a humid atmosphere with 5% CO2 at 37°C. The
miR-451 mimics and negative control miRNA (miR-NC), siRNA-c-Myc and
negative control siRNA (siR-NC) were synthesized by RioBio
(Guangzhou, China). Cells were plated to 50–60% confluency in
medium and then transfected with oligofectamine reagent (Guangzhou,
China) based on the manufacturer's protocol. The miRNAs were used
at a final concentration of 100 nM and transfection efficiency
detected by real-time RT-PCR.
RNA extraction and quantitative real-time
PCR (qPCR)
Total RNAs were extracted from human tissues and
cell lines with TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
following the protocol as previously described (10). RNA purity and concentrations were
assessed by measuring the absorbance at 260 and 280 nm using a
NanoDrop® ND-1000 spectrophotometer (Thermo Scientific,
Waltham, MA, USA). Samples at optical density (OD) 260/280 nm
ratios between 1.80–2.00 were used for further analysis. Total RNA
was reverse-transcribed into cDNA with One Step PrimeScript miRNA
cDNA Synthesis kit (Takara, Otsu, Japan). qRT-PCR reactions were
conducted using the ABI 7500 Fast Real-Time PCR system (Applied
Biosystems, Carlsbad, CA, USA), followed by 95°C for 30 sec and 40
thermal cycles of 95°C for 5 sec and 60°C for 30 sec. The U6 or
GAPDH were used as the internal endogenous controls for miR-451 or
siR-c-Myc and the 2−ΔΔCt method was used to calculate
the relative expression levels. The sequences of the primers are
shown in Table II.
 | Table IIPrimer sequences. |
Table II
Primer sequences.
| Name | Primes (5′-3′) |
|---|
| miR-451 | F
AAACCGTTACCATTACTGAGTT |
| R
GCGAGCACAGAATTAATACGAC |
| U6 | F
CTCGCTTCGGCAGCACATATACT |
| R
ACGCTTCACGAATTTGCGTGTC |
| c-Myc | F
TGCTAAGAAGATTGGTGCTGTA |
| R
GCGAAGGGCTGAGACATTTAC |
| GAPDH | F
GAAATCCCATCACCATCTTC |
| R
GAGCCCCAGCCTTCTCCATG |
Cell proliferation assay
Cell proliferation was measured using Cell Counting
Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). Cells were seeded into
96-well plates at 5,000 cells/well. After 24 h cultivation, cells
were transfected with miR-451 mimics, miR-NC, si-c-Myc and siR-NC.
CCK-8 reagent 10 μl was added into each plate after cells
were transfected 0, 24, 48, 72 and 96 h. The cells were cultured
for another 1 h and the absorbance at 450 nm was measured with a
spectrophotometric plate reader (Bio-Rad, Richmond, CA, USA).
Cell Transwell assay
Cell Transwell assay was performed in 24-well plates
with Transwell chamber (8.0-μm pore size polycarbonate
filter) as previously described (11). Briefly, cells (5×104) in
100 μl serum-free medium were placed into the upper chamber
with or without Matrigel (BD Biosciences, Bedfold, MA, USA).
RPMI-1640 medium (1 ml) containing 10% FBS was added in the lower
chamber. After cultured for 48 h, cells on the upper chamber were
washed, fixed and stained with methanol and crystal violet, then
counted by inverted microscopy at a magnification of ×100 (Sony,
Tokyo, Japan).
Dual-luciferase reporter assay
The luciferase reporter assay was carried out as
previously described (12). In
brief, the recombinant vectors (HaoGe, Shanghai, China) were
constructed with wild-type (WT)-c-Myc sequences or mutant
(MUT)-c-Myc sequences inserted between the hRluc and the hLuc gene.
Cells (2×104) were seeded into 96-well plates, after
growth to 70–80% confluence, and cotransfected with miR-NC or
miR-451 mimics and the WT c-Myc 3′-UTR vector and the MUT c-Myc
3′-UTR vector using Lipofectamine 2000 (Invitrogen). After
transfecting for 24 h, the firefly and Renilla luciferase
activities were measured consecutively through a Dual-Luciferase
Reporter Assay system (Promega, Madison, WI, USA). Renilla
luciferase intensity (Renilla/firefly) was used as
normalized data.
Western blotting
Total proteins were extracted from cells by PARIS
kit (Ambion, Carlsbad, CA, USA), and protein concentration was
detected by BCA protein assay kit (Beyotime, China). Then, proteins
were separated using 10% SDS-PAGE gels, and transferred to
polyvinylidene fluoride (PVDF) membranes. Next, the membranes were
blocked with 5% non-fat dried milk for 2 h and incubated with
rabbit anti-MMP-7 (1:1,000; Cell Signaling Technology, Danvers, MA,
USA), anti-PARP1 antibody (1:1,000; ProteinTech Group, Inc.,
Chicago, IL, USA), anti-c-Myc antibody (1:500; Santa Cruz
Biotechnology, Santa Cruz, CA, USA) and anti-GAPDH antibody
(ProteinTech) at a 1:2,000 dilution overnight at 4°C. Membranes
were washed three times in PBST, and incubated with a goat
anti-rabbit antibody at a 1:2,000 dilution for 2 h. Blots were
detected by an enhanced chemiluminescence (ECL) system (Pierce
Biotechnology Inc., Rockford, IL, USA).
Cell apoptosis analysis
The transfected cells were harvested and washed
twice by ice cold PBS. Cells, were then resuspended at a density of
1×106 cells/ml in 1X binding buffer, and stained with an
Annexin V-FITC apoptosis detection kit (Clontech, Beijing, China).
Cell apoptosis was detected using a FACSCalibur flow cytometer (BD
Biosciences, San Jose, CA, USA).
Statistical analysis
Data are expressed as means ± standard deviation
(SD). Statistical significance was evaluated by Student's t-test or
one-way analysis of variance (ANOVA). Differences were considered
statistically significant at p<0.05. All experiments were
repeated three times.
Results
Expression of miR-451 is downregulated in
bladder cancer tissues
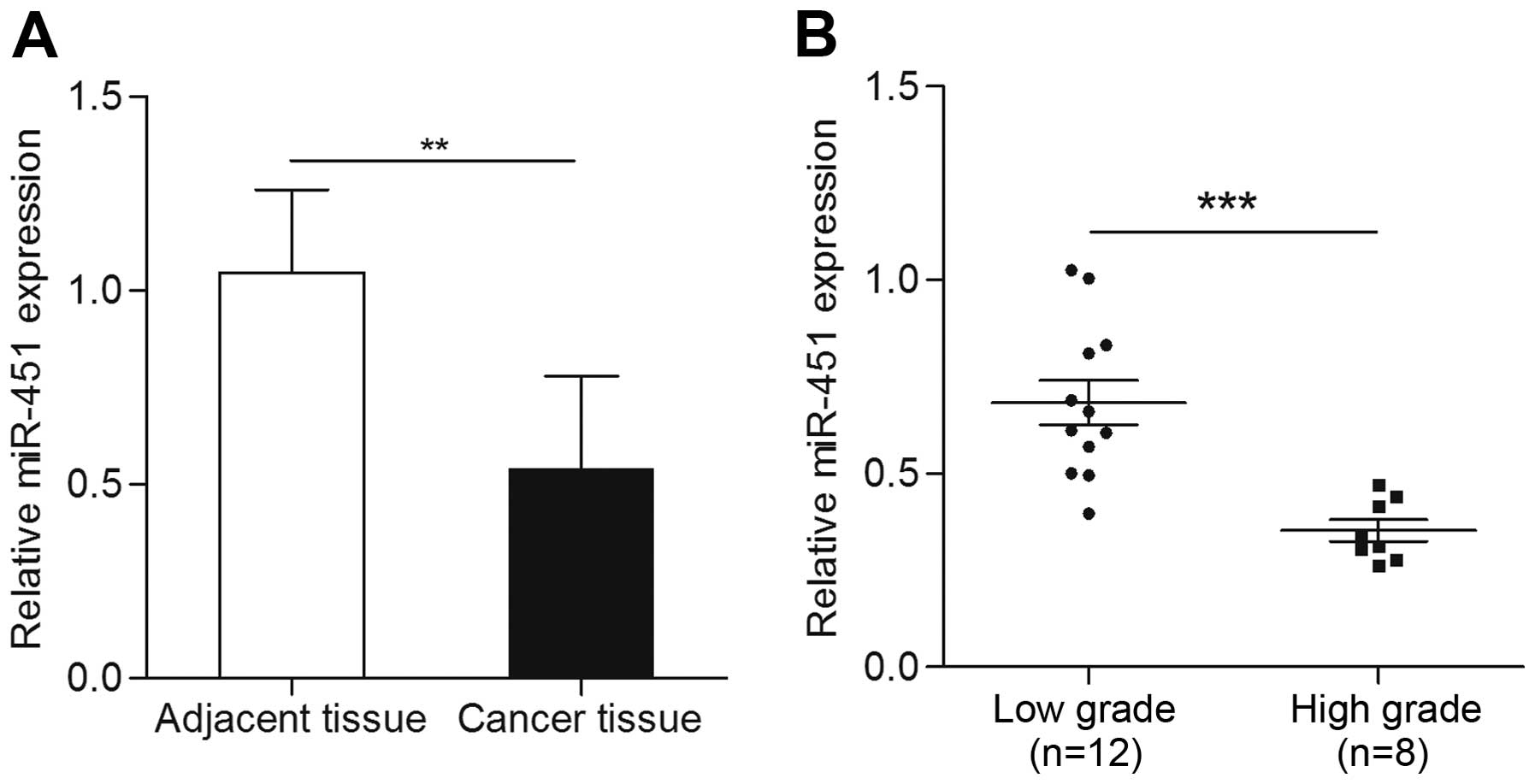
The expression of miR-451 was detected by qRT-PCR in
paired 20 bladder cancer tissues and matched adjacent non-cancer
tissues in the present study. The results showed that expression of
miR-451 in bladder cancer tissues was significantly decreased when
compared with adjacent non-cancerous tissues (p<0.01) (Fig. 1A). In order to further determine the
relation of miR-451 and bladder cancer, we analyzed miR-451
expression disparity in different grade bladder cancer tissues. The
expression of miR-451 in low grade bladder cancer tissues was found
noticeable higher than that of the high grade tissues (p<0.001)
(Fig. 1B). These results suggested
that miR-451 may be associated with bladder cancer.
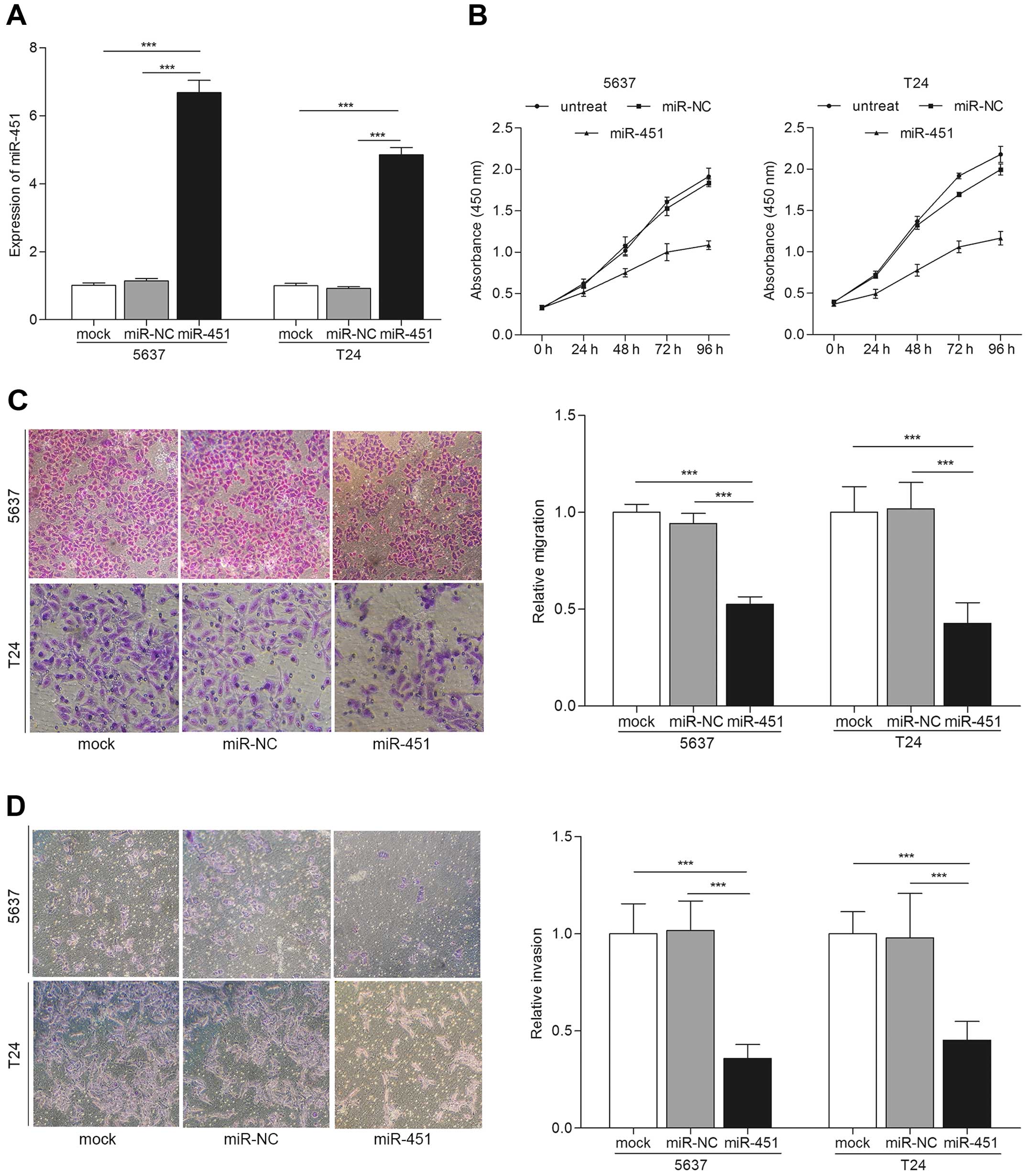
miR-451 overexpression suppresses the
proliferation, migration and invasion of bladder cancer cells
To study the tumor suppressive role of miR-451 in
bladder cancer, we examined the proliferation, migration and
invasion of bladder cancer cells after the different transfections.
qRT-PCR was carried out for miR-451 expression evaluation after
transfections. The results showed the expression of miR-451 was
increased in miR-451 transfected cells compared with mock or miR-NC
(p<0.001) (Fig. 2A). Next, we
performed CCK-8 cell proliferation assay. The results indicated
that bladder cancer cells transfected with miR-451 showed
significantly low proliferation rate than controls (Fig. 2B). Cell migration and invasion are
key steps during tumor progression and metastasis (17). We detected the migration and
invasion capability of bladder cancer cells after transfection with
miR-451 by Transwell assay. Fig. 2C
showed that overexpression of miR-451 significantly induced the
migration ability of bladder cancer cells compared with the
controls by Transwell migration assay (p<0.001). Similarly
results were observed in Transwell invasion assay, cells
transfected with miR-451 showed attenuated invasiveness compared to
the controls (p<0.001) (Fig.
2D). These results suggest that overexpression of miR-451
suppresses biological behavior of bladder cancer in
vitro.
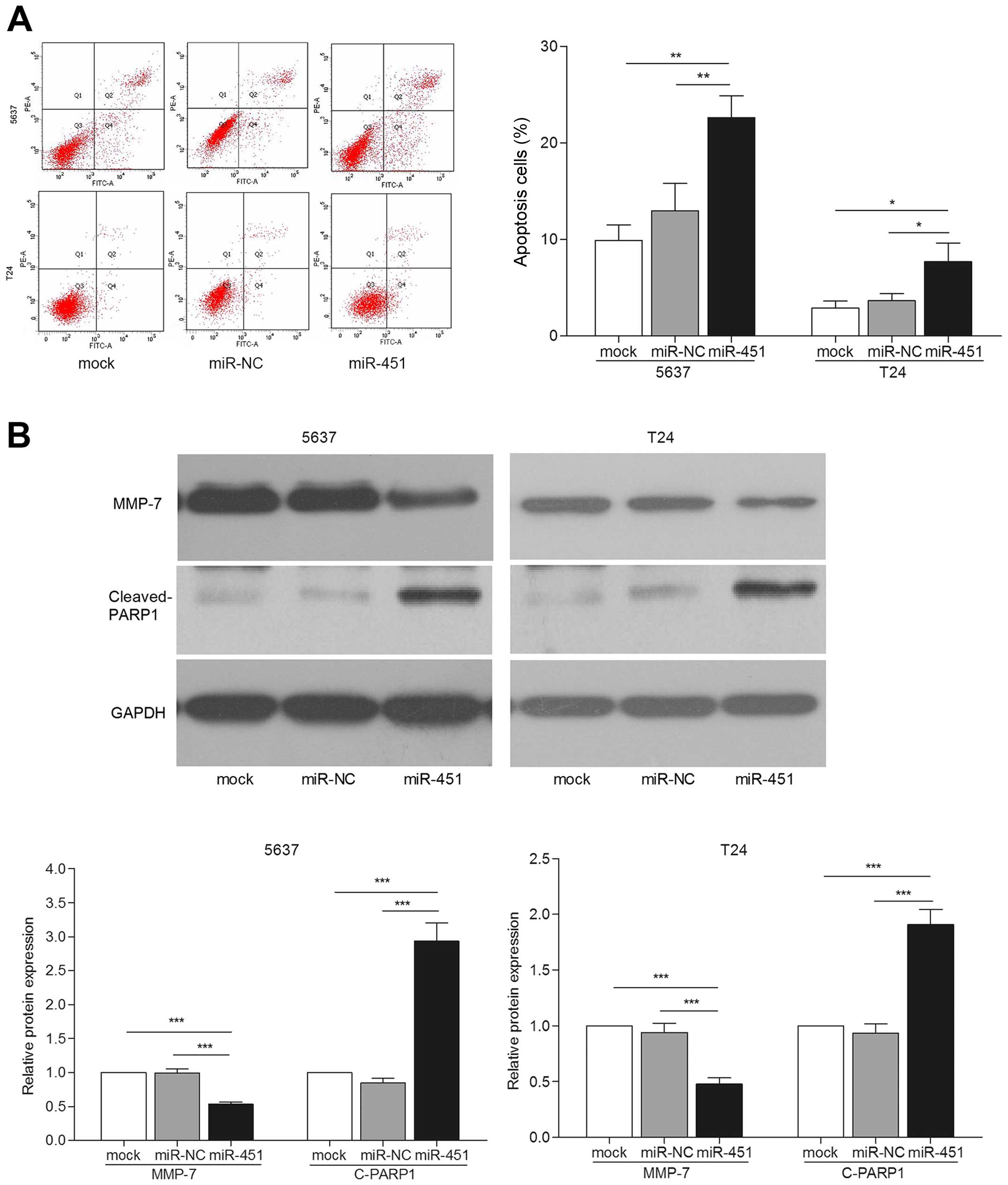
miR-451 reduces cell migration and
invasion by increasing apoptosis
Previous studies implied that apoptosis represents
one of the important processes in cancer metastasis (18). Thus, we investigated the possible
mechanism of miR-451 involved in inhibiting bladder cancer cell
migration and invasion. Significantly increased apoptotic
proportions were observed in bladder cancer cells transfected with
miR-451 (p<0.05) (Fig. 3A). We
also detected the expression of key apoptosis related protein, PARP
(19). The results showed that
cleaved PARP1 protein expression was apparently increased in
miR-451 transfected cells compared with the controls by western
blot assay (p<0.001) (Fig. 3B).
A previous study showed that MMPs served as metastasis-related
factors in malignancy mediated tumor cell apoptosis (18). Then, expression of MMP-7 was
detected by western blotting. We observed that miR-451
overexpression led to lower expression level of MMP-7 protein
(Fig. 3B). These results suggest
that apoptosis may mediate bladder cancer cell migration and
invasion.
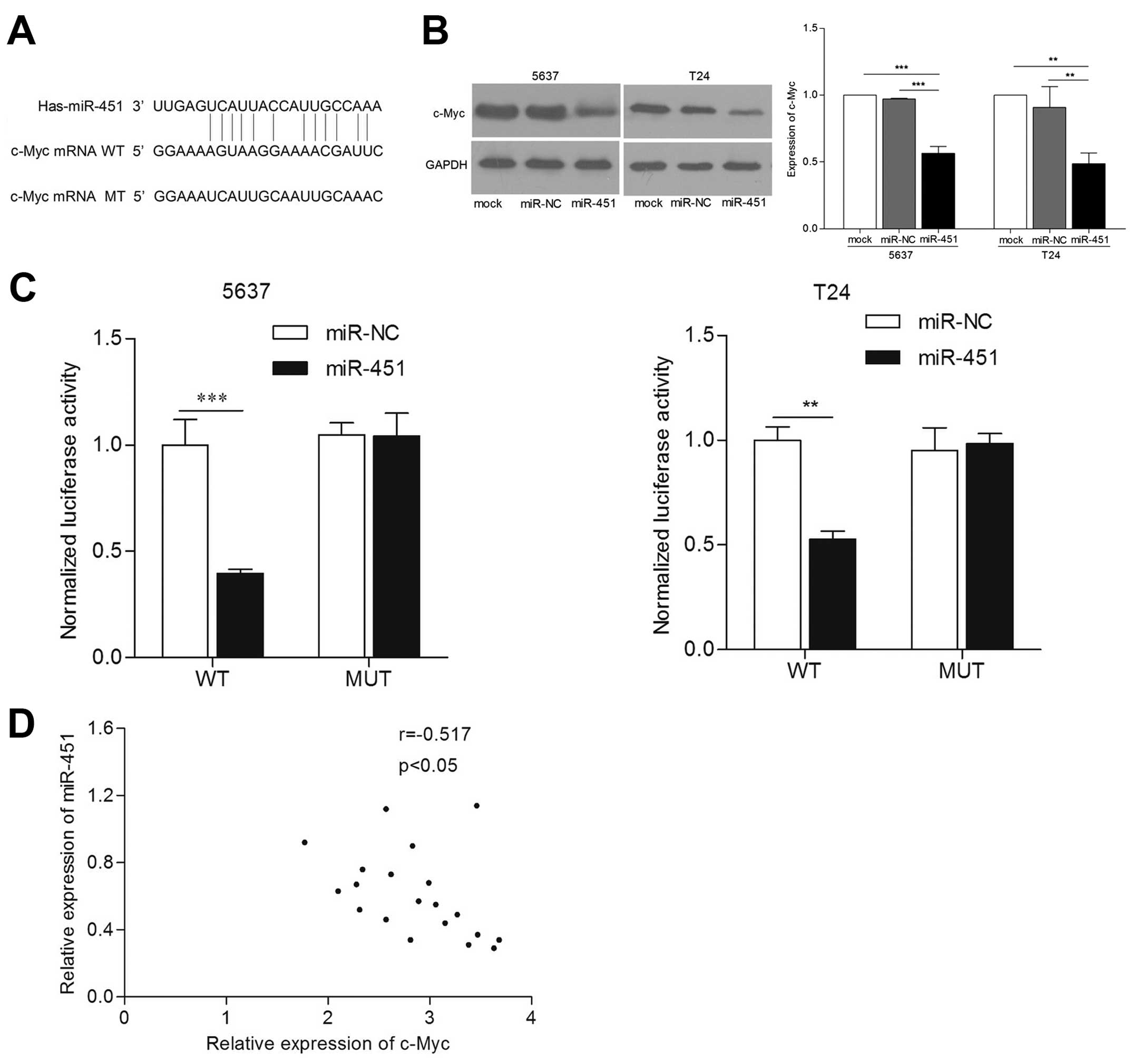
miR-451 regulates c-Myc expression by
directly targeting its 3′-UTR
To understand the details of antitumorigenic
mechanism of miR-451 in bladder cancer, we explored target gene of
miR-451 by multi-bioinformatics analysis, such as TargetScan,
MicroRNA and PicTar. After analysis, we found that 3′-UTR of c-Myc
mRNA contained one conserved complimentary binding site with
miR-451 (Fig. 4A). This indicated
that c-Myc may be a putative target gene of miR-451. In order to
validate whether c-Myc was directly regulated by miR-451 with its
3′-UTR, we co-transfected the luciferase plasmid harboring the
wild- or mutant-type 3′-UTR of c-Myc, and then carried out
luciferase reporter assay. The results confirmed that luciferase
activity of c-Myc 3′-UTR containing wild-type was suppressed in
miR-451 re-expressed cells, but not containing mutant type
(p<0.01) (Fig. 4B). Next, we
detected expression levels of c-Myc in 5637 and T24 cell lines by
western blot assay. The results showed that c-Myc protein levels
are markedly reduced in cells transfected with miR-451 compared
with that in control cells (p<0.01) (Fig. 4C). Moreover, we observed an inverse
correlation between miR-451 and c-Myc protein level in bladder
cancer tissues (Fig. 4D). The above
demonstrates that c-Myc is a direct downstream target gene of
miR-451 in bladder cancer cells.
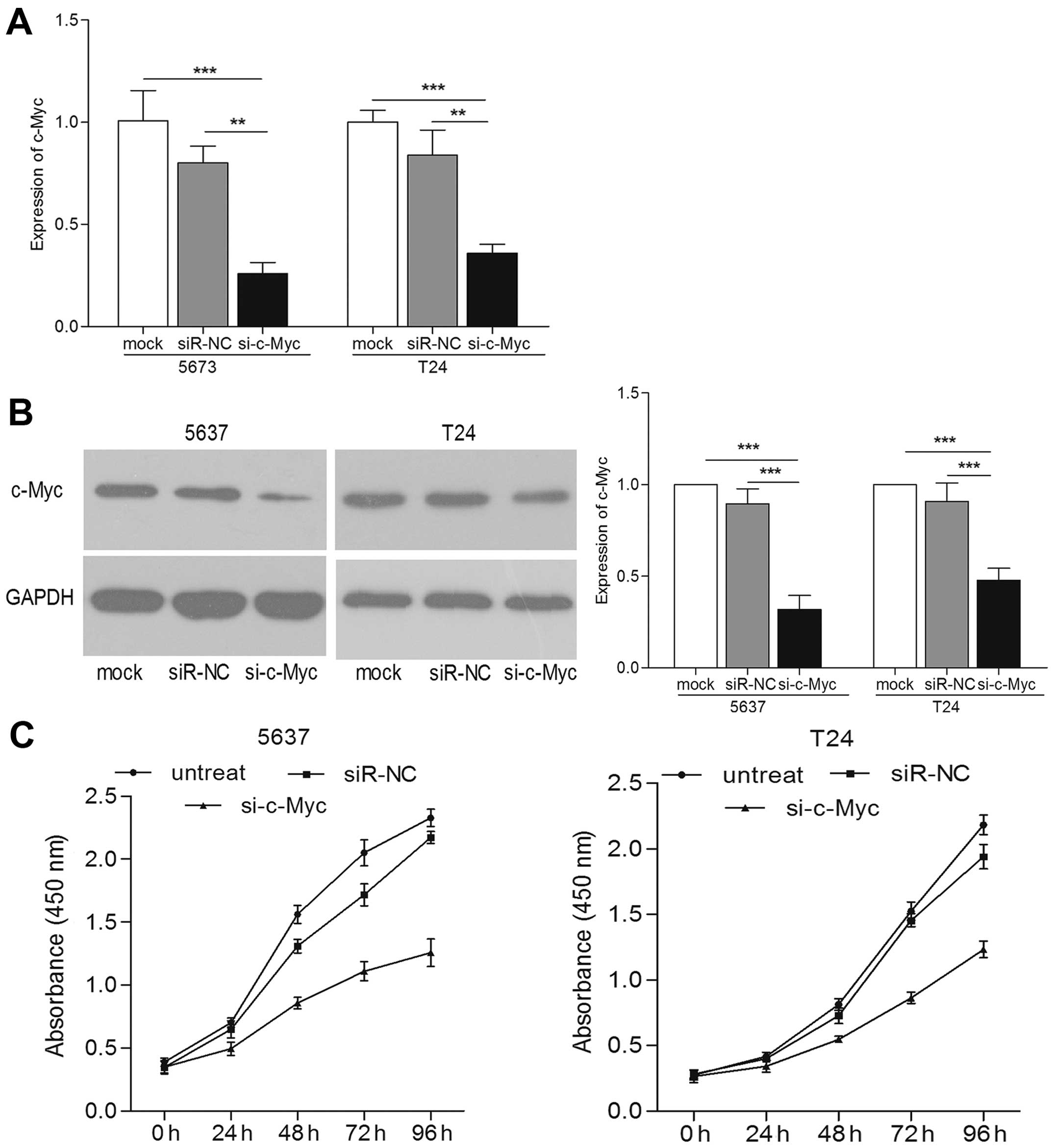
To further confirm that the role of miR-451 on the
biological behavior of bladder cancer cells was through targeting
c-Myc, we knocked down the expression of c-Myc in bladder cancer
cells. We transfected cells with c-Myc siRNA. Both the expression
levels of c-Myc mRNA and protein were reduced compared with those
in the controls (Fig. 5A and B).
Using cell proliferation technique, we observed the significantly
reduced cell proliferation in si-c-Myc transfected bladder cancer
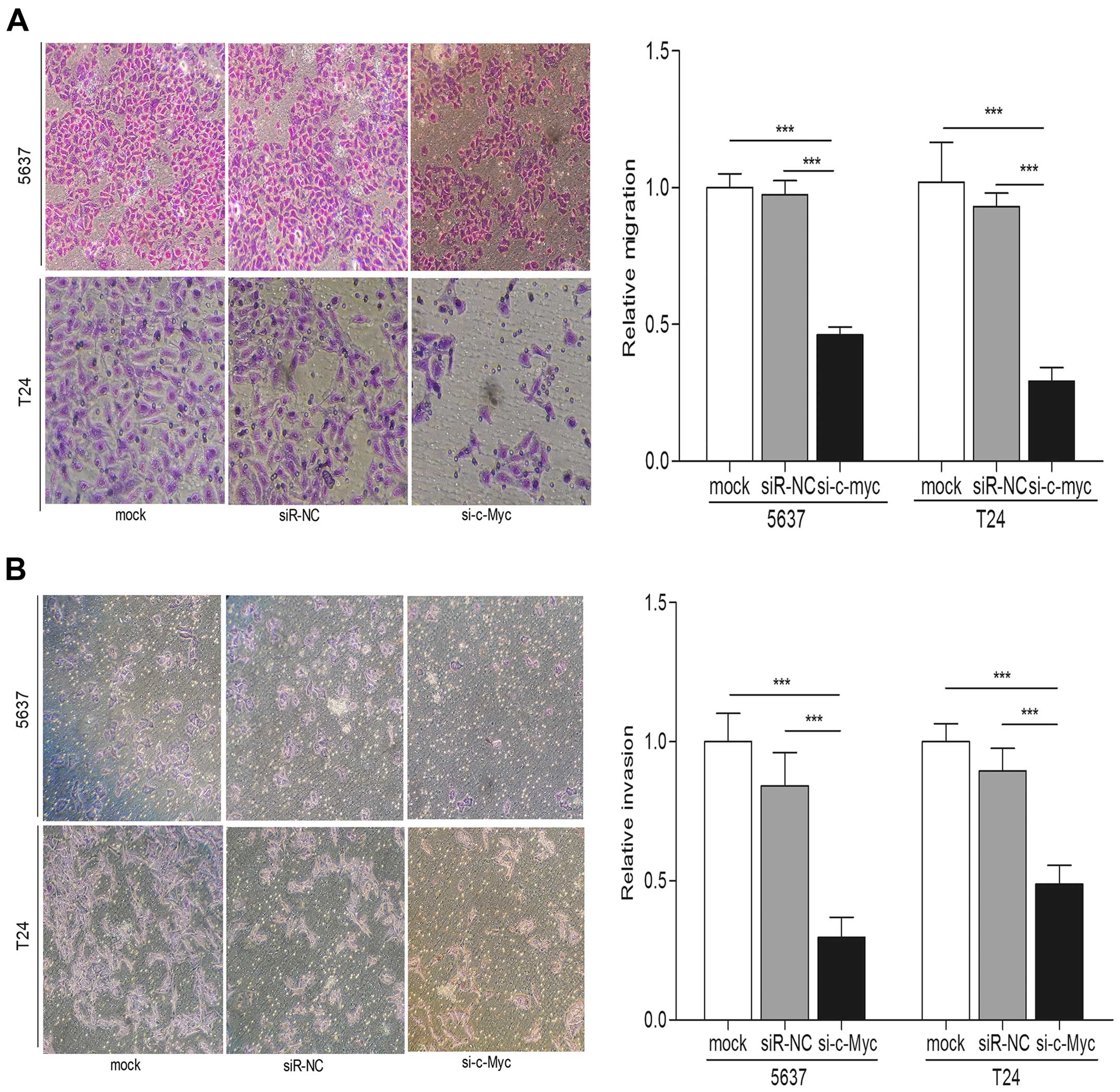
cells (Fig. 5C). Both Transwell
migration and invasion assays indicated that c-Myc knockdown played
an inhibitory effect on the bladder cancer cell migration and
invasion compared with control groups (p<0.001) (Fig. 6A and B). In addition, protein levels
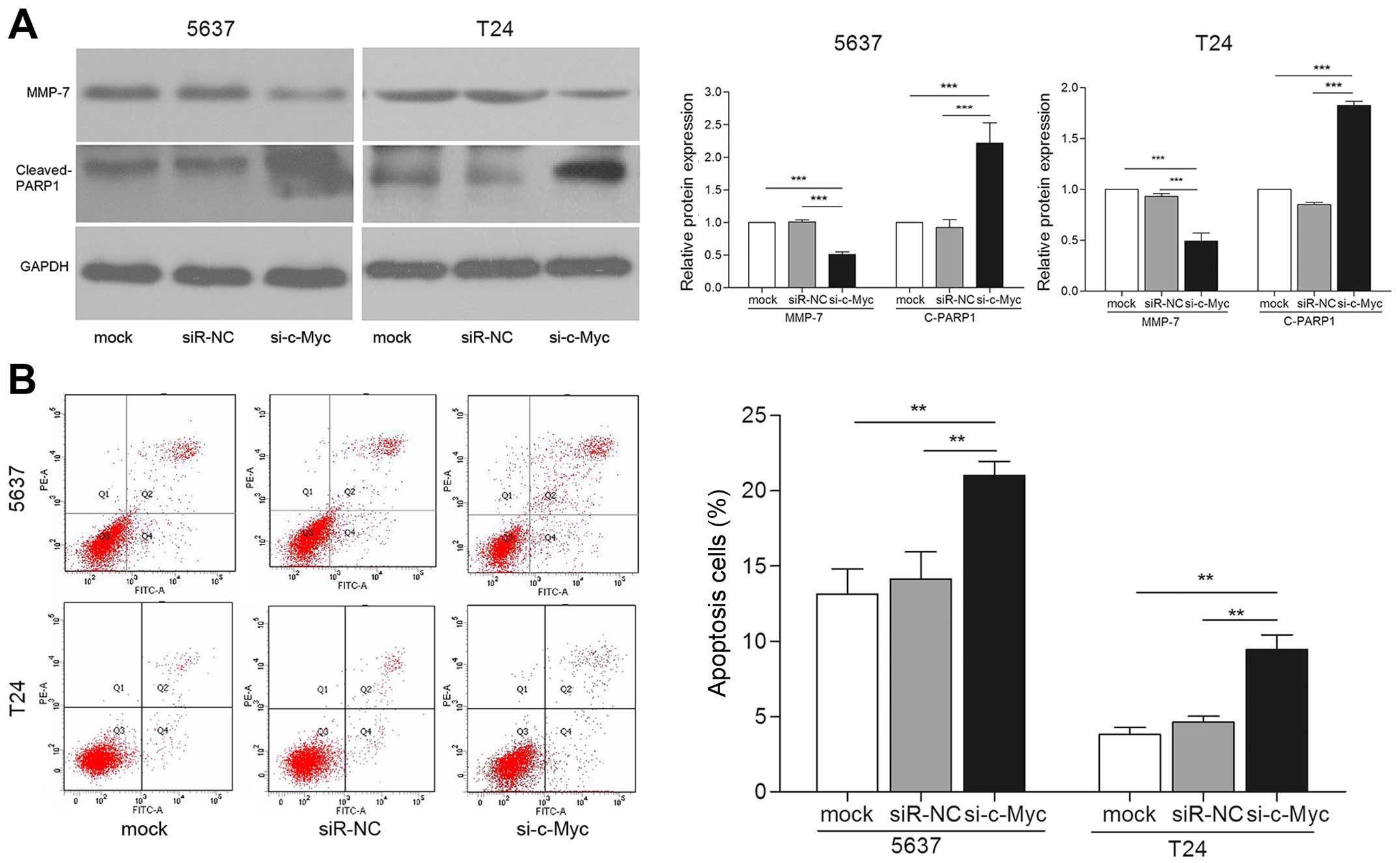
of MMP-7 were significantly repressed in cells transfected with
si-c-Myc (Fig. 7A). Furthermore,
cells transfected with si-c-Myc showed increased apoptosis
(Fig. 7A and B). Taken together,
these effects induced by knockdown of c-Myc are consistent with
miR-451 overexpression in bladder cancer cells. Thus, we concluded
that c-Myc may be a downstream functional regulator of miR-451.
Discussion
It is well known that miRNAs are pivotal players in
the highly complex world of gene regulation; they regulate over 30%
of protein-coding genes, play a critical role in normal cellular
processes such as proliferation, apoptosis, migration, invasion and
metabolism (20). In recent years,
numerous studies found that dysregulation of miRNAs serve as
modulator and lead to the development of human cancers such as
bladder cancer (21). Various
miRNAs (e.g., miR-137, miR-424 and miR-195/497) present
differential expression in bladder cancer. In addition, they also
functionally regulate biological behavior of bladder cancer
(6,22,23).
Altered expression of miR-451 has been reported in some types of
cancers and involved in cancer progression. For example, the
expression of miR-451 in glioblastoma tissues is higher than that
in matched normal adjacent brain tissue, and miR-451 suppressed the
migration of glioblastoma cells by CAB39 (14). Using a T cell acute lymphoblastic
leukemia xenograft model, it was observed that tumor development
was delayed in miR-451-treated mice (24). Kovalchuk et al showed that
increased miR-451 expression level enhanced sensitivity of
resistant MCF-7 cells to doxorubicin (25). These results suggest that miR-451
may be a critical regulator in neoplasm formation and progress. Our
preliminary study found that miR-451 expression was significantly
upregulated in arsenic trioxide treatment in bladder cancer cells
(Fig. 8). Therefore, the relation
between miR-451 with development and progress of bladder cancer was
evaluated. In the present study, we found miR-451 was downregulated
in bladder cancer tissues compared with adjacent non-tumor tissues.
This result is consistent with the result of Zeng et al
(26). Furthermore, miR-451 was
proved negatively correlated with cancer grade, miR-451 in high
grade of bladder cancer was markedly lower. Then, we focused on and
explored potential function of miR-451 by restored expression of
miR-451 in bladder cancer cell lines. The results revealed that
overexpressed miR-451 markedly inhibited proliferation, migration
and invasion of bladder cancer cells. These results predicted that
miR-451 may act as an anti-oncogene and regulate bladder cancer
formation and progression.
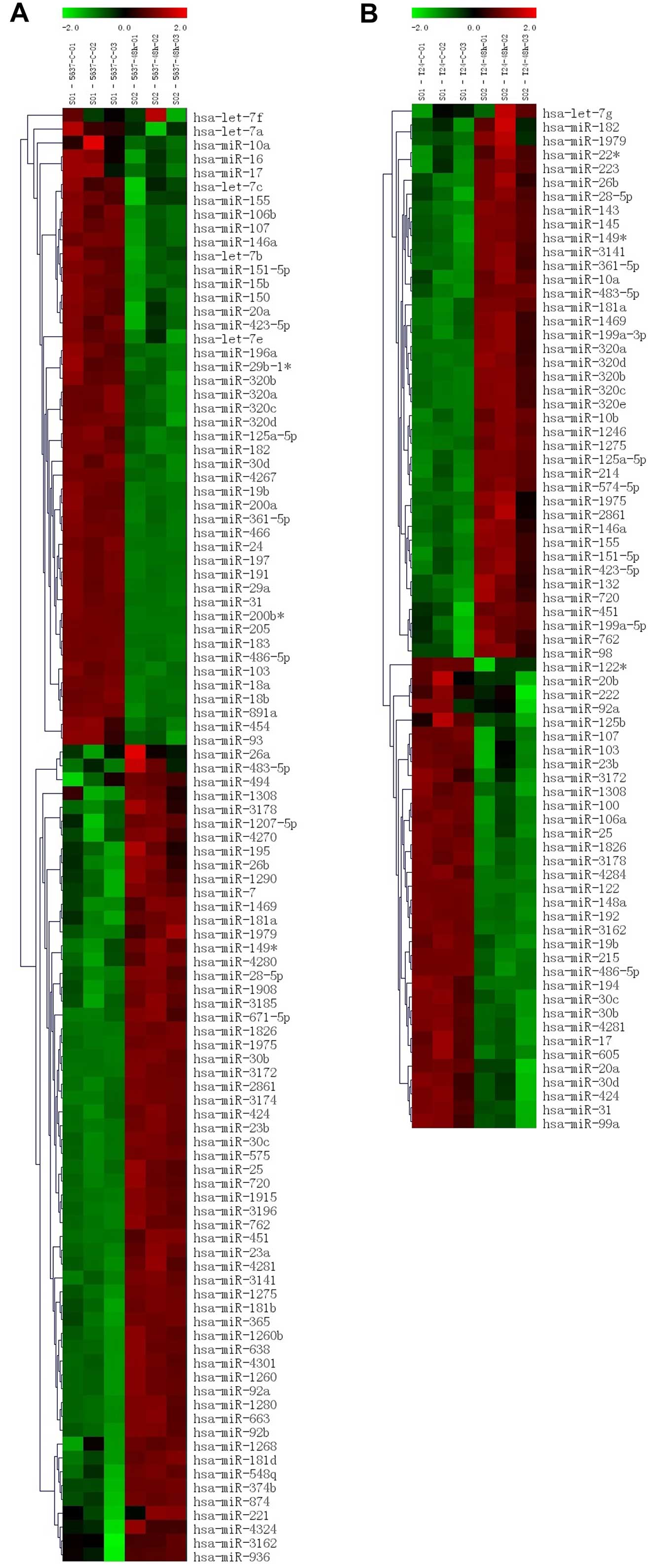 | Figure 8Heatmap illustrating expression of
miRNAs in response to arsenic trioxide in bladder cancer cells,
miRNAs from each time point were labeled with Cy5. (A and B)
Heatmap of 5637 and T24 cells after arsenic trioxide treatment by
miRNA microarray analysis, 5637-c-01,02,03 and T24-c-01,02,03
represent result of untreated 5637 cells, 5637-48h-01,02,03 and
T24-48h-01,02,03 represent results of arsenic trioxide treated 5637
cells. |
Apoptosis is the common form of programmed cell
death, as an important mechanism that negatively regulates cancer
development. Mehlen et al found that apoptosis plays a key
role in inhibiting process of cancer metastasis (18). MMPs are a class of critical protein
usually involved in tumor cell invasion and migration (27). However, evidence also showed that
MMPs play a role in the regulation of cell apoptosis. The ability
of tumor cells to resist apoptosis was induced by MMP-7 in early
tumor development though cleaving CD95 (28). Abraham et al found that
MMP-15 knockdown led to an increase of the cancer cell apoptosis
(29). Based on this, we detected
whether miR-451 attenuate bladder cancer migration and invasion
through activation of apoptosis. Our results showed that miR-451
overexpression induced cells apoptosis, increased expression of
cleaved PARP and decreased the level of MMP-7, which are associated
with tumor apoptosis and metastasis. These results indicated that
overexpression of miR-451 promoted apoptosis, which may represent a
way for inhibiting migration and invasion of bladder cancer.
The region of 5′-end of miRNA can perfectly or
near-perfectly bind complementary sequences with 3′-UTR of mRNA.
Such binding induced target mRNA silencing or translation-repressed
at post-transcriptional level. A dysfunctional miRNA-guided mRNA
silencing pathway would lead to altered expression of target genes
involved in cellular processes of tumorigenesis (30). Every miRNA has a large amount of
downstream target genes, each target gene is also modulated by
numerous miRNAs. Arduous function of miRNAs in cancer not only
correlated with the tumor type, but also associated with diverse
target genes. Therefore, exploration of more target genes of miRNAs
may help us further understand the mechanisms of miRNA in cancer
development. Several target genes of miR-451 have been found in
certain cancers in previous research, such as ESDN/DCBLD2 (10), PSMB8 (11) and CAB39 (14). In the present study, c-Myc was
predicted as a direct target gene of miR-451 by bioinformatics
analysis, and then identified in bladder cancer cells using RT-PCR,
western blot and luciferase reporter assays.
c-Myc, a central gene in mammalian cells, is a
member of the family of Myc genes involved in diverse physiological
processes, embryonic development, cell cycle, cell proliferation,
apoptosis and protein synthesis (31). In normal cells, expression and
function of c-Myc are regulated by developmental or mutagenic
signals. Thus, the mRNA and protein levels of c-Myc are low, c-Myc
is one of the most highly amplified oncogenes among many different
human cancers such as colon cancer, T-cell leukemia, lung and
bladder cancer (32). It has been
reported that altered expression of c-Myc is induced by upstream
signal pathways (33). In addition,
various studies found miRNAs may be upstream regulators of c-Myc by
targeting its mRNA, then inhibiting its translation (34,35).
In addition, miRNA has been proved to be able to regulate
chemoresistance of cancer cells by inhibiting c-Myc (36,37).
The present study confirmed that c-Myc was directly targeted by
miR-451 in bladder cancer. Overexpression of miR-451 inhibited mRNA
and protein of c-Myc. The results of luciferase reporter assay
demonstrated that c-Myc had a binding site with miR-451 at its
3′-UTR. The expression of c-Myc is reduced in miR-451 overexpressed
cells. Further study showed that c-Myc knockdown could repress
bladder cancer cell proliferation, migration and invasion
indicating that the reduced c-Myc expression exert similar effects
of miR-451 overexpression in bladder cancer cells. Taken together,
these findings may suggest that miR-451 suppresses the biologic
activity of bladder cancer by negatively regulating c-Myc.
In conclusion, our findings provide evidence that
miR-451 is downregulated in bladder cancer tissues and is involved
in multiple cellular biological behaviors. We also confirmed a
mechanism that miR-451 reduced migration and invasion of bladder
cancer cells probably by inducing apoptosis. Furthermore, miR-451
is a tumor-suppressor by directly inhibiting c-Myc expression in
bladder cancer. These data enrich our knowledge of miR-451 that
plays a significant role in bladder cancer formation and
progression. It may provide novel therapeutic targets for bladder
cancer at gene level.
Acknowledgments
The present study was supported by the Natural
Science Youth Foundation of Heilongjiang Province (no. QC2010010)
and the Postdoctoral Science-Research Foundation of Heilongjiang
(no. Q11067).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zuiverloon TC, Nieuweboer AJ, Vékony H,
Kirkels WJ, Bangma CH and Zwarthoff EC: Markers predicting response
to bacillus Calmette-Guérin immunotherapy in high-risk bladder
cancer patients: A systematic review. Eur Urol. 61:128–145. 2012.
View Article : Google Scholar
|
|
3
|
Luke C, Tracey E, Stapleton A and Roder D:
Exploring contrary trends in bladder cancer incidence, mortality
and survival: Implications for research and cancer control. Intern
Med J. 40:357–362. 2010. View Article : Google Scholar
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lages E, Ipas H, Guttin A, Nesr H, Berger
F and Issartel JP: MicroRNAs: Molecular features and role in
cancer. Front Biosci. 17:2508–2540. 2012. View Article : Google Scholar
|
|
6
|
Xiu Y, Liu Z, Xia S, Jin C, Yin H, Zhao W
and Wu Q: Micro-RNA-137 upregulation increases bladder cancer cell
proliferation and invasion by targeting PAQR3. PLoS One.
9:e1097342014. View Article : Google Scholar
|
|
7
|
Price C and Chen J: MicroRNAs in cancer
biology and therapy: Current status and perspectives. Genes Dis.
1:53–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tölle A, Ratert N and Jung K: miRNA panels
as biomarkers for bladder cancer. Biomarkers Med. 8:733–746. 2014.
View Article : Google Scholar
|
|
9
|
Pan X, Wang R and Wang ZX: The potential
role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol
Cancer Ther. 12:1153–1162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fukumoto I, Kinoshita T, Hanazawa T,
Kikkawa N, Chiyomaru T, Enokida H, Yamamoto N, Goto Y, Nishikawa R,
Nakagawa M, et al: Identification of tumour suppressive
microRNA-451a in hypopharyngeal squamous cell carcinoma based on
microRNA expression signature. Br J Cancer. 111:386–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin P, Peng R, Peng H, Yao L, Sun Y, Wen
L, Wu T, Zhou J and Zhang Z: MiR-451 suppresses cell proliferation
and metastasis in A549 lung cancer cells. Mol Biotechnol. 57:1–11.
2015. View Article : Google Scholar
|
|
12
|
Lv G, Hu Z, Tie Y, Du J, Fu H, Gao X and
Zheng X: MicroRNA-451 regulates activating transcription factor 2
expression and inhibits liver cancer cell migration. Oncol Rep.
32:1021–1028. 2014.PubMed/NCBI
|
|
13
|
Xu H, Mei Q, Shi L, Lu J, Zhao J and Fu Q:
Tumor-suppressing effects of miR451 in human osteosarcoma. Cell
Biochem Biophys. 69:163–168. 2014. View Article : Google Scholar
|
|
14
|
Godlewski J, Nowicki MO, Bronisz A, Nuovo
G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA
and Lawler SE: MicroRNA-451 regulates LKB1/AMPK signaling and
allows adaptation to metabolic stress in glioma cells. Mol Cell.
37:620–632. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen MB, Wei MX, Han JY, Wu XY, Li C, Wang
J, Shen W and Lu PH: MicroRNA-451 regulates AMPK/mTORC1 signaling
and fascin1 expression in HT-29 colorectal cancer. Cell Signal.
26:102–109. 2014. View Article : Google Scholar
|
|
16
|
Eble JN, Sauter G, Epstein JI and
Sesterhenn IA: World Health Organization classification of Tumours:
Pathology and Genetics of Tumours of the Urinary System and Male
Genital Organs. IARC Press; Lyon: 2004
|
|
17
|
Kawahara T, Kashiwagi E, Ide H, Li Y,
Zheng Y, Miyamoto Y, Netto GJ, Ishiguro H and Miyamoto H:
Cyclosporine A and tacrolimus inhibit bladder cancer growth through
down-regulation of NFATc1. Oncotarget. 6:1582–1593. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mehlen P and Puisieux A: Metastasis: A
question of life or death. Nat Rev Cancer. 6:449–458. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Raghavan A and Shah ZA: Withania somnifera
improves ischemic stroke outcomes by attenuating PARP1-AIF-mediated
caspase-independent apoptosis. Mol Neurobiol. 52:1093–1105. 2015.
View Article : Google Scholar
|
|
20
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Costa PM and Pedroso de Lima MC: MicroRNAs
as molecular targets for cancer therapy: On the modulation of
microRNA expression. Pharmaceuticals. 6:1195–1220. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu CT, Lin WY, Chang YH, Lin PY, Chen WC
and Chen MF: DNMT1-dependent suppression of microRNA424 regulates
tumor progression in human bladder cancer. Oncotarget.
6:24119–24131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Itesako T, Seki N, Yoshino H, Chiyomaru T,
Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa
M, et al: The microRNA expression signature of bladder cancer by
deep sequencing: The functional significance of the miR-195/497
cluster. PLoS One. 9:e843112014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Sanda T, Look AT, Novina CD and von
Boehmer H: Repression of tumor suppressor miR-451 is essential for
NOTCH1-induced oncogenesis in T-ALL. J Exp Med. 208:663–675. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kovalchuk O, Filkowski J, Meservy J,
Ilnytskyy Y, Tryndyak VP, Chekhun VF and Pogribny IP: Involvement
of microRNA-451 in resistance of the MCF-7 breast cancer cells to
chemotherapeutic drug doxorubicin. Mol Cancer Ther. 7:2152–2159.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zeng T, Peng L, Chao C, Fu B, Wang G, Wang
Y and Zhu X: miR-451 inhibits invasion and proliferation of bladder
cancer by regulating EMT. Int J Clin Exp Pathol. 7:7653–7662.
2014.
|
|
27
|
Hofmann UB, Houben R, Bröcker EB and
Becker JC: Role of matrix metalloproteinases in melanoma cell
invasion. Biochimie. 87:307–314. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Strand S, Vollmer P, van den Abeelen L,
Gottfried D, Alla V, Heid H, Kuball J, Theobald M, Galle PR and
Strand D: Cleavage of CD95 by matrix metalloproteinase-7 induces
apoptosis resistance in tumour cells. Oncogene. 23:3732–3736. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abraham R, Schäfer J, Rothe M, Bange J,
Knyazev P and Ullrich A: Identification of MMP-15 as an
anti-apoptotic factor in cancer cells. J Biol Chem.
280:34123–34132. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Perron MP and Provost P: Protein
interactions and complexes in human microRNA biogenesis and
function. Front Biosci. 13:2537–2547. 2008. View Article : Google Scholar
|
|
31
|
Chen BJ, Wu YL, Tanaka Y and Zhang W:
Small molecules targeting c-Myc oncogene: Promising anti-cancer
therapeutics. Int J Biol Sci. 10:1084–1096. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seo HK, Ahn KO, Jung NR, Shin JS, Park WS,
Lee KH, Lee SJ and Jeong KC: Antitumor activity of the c-Myc
inhibitor KSI-3716 in gemcitabine-resistant bladder cancer.
Oncotarget. 5:326–337. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin F, Ding R, Zheng S, Xing D, Hong W,
Zhou Z and Shen J: Decreased expression of microRNA-744 promotes
cell proliferation by targeting c-Myc in human hepatocellular
carcinoma. Cancer Cell Int. 14:582014. View Article : Google Scholar
|
|
35
|
Liu Z, Zhang G, Li J, Liu J and Lv P: The
tumor-suppressive microRNA-135b targets c-myc in osteoscarcoma.
PLoS One. 9:e1026212014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang X, Cai H, Liang Y, Chen L, Wang X, Si
R, Qu K, Jiang Z, Ma B, Miao C, et al: Inhibition of c-Myc by
let-7b mimic reverses mutidrug resistance in gastric cancer cells.
Oncol Rep. 33:1723–1730. 2015.PubMed/NCBI
|
|
37
|
Lü M, Ding K, Zhang G, Yin M, Yao G, Tian
H, Lian J, Liu L, Liang M, Zhu T, et al: MicroRNA-320a sensitizes
tamoxifen-resistant breast cancer cells to tamoxifen by targeting
ARPP-19 and ERRγ. Sci Rep. 5:87352015. View Article : Google Scholar
|