Introduction
As one of the major causes of cancer-related
mortality worldwide, colorectal cancer (CRC) is surgically curable
at early stages, but advanced disease at the metastatic stage is
associated with high mortality rates (1,2). CRCs
progress from local adenomas in the intestinal epithelium to
invasive carcinomas that typically metastasize to the liver
(3). Approximately half of all
patients with local CRCs may develop metastases. Current therapies
show a 5-year survival of less than 5% for metastatic disease
(4).
The forkhead box (Fox) gene family encodes a large
and diverse group of transcription factors that share certain
characteristics in a conserved, ~100 amino acid DNA-binding motif
known as the forkhead or winged helix domain; over 150 proteins
with forkhead domains have been identified, comprising at least 17
subclasses, to date (5,6). Through the transcriptional control of
gene expression, many FOX protein members reportedly play important
roles in embryonic development and organogenesis and in the
regulation of various physiological processes, such as the cell
cycle (7), progression (8), cell survival (9), cellular metabolism (10), life span (11) and immune responses (12). Consequently, dysregulation of the
function, subcellular localization and expression of FOX
transcription factors leads to the development and progression of
diseases, particularly cancer (13,14).
Several other FOX family proteins have been shown to have either a
tumor-promoting [FOXA1 (15), FOXQ1
(16), FOXM1 (17) and FOXC2 (18)] or -suppressing [FOXD3 (19), FOXP1 (20), FOXO1 (21) and FOXP2 (22)] role in the cellular signaling that
is associated with proliferation and/or suppression.
The human forkhead box K1 (FOXK1) gene encodes
predicted proteins that are most homologous to the mouse myocyte
nuclear factor MNF/Forkhead box K1 (Foxk1) protein. FOXK1 is
predominantly expressed in many malignant tissues and in the brain,
colon and lymph nodes (23). This
gene has been implicated in normal and neoplastic developmental
processes. Wang et al demonstrated that FOXK1 and FOXK2
positively regulate Wnt/β-catenin signaling by translocating
Dishevelled (DVL) into the nucleus. Moreover, FOXK1 and FOXK2
protein levels are elevated in human CRCs (24). However, the underlying mechanism of
FOXK1 induction in multiple cellular events, such as growth,
apoptosis and chemotherapy resistance, remains unclear.
In the present study, we demonstrated a novel and
effective way to knock down FOXK1, thereby significantly inducing
apoptosis, inhibiting tumorigenesis and tumor growth and strongly
enhancing the antitumor activity of 5-fluorouracil (5-FU) in
vitro and in vivo. Our results showed that
lenti-shRNA-FOXK1 is a promising candidate for the gene therapy of
colon cancer.
Materials and methods
Reagents and cell culture
5-FU was purchased from Sigma-Aldrich (St. Louis,
MO, USA). Mouse antibodies against FOXK1 (G-4), cyclin D1 (A-12),
CDK4 (H-22), CDK6 (C-21), caspase 3 p11 (C-6), caspase 8 p18 (E-8),
caspase 9 p35 (A-9), PARP-1 (H-300), GAPDH (G-9), horseradish
peroxidase-conjugated anti-goat IgG, and anti-mouse IgG were
purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Culture reagents were purchased from Invitrogen (Carlsbad, CA,
USA). The colon cancer cell lines SW480 and SW1116 were obtained
from the American Type Culture Collection (ATCC; Rockville, MD,
USA) and were cultured as previously described (25). The cells were maintained at 37°C
with 5% CO2 in a humidified incubator and were
subcultured using 0.25% trypsin every 2–3 days, before confluence
was reached.
Constructs and establishment of stable
transfectants
Complementary DNA (cDNA) corresponding to
full-length FOXK1 was obtained by RT-PCR amplification of normal
human testis cDNA with primers specific to FOXK1. The PCR products
were subcloned into the mammalian expression vector pcDNA3.1
(Invitrogen).
To establish stable cell lines, cells transfected
with the empty pcDNA3.1 vector or with pcDNA3.1-FOXK1 were passaged
at 1:15 (vol/vol) and were cultured in RPMI-1640 medium
supplemented with geneticin (G418; Calbiochem, Darmstadt, Germany)
at 1,000 µg/ml for 4 weeks. Stably transfected clones were
selected by immunoblotting for FOXK1 expression and were maintained
in medium containing 800 µg/ml G418 for additional
studies.
RNA isolation and reverse
transcription-PCR analysis
Cells were harvested, and total RNA was extracted
using TRIzol reagent (Gibco-BRL and Life Technologies) (25). RNA was reverse transcribed to cDNA
using the ThermoScript RT system reagent (Gibco-BRL) according to
the manufacturer's instructions. PCR was performed using 2
µl of the resulting cDNA and 0.3 U of hot start DNA
polymerase. Hot start PCR was performed for 32 cycles, with an
initial 95°C denaturation for 3 min before cycles of 94°C
denaturation for 45 sec, 55°C annealing for 45 sec, and 72°C
elongation for 45 sec and a final extension of 10 min.
Transient siRNA transfection
Ablation of FOXK1 was performed by transfection with
small interfering RNA (siRNA) duplex oligos, which were synthesized
by GenePharma (Shanghai, China). Control siRNA (scrambled RNA) and
FOXK1-specific siRNA [sense, 889-GAGACAGCCCCAAGGAUGA (dTdT)-908 and
antisense, 908-UCAUCCUUGGGGCUGUCUC(dTdT)-889] were transfected
using Lipofectamine 2000 (Invitrogen). Forty-eight hours after
transfection, western blot analysis was performed.
Western blot analysis
For western blot analysis, cells were harvested 48 h
after transfection and lysed in lysis buffer [10 mM Tris-HCl (pH
7.4), 1% SDS, 10% glycerol, 5 mM MgCl2, 1 mM
phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 5
µg/ml leupeptin and 21 µg/ml aprotinin). A total of
30 µg of each protein lysate was separated by SDS-PAGE and
transferred onto a polyvinylidene fluoride (PVDF) membrane. The
primary antibodies were diluted according to the manufacturer's
recommendations. The antibodies were then visualized using an
enhanced chemiluminescence detection system.
Flow cytometric analysis
For cell cycle analysis, SW480 cells were reverse
transfected into 6-well dishes and were incubated for 48 h under
standard growth conditions. Cells were fixed for 90 min with 70%
(v/v) pre-cooled ethanol (−20°C) and harvested by centrifugation
(4°C, 5 min, 1,000 × g). Staining was performed using a
69-µM propidium iodide (PI) solution in phosphate-buffered
saline (PBS) containing RNase A (0.6 µg/ml) for 30 min at
37°C. The cell cycle distribution was calculated from the resultant
DNA histograms using FlowJo software.
Apoptosis was detected using the Annexin V-FITC kit
(Trevigen, Inc., Gaithersburg, MD, USA) according to the
manufacturer's instructions. Briefly, cells with various treatments
were collected and stained with Annexin V-FITC and PI in the dark
for 15 min before being subjected to flow cytometry and analyzed
using FlowJo software.
Cell growth assay
Cells were seeded at 5.0×104/well into
96-well plates. Cell proliferation was determined by measuring the
absorption of the cell proliferation reagent WST-1 (Roche Molecular
Biochemicals, Basel, Switzerland) according to the manufacturer's
protocol. The ratio of the absorbance of treated cells at 450 nm
relative to that of the untransfected cells was calculated and
expressed as the proliferation index. Each treatment was performed
in quadruplicate, and the proliferation values were expressed as
the mean ± SEM. The anchorage-independent cell growth assay was
performed as previously described (25). Briefly, stable transfectants were
seeded at 2×103/well into 6-well plates and cultured for
14 days. Cell colonies were microscopically visualized after
staining with 0.005% crystal violet. Each treatment was conducted
in triplicate, and 3 independent experiments were performed.
Morphological detection of apoptosis
Apoptotic cell death was morphologically evaluated
as previously described, with some modifications. Cells were fixed
for 5 min in 3% paraformaldehyde in PBS. After air drying, the
cells were stained for 10 min in Hoechst 33258 (10 µg/ml),
mounted in 50% glycerol containing 20 mM citric acid and 50 mM
orthophosphate, and stored at −20°C before analysis; nuclear
morphology was evaluated using a Zeiss IM 35 fluorescence
microscope.
Construction of lentiviral vectors with
FOXK1 short hairpin RNA
To further investigate the effect of siRNA-induced
downregulation of FOXK1 expression on the in vivo tumor
growth of CRC, a FOXK1-RNAi lentiviral vector (GV208-FOXK1-shRNA)
was constructed by Shanghai GeneChem Co., Ltd. (Shanghai, China).
Double-stranded oligonucleotides encoding human FOXK1-vshRNA
(NM_001037165;
CCGGGAGACAGCCCCAAGGATGATCAAGAGTCATCCTTGGGGCTGTCTCTTTTTG) were
annealed and inserted into the short hairpin RNA (shRNA) expression
vector GV208-GFP (Shanghai GeneChem Co, Ltd.). A GFP lentiviral
vector (GV208-GFP) was used as a negative control. Clone identity
was verified by sequencing.
Recombinant lentiviral vector was produced by
co-transfecting HEK293FT cells with the lentiviral expression
vector and packing the plasmid mix using Lipofectamine™ 2000
according to the manufacturer's instructions. Infectious lentiviral
particles were harvested at 48 h post-transfection and were then
filtered through 0.45 µm cellulose acetate filters. The
virus was concentrated, and the titer was determined by serial
dilution of 293T cells.
For lentivirus transduction, the SW480 cells were
subcultured at 1×105 cells/well into 6-well culture
plates. Cells were transduced with the FOXK1-siRNA-expressing
(FOXK1-siRNA) or scramble (scr)-siRNA-expressing lentivirus at a
multiplicity of infection (MOI) of 50. Cells were then harvested 72
h after infection, and the transduction efficiency was evaluated by
counting the percentage of GFP-positive cells.
In vivo tumor growth assay
The present study was conducted in strict accordance
with the recommendations in the Guide for the Institutional animal
care and Use committee (IACUC), and the protocol was approved by
the committee on the Ethics of Animal Experiments of Nanfang
Hospital. All of the surgeries were performed under sodium
pentobarbital anesthesia, and all efforts were made to minimize
suffering. After the surgery, all of the nude mice were euthanized
by sodium pentobarbital anesthesia.
SW480 CRC cells were transduced in vitro at
an MOI of 50 with lenti-scr-shRNA or lenti-FOXK1-shRNA infected
with NS. Twelve hours after transduction, 5×106 viable
cells expressing lenti-scr-shRNA or lenti-FOXK1-shRNA were injected
into the right flanks of 6-week-old female nude mice, respectively.
On day 28 after inoculation, the mice were sacrificed, and the
tumors were dissected and weighed. Immunohistochemical analysis was
performed using anti-Ki-67 and anti-CD105 antibodies.
For the other 3 groups of mice, lenti-scr-shRNA,
lenti-scr-shRNA + 5-FU or lenti-FOXK1 + 5-FU were directly injected
into the tumors at 3 different locations using a 10-ml
micro-syringe (Hamilton, Reno, NV, USA). Three mice (3 injections)
were included in each group. The volume of each tumor was
calculated as follows: V = (1/2) R12R2, where R1 is
radius 1, R2 is radius 2, and R1<R2. The mice were sacrificed,
and the tumors were dissected and weighed on day 28 after
inoculation. The tumors were then removed, fixed in formalin,
embedded in paraffin and subjected to terminal
deoxynucleotidyltransferase-mediated deoxyuridine
triphosphate-digoxigenin nick-end labeling (TUNEL).
In situ detection of apoptotic cells by
the TUNEL assay
Cell apoptosis in tumor xenograft tissues was
detected by TUNEL staining. Tumor tissues from xenografts were
excised and immediately formalin-fixed. TUNEL staining was
performed using an ApoAlert DNA Fragmentation Assay kit (Clontech,
Palo Alto, CA, USA) according to the manufacturer's instructions,
with apoptotic cells exhibiting green nuclear fluorescence
(26). Tumor tissue sections were
counterstained with PI for 5–10 min to stain cell nuclei. The
percentage of apoptotic cells was assessed in 10 randomly selected
fields at a magnification of ×40. The apoptotic index was
calculated as the number of apoptotic cells/total number of
nucleated cells × 100%.
Statistical analysis
The results obtained from cell growth experiments
are expressed as the mean ± SD. The results from different
treatments were compared using two-tailed Student's t-tests and
were considered significant at a threshold of p<0.05.
Results
Ectopic overexpression of FOXK1 increases
the expression of multiple oncogenes in vitro
We developed in vitro models to examine the
mechanistic role of FOXK1 in CRC biology, and we established stable
transfectants with FOXK1 sense and vector (pcDNA3.1) plasmids. The
overexpression of FOXK1 was confirmed by western blot analysis
(Fig. 1A).
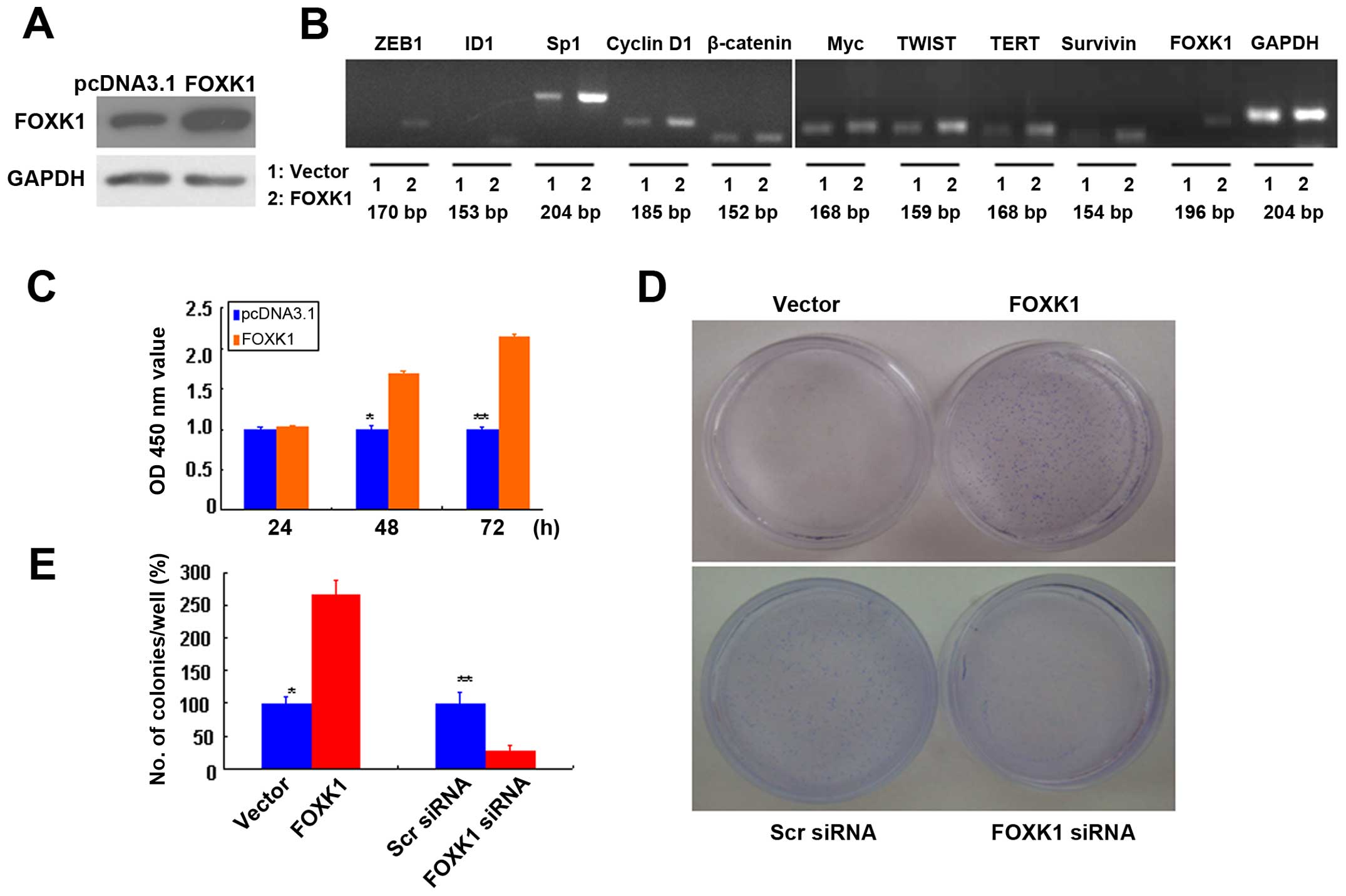 | Figure 1FOXK1 regulates multiple oncogenes
and promotes the proliferation and anchorage-independent growth of
CRC cells. (A) FOXK1 expression in stable transfectants of SW480
cells harboring the empty vector or expressing FOXK1, as detected
by western blotting. GAPDH was used as the internal control. (B)
The mRNA expression levels of ZEB1, ID1, Sp1, cyclin D1, β-catenin,
Myc, TWIST, TERT, survivin and FOXK1 were upregulated in stable
transfectants of FOXK1. (C) Significant differences between FOXK1-
and empty vector-transfected cells were found at 48 and 72 h
(p<0.05). (D) SW480 and empty vector-transfected cells formed
colonies in soft agar as expected, while forced expression of FOXK1
markedly enhanced colony formation in soft agar. (E) FOXK1
expression increased the clonogenicity of SW480 cells by 155%. |
Next, we screened for potential FOXK1 target genes
by ectopically expressing FOXK1 in SW480 cells and examining the
subsequent changes in the expression levels of 10 major oncogenes
that are reportedly involved in proliferation, transformation or
apoptosis inhibition (27). The
mRNA expression levels of ZEB1, ID1, Sp1, cyclin D1, β-catenin,
Myc, TWIST, TERT, survivin and FOXK1 were upregulated in the stable
transfectants of FOXK1 (Fig.
1B).
Our results suggest that FOXK1 activates the
expression of multiple oncogenes in CRC.
FOXK1 promotes cell growth by stimulating
cell proliferation
To analyze the effect of FOXK1 on cell growth, we
assessed the proliferation of the stable transfectants cultured in
complete cell culture medium at various time points. The cell
growth percentages relative to those of transfectants carrying an
empty vector were calculated. We showed that the growth rates of
the SW480-FOXK1 transfectants were 103.1±1.5, 169.6±3.1 and
214.3±4.0% after culture for 24, 48 and 72 h, respectively
(Fig. 1C). Significant differences
between the FOXK1- and empty vector-transfected cells were found at
48 and 72 h (p<0.05).
Anchorage-independent growth is one pivotal
characteristic of malignant transformation. To determine whether
forced expression of FOXK1 may alter the oncogenicity of CRC cells,
we examined the anchorage-independent growth of the stable
transfectants with the empty vector or with FOXK1 in soft agar
after 14 days. SW480 and empty vector-transfected cells formed
colonies in soft agar as expected, while forced expression of FOXK1
markedly enhanced colony formation in soft agar (Fig. 1D). FOXK1 expression increased the
clonogenicity of the SW480 cells by 155% (Fig. 1E). Conversely, we also tested the
effect of RNAi-mediated FOXK1 knockdown in the SW480 cells.
FOXK1-siRNA knockdown caused a significant loss of colony-forming
capacity. The clonogenicity of the suppressed SW480 cells was
reduced by 71.4% (Fig. 1E).
Therefore, the overexpression of FOXK1 markedly
promoted the proliferation of CRC cells.
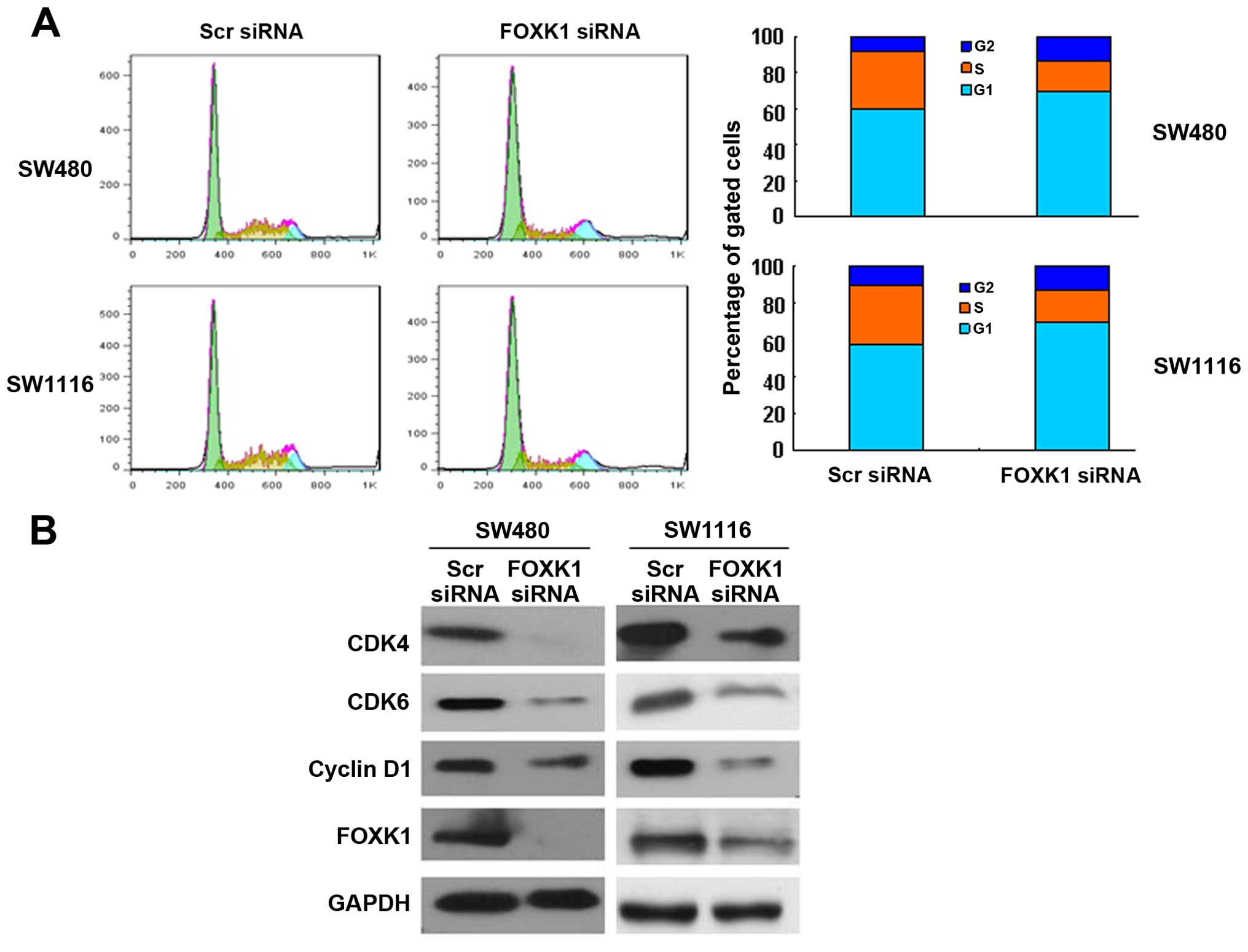
Knockdown of FOXK1 induces G0/G1 cell
cycle arrest
To assess in more detail the effect of constitutive
FOXK1 expression on the growth characteristics of CRC cells in
vitro, we evaluated the cell cycle distribution of the cells by
flow cytometry. FOXK1 knockdown increased the proportion of cells
in the G0/G1 phase and concomitantly decreased the proportion of
cells in the G2/M phase of the cell cycle (Fig. 2A).
Furthermore, cell cycle-related protein expression
was assessed by western blotting. Consistent with the accumulation
of cells in G0/G1 phase, cyclin D1, CDK4 and CDK6 were
significantly decreased in the FOXK1-siRNA-treated SW480 and SW1116
cells (Fig. 2B). The
cyclin-dependent kinases (CDKs) CDK4 and CDK6, along with cyclin D1
proteins, are involved in the progression of cells through G1 phase
and their entry into S phase. Collectively, these findings indicate
that reduced expression of FOXK1 slowed cell cycle progression
through the G1 phase.
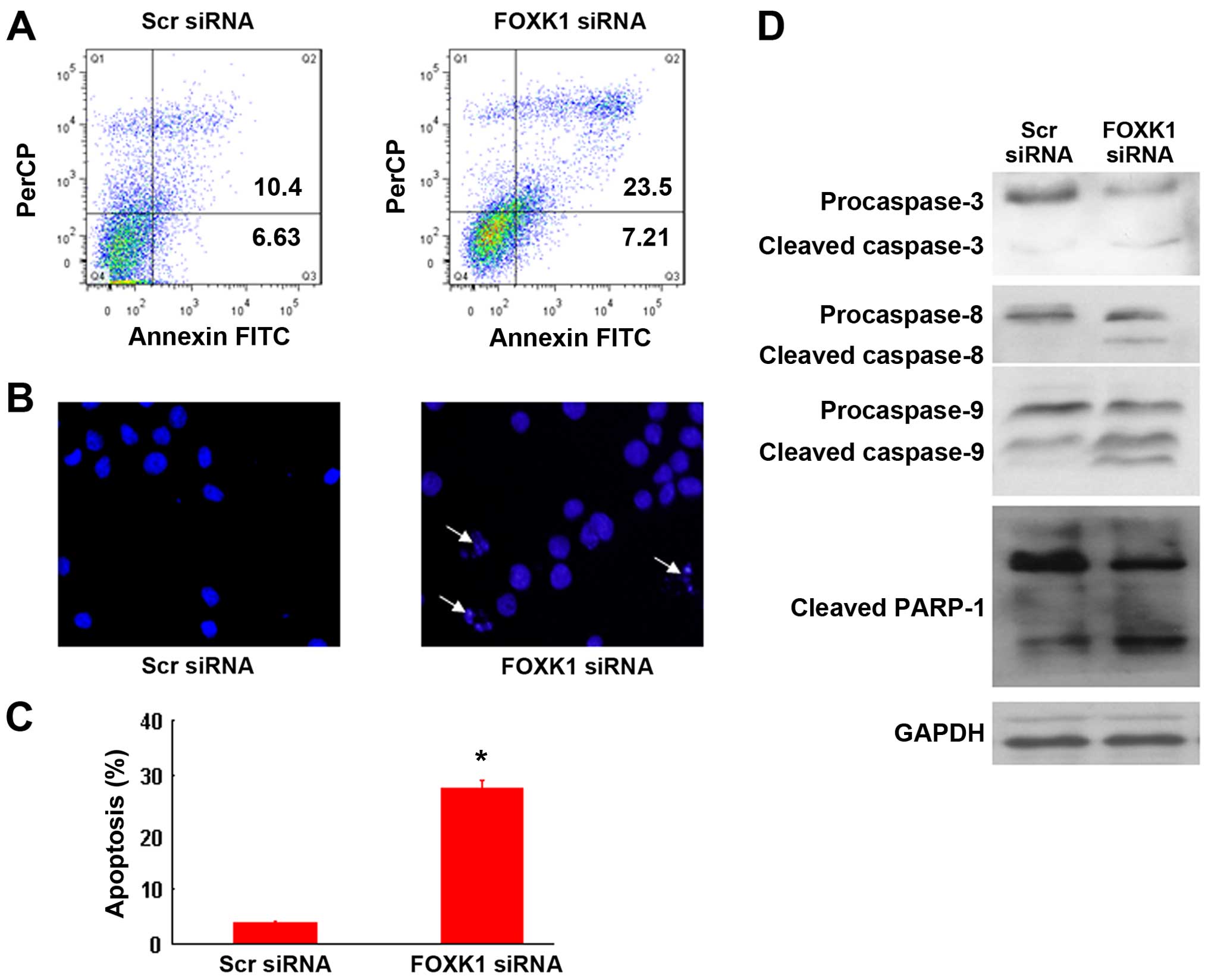
Knockdown of FOXK1 induces CRC cell
apoptosis
To investigate the mechanism by which FOXK1-siRNA
induces growth suppression, we performed an apoptosis assay.
Apoptosis was detected by FACS analysis after double staining with
PI and Annexin V-FITC. As shown in Fig.
3A, FOXK1-siRNA induced more apoptosis than scr-siRNA in the
SW480 cells, indicating that FOXK1-siRNA inhibited cell growth by
inducing apoptosis. Apoptotic induction was further confirmed by
Hoechst 33258 staining at the single-cell level (Fig. 3B and C). Moreover, FOXK1-siRNA
activated caspases 3, 8 and 9, as evidenced by the increased
protein level of cleaved caspases compared with that in the
scr-siRNA-treated cells (Fig. 3D).
Taken together, these results support the idea that knockdown of
FOXK1 promotes the apoptosis of CRC cells.
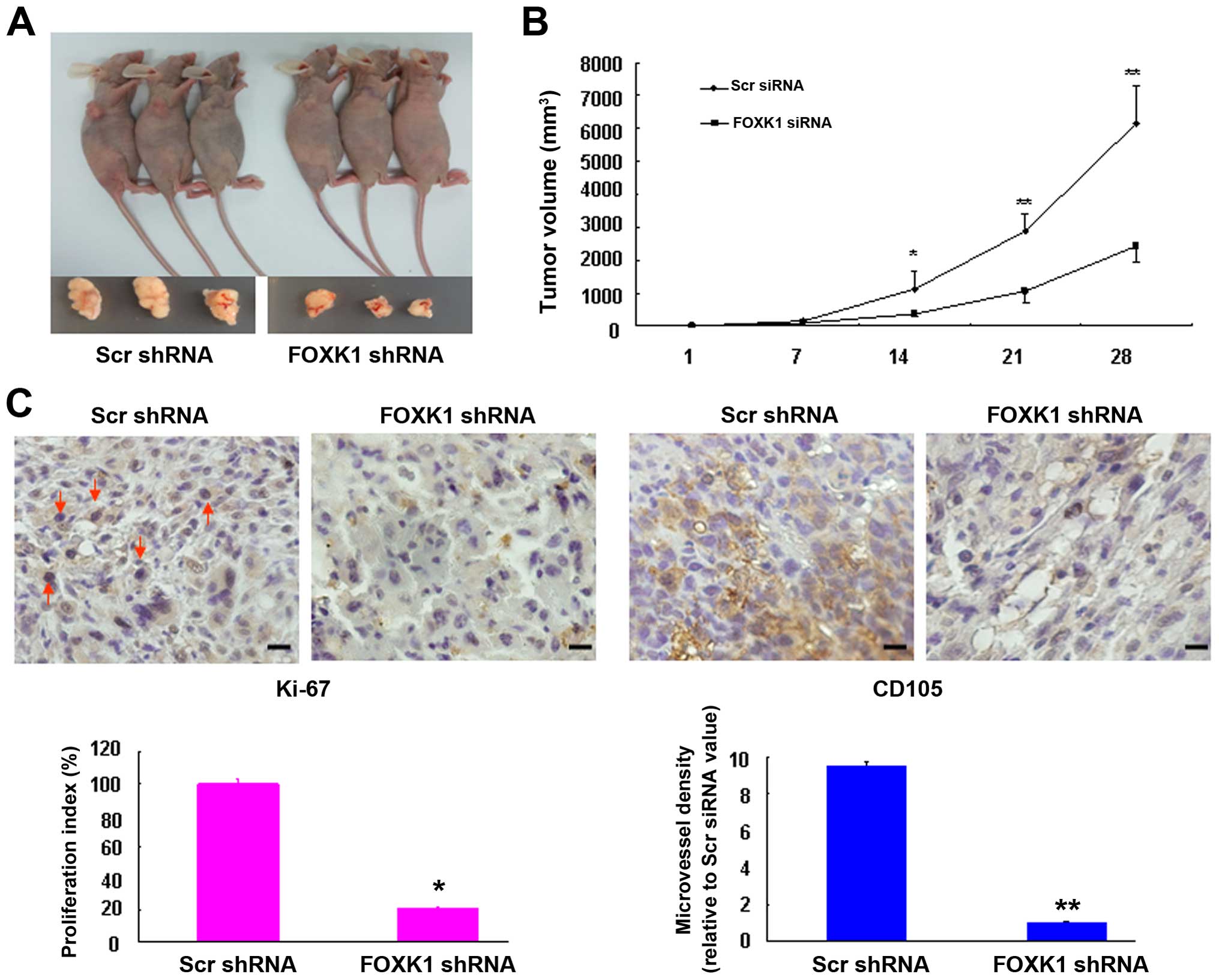
Silencing of FOXK1 suppresses the growth
of xenograft tumors in nude mice
To explore the effects of FOXK1 knockdown in
vivo, xenograft tumors were generated by injecting SW480 cells
that were stably infected with either scr-shRNA or FOXK1-shRNA. The
cells were subcutaneously injected into the flanks of nude mice
(Fig. 4A). As shown in Fig. 4B, compared with the scr-shRNA group,
the tumors derived from FOXK1-shRNA-infected cells grew much more
slowly throughout the experiment, suggesting that knockdown of
FOXK1 markedly impaired the tumorigenic growth of the SW480
cells.
We next examined the protein expression of cell
proliferation (Ki-67) and angiogenesis markers (CD105) in xenograft
tumors. Representative immunohistochemical images of the tumors are
shown in Fig. 4C. The knockdown of
FOXK1 in the FOXK1 group showed significantly inhibited
proliferation rates and lower tumor vessel density relative to the
scr-shRNA group. These findings suggest that FOXK1-shRNA may have
an inhibitory effect on tumorigenesis in vivo.
Knockdown by FOXK1-shRNA increases cell
susceptibility to apoptotic stimuli in vitro and in vivo
To evaluate the role of FOXK1-siRNA-mediated
knockdown in the susceptibility of cells to chemotherapy-induced
apoptosis, SW480 cells were treated with or without 5-FU (50
µg/ml, using NS as the vehicle) for 48 h in vitro.
Double staining with Annexin V and PI was then performed, followed
by flow cytometric analysis to determine the apoptosis rate. As
shown in Fig. 5A, the apoptotic
indices of the cells transfected with FOXK1-siRNA + 5-FU and
scr-siRNA + 5-FU were significantly increased relative to the
scr-siRNA controls.
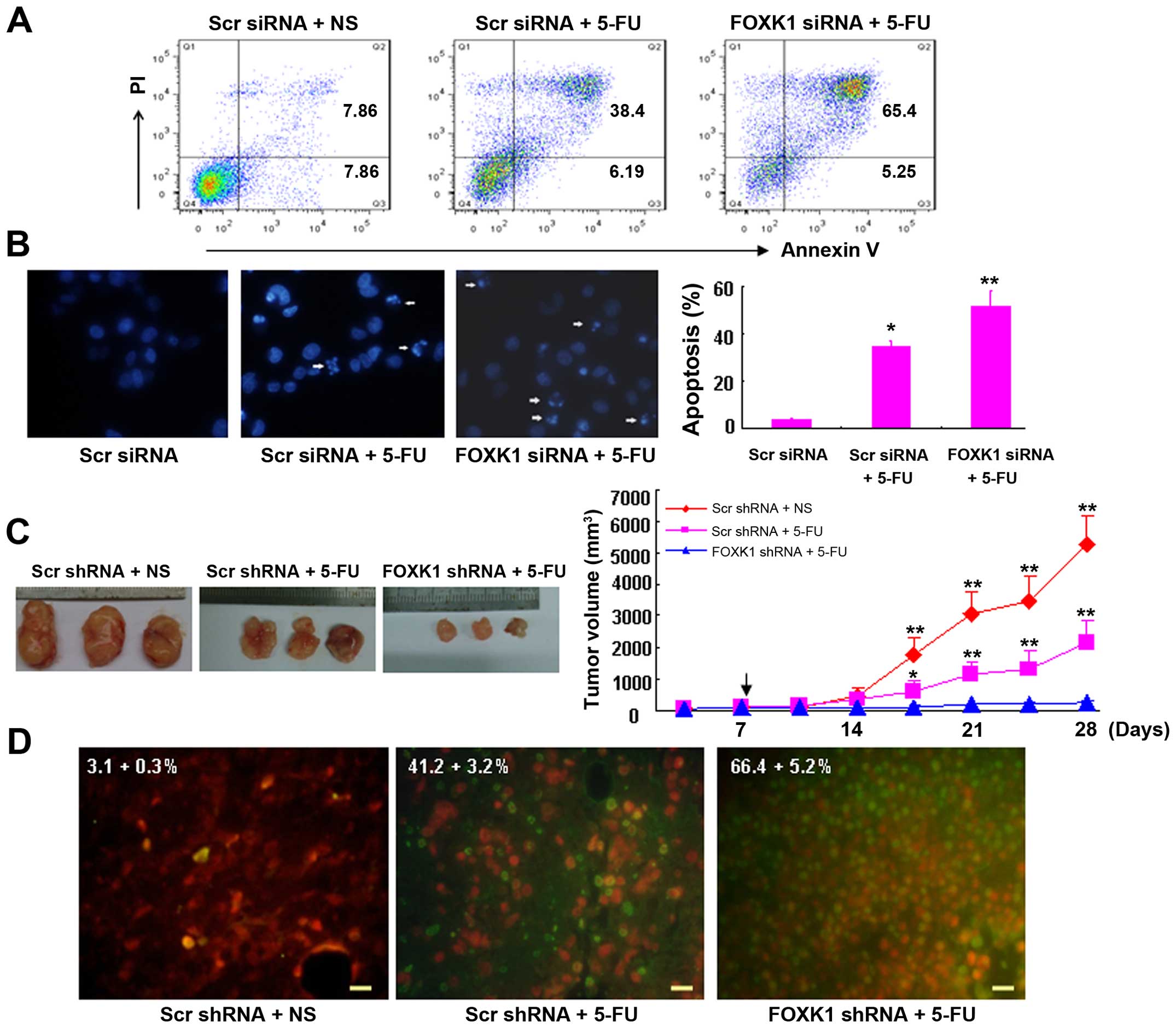 | Figure 5Knockdown of FOXK1 with shRNA
sensitizes cancer cells to 5-FU-induced apoptosis in vitro
and in vivo. (A) Knockdown of cells with FOXK1-siRNA
increased the cell susceptibility to apoptotic stimuli. Cells
transfected with scr-siRNA or FOXK1-siRNA were treated with 5-FU
for 48 h, double-stained with annexin V-FITC and PI, and subjected
to flow cytometric analysis to detect apoptosis. (B) By Hoechst
33258 staining, the percentages of condensed nuclei-positive cells
were higher in the SW480 cells transfected with FOXK1-siRNA + 5-FU
(**p<0.001 compared with FOXK1-siRNA + 5-FU) and
scr-siRNA + 5-FU (*p<0.01 compared with scr-siRNA +
5-FU) when compared with this percentage in the SW480 cells
transfected with scr-siRNA. (C) Lentivirus-transduced SW480 cells
were subcutaneously injected into mice. When the tumor nodules
became visible (7 days later), 5-FU (50 µg/kg, once/2 days)
was administered by intraperitoneal injection. Tumor growth was
monitored in 3 dimensions and was expressed as the tumor volume in
cubic millimeters. The arrow indicates the time at which
intraperitoneal injections were conducted. The data are the pooled
averages ± standard errors of the means of the tumor volumes for
each of the 3 animals/group; *p<0.01 and
**p<0.001, scr-shRNA + 5-FU vs. scr-shRNA;
**p<0.001, scr-shRNA vs. FOXK1-shRNA + 5-FU. (D)
Spontaneous apoptosis in tumor tissues as visualized by TUNEL
staining as a marker of apoptosis (apoptotic cells are green). The
tumor section was counterstained with PI for 5–10 min. The number
of apoptotic cells was assessed in 3 randomly selected fields,
viewed at a magnification of ×40, on each slide. The apoptotic
index was calculated as the number of apoptotic cells/total number
of nucleated cells × 100%. NS was used as the vehicle. Scale bars,
100 µm. |
Similarly, by Hoechst 33258 staining, the proportion
of condensed nuclei-positive cells was higher in the SW480 cells
transfected with FOXK1-siRNA + 5-FU (p<0.001 compared with
FOXK1-siRNA + 5-FU) and scr-siRNA + 5-FU (p<0.01 compared with
scr-siRNA + 5-FU) than in SW480 cells transfected with the
scr-siRNA (Fig. 5B).
Moreover, the antitumor effect of FOXK1-shRNA and/or
5-FU in vivo was demonstrated by a xenograft model in nude
mice (Fig. 5C). We subcutaneously
injected SW480 cells that were stably infected with either
scr-shRNA or FOXK1-shRNA into the right flanks of nude mice,
respectively. When tumor nodules became visible (~3–5 mm in
diameter), 5-FU was intraperitoneally injected as a co-treatment.
The tumor sizes were then monitored weekly. As shown in Fig. 5C, the tumor volumes of the
scr-shRNA-treated mice were markedly greater than those of the
FOXK1-shRNA + 5-FU- and scr-shRNA + 5-FU-treated mice 4 weeks after
injection of the SW480 cells.
Tumors injected with the combination of FOXK1-shRNA
and/or 5-FU exhibited the greatest percentage of apoptotic cells.
In contrast, tumors injected with scr-shRNA exhibited relatively
few TUNEL-positive cells (Fig. 5D).
These findings suggest that FOXK1-shRNA enhances the susceptibility
of cancer cells to apoptotic triggers that are induced by 5-FU.
Discussion
In the present study, we found that the
overexpression of FOXK1 increased the expression of multiple
oncogenes. We also demonstrated that the antitumor efficacy of
shRNA-mediated FOXK1 inhibition in cells and nude mouse xenograft
models was due to the induction of cell cycle arrest and apoptosis.
These results indicate that FOXK1-shRNA may represent a novel and
potent therapeutic agent, on its own or in conjunction with 5-FU,
for the treatment of colon cancer.
Members of the Fox transcription factor family have
been identified in several vertebrate cell lineages. Fox proteins
have similar binding specificity for a core DNA sequence,
(T/C)(A/C)AA(C/T)A (28,29), and they display conserved amino acid
sequences in the putative recognition helix. There is evidence that
FOX proteins have a central function in established cancers. For
example, FOXA1, the most extensively studied member of the family,
is upregulated in nearly all cancers, including breast (30), bladder (31), prostate (32) and pancreatic cancers (33). This protein contributes to many of
the typical characteristics of cancer, including increased
proliferation, resistance to cell death, and increased invasion and
metastasis. Wang et al showed that FOXK1 protein levels are
elevated in human CRCs (24).
However, the use of FOXK1 as a target in CRC biological therapy has
yet to be established. The present study showed that ectopic FOXK1
expression significantly induced the RNA levels of multiple
oncogenes that are involved in neoplastic transformation,
apoptosis, the cell cycle and proliferation, including ZEB1, ID1,
Sp1, cyclin D1, β-catenin, Myc, TWIST, c-Jun and survivin. Among
these targets, Myc and cyclin D1 are involved in cell cycle
regulation. Survivin and β-catenin are involved in neoplastic
transformation. The transcription factors TWIST, AP-1, Sp1, ZEB1
and ID1, as well as epithelial-to-mesenchymal transition inducers,
underlie the phenotypic conversion from epithelial cells to
mesenchymal cells. Thus, FOXK1 acts as an oncogene in CRC.
The oncogenic function of FOXK1 in colon cancer was
further investigated using in vitro and in vivo
assays. siRNA-mediated FOXK1 knockdown had a marked growth
suppression effect under both anchorage-dependent and -independent
culture conditions, and it also reduced tumor size in nude mice.
Flow cytometry revealed that FOXK1 knockdown caused significant
G1/G0 arrest, and a concomitant reduction of CDK4/CDK6 expression
was apparent. Furthermore, we found that FOXK1 knockdown increased
apoptosis, a finding further confirmed by staining with Hoechst
33258. Notably, apoptotic induction by FOXK1-siRNA was mediated
through the intrinsic and extrinsic caspase-dependent pathways.
Taken together, these results indicate that FOXK1 is involved in
cell growth and apoptosis in CRC.
The drug 5-FU is an important component of standard
chemotherapy protocols for various solid tumors, including CRCs
(34,35). In the present study, we further
investigated the in vitro and in vivo effects of
combination treatment with FOXK1-shRNA and 5-FU on colon cancer
growth. According to flow cytometric analysis and Hoechst 33258
staining, knockdown of FOXK1 resulted in a greater proportion of
5-FU-induced apoptotic cells. We also found that the volumes of
xenograft tumors were significantly reduced when treated with a
combination of FOXK1-shRNA and 5-FU. Furthermore, TUNEL staining
demonstrated that FOXK1-shRNA could further enhance 5-FU-induced
apoptosis. To the best of our knowledge, no studies have yet shown
the therapeutic value of RNAi-mediated FOXK1 knockdown in
conjunction with 5-FU in CRC.
The present study demonstrates that FOXK1 acts as an
oncogene in CRC. FOXK1 knockdown inhibits cell growth and promotes
apoptosis in CRC cells in vitro. Furthermore, FOXK1-shRNA
hinders tumor progression and results in outright regression when
combined with 5-FU in vivo. Thus, our current data provide
direct evidence that a combination of RNAi-mediated FOXK1 knockdown
and 5-FU may be a potent strategy for colon cancer therapy.
References
|
1
|
Finlay IG and McArdle CS: Effect of occult
hepatic metastases on survival after curative resection for
colorectal carcinoma. Gastroenterology. 85:596–599. 1983.PubMed/NCBI
|
|
2
|
Kemp Z, Thirlwell C, Sieber O, Silver A
and Tomlinson I: An update on the genetics of colorectal cancer.
Hum Mol Genet. 13(Suppl 2): R177–R185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Konishi M, Kikuchi-Yanoshita R, Tanaka K,
Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, et
al: Molecular nature of colon tumors in hereditary nonpolyposis
colon cancer, familial polyposis, and sporadic colon cancer.
Gastroenterology. 111:307–317. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Erning FN, Crolla RM, Rutten HJ,
Beerepoot LV, van Krieken JH and Lemmens VE: No change in lymph
node positivity rate despite increased lymph node yield and
improved survival in colon cancer. Eur J Cancer. 50:3221–3229.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mazet F, Yu JK, Liberles DA, Holland LZ
and Shimeld SM: Phylogenetic relationships of the Fox (Forkhead)
gene family in the Bilateria. Gene. 316:79–89. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bieller A, Pasche B, Frank S, Gläser B,
Kunz J, Witt K and Zoll B: Isolation and characterization of the
human forkhead gene FOXQ1. DNA Cell Biol. 20:555–561. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ackermann S, Kocak H, Hero B, Ehemann V,
Kahlert Y, Oberthuer A, Roels F, Theißen J, Odenthal M, Berthold F,
et al: FOXP1 inhibits cell growth and attenuates tumorigenicity of
neuroblastoma. BMC Cancer. 14:8402014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stoll SW, Stuart PE, Swindell WR, Tsoi LC,
Li B, Gandarillas A, Lambert S, Johnston A, Nair RP and Elder JT:
The EGF receptor ligand amphiregulin controls cell division via
FoxM1. Oncogene. 35:2075–2086. 2016. View Article : Google Scholar
|
|
9
|
Dai B, Gong A, Jing Z, Aldape KD, Kang SH,
Sawaya R and Huang S: Forkhead box M1 is regulated by heat shock
factor 1 and promotes glioma cells survival under heat shock
stress. J Biol Chem. 288:1634–1642. 2013. View Article : Google Scholar :
|
|
10
|
Kikuchi K, Hettmer S, Aslam MI, Michalek
JE, Laub W, Wilky BA, Loeb DM, Rubin BP, Wagers AJ and Keller C:
Cell-cycle dependent expression of a translocation-mediated fusion
oncogene mediates checkpoint adaptation in rhabdomyosarcoma. PLoS
Genet. 10:e10041072014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burgering BM: A brief introduction to
FOXOlogy. Oncogene. 27:2258–2262. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ouyang W, Beckett O, Flavell RA and Li MO:
An essential role of the Forkhead-box transcription factor Foxo1 in
control of T cell homeostasis and tolerance. Immunity. 30:358–371.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schimmel J, Eifler K, Sigurðsson JO,
Cuijpers SA, Hendriks IA, Verlaan-de Vries M, Kelstrup CD,
Francavilla C, Medema RH, Olsen JV, et al: Uncovering SUMOylation
dynamics during cell-cycle progression reveals FoxM1 as a key
mitotic SUMO target protein. Mol Cell. 53:1053–1066. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Crose LE, Galindo KA, Kephart JG, Chen C,
Fitamant J, Bardeesy N, Bentley RC, Galindo RL, Chi JT and Linardic
CM: Alveolar rhabdomyosarcoma-associated PAX3-FOXO1 promotes
tumorigenesis via Hippo pathway suppression. J Clin Invest.
124:285–296. 2014. View
Article : Google Scholar
|
|
15
|
Nucera C, Eeckhoute J, Finn S, Carroll JS,
Ligon AH, Priolo C, Fadda G, Toner M, Sheils O, Attard M, et al:
FOXA1 is a potential oncogene in anaplastic thyroid carcinoma. Clin
Cancer Res. 15:3680–3689. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
VersicanV1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar
|
|
17
|
Wang Z, Banerjee S, Kong D, Li Y and
Sarkar FH: Downregulation of Forkhead Box M1 transcription factor
leads to the inhibition of invasion and angiogenesis of pancreatic
cancer cells. Cancer Res. 67:8293–8300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui YM, Jiao HL, Ye YP, Chen CM, Wang JX,
Tang N, Li TT, Lin J, Qi L, Wu P, et al: FOXC2 promotes colorectal
cancer metastasis by directly targeting MET. Oncogene.
34:4379–4390. 2015. View Article : Google Scholar
|
|
19
|
Li D, Mei H, Qi M, Yang D, Zhao X, Xiang
X, Pu J, Huang K, Zheng L and Tong Q: FOXD3 is a novel tumor
suppressor that affects growth, invasion, metastasis and
angiogenesis of neuroblastoma. Oncotarget. 4:2021–2044. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Zhang S, Wang X, Liu J, Yang L,
He S, Chen L and Huang J: Prognostic significance of FOXP1 as an
oncogene in hepatocellular carcinoma. J Clin Pathol. 65:528–533.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ko YS, Cho SJ, Park J, Kim Y, Choi YJ, Pyo
JS, Jang BG, Park JW, Kim WH and Lee BL: Loss of FOXO1 promotes
gastric tumour growth and metastasis through upregulation of human
epidermal growth factor receptor 2/neu expression. Br J Cancer.
113:1186–1196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan X, Zhou H and Zhang T, Xu P, Zhang S,
Huang W, Yang L, Gu X, Ni R and Zhang T: Downregulation of FOXP2
promoter human hepatocellular carcinoma cell invasion. Tumour Biol.
36:9611–9619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang JT and Lee V: Identification and
characterization of a novel human FOXK1 gene in silico. Int J
Oncol. 25:751–757. 2004.PubMed/NCBI
|
|
24
|
Wang W, Li X, Lee M, Jun S, Aziz KE, Feng
L, Tran MK, Li N, McCrea PD, Park JI, et al: FOXKs promote
Wnt/β-catenin signaling by translocating DVL into the nucleus. Dev
Cell. 32:707–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J, Yang Y, Xia HH, Gu Q, Lin MC,
Jiang B, Peng Y, Li G, An X, Zhang Y, et al: Suppression of FHL2
expression induces cell differentiation and inhibits gastric and
colon carcinogenesis. Gastroenterology. 132:1066–1076. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tu SP, Liston P, Cui JT, Lin MC, Jiang XH,
Yang Y, Gu Q, Jiang SH, Lum CT, Kung HF, et al: Restoration of XAF1
expression induces apoptosis and inhibits tumor growth in gastric
cancer. Int J Cancer. 125:688–697. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng Y, Geng H, Cheng SH, Liang P, Bai Y,
Li J, Srivastava G, Ng MH, Fukagawa T, Wu X, et al: KRAB zinc
finger protein ZNF382 is a proapoptotic tumor suppressor that
represses multiple oncogenes and is commonly silenced in multiple
carcinomas. Cancer Res. 70:6516–6526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Biggs WH III, Cavenee WK and Arden KC:
Identification and characterization of members of the FKHR (FOX O)
subclass of winged-helix transcription factors in the mouse. Mamm
Genome. 12:416–425. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lalmansingh AS, Karmakar S, Jin Y and
Nagaich AK: Multiple modes of chromatin remodeling by Forkhead box
proteins. Biochim Biophys Acta. 1819:707–715. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Badve S, Turbin D, Thorat MA, Morimiya A,
Nielsen TO, Perou CM, Dunn S, Huntsman DG and Nakshatri H: FOXA1
expression in breast cancer - correlation with luminal subtype A
and survival. Clin Cancer Res. 13:4415–4421. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reddy OL, Cates JM, Gellert LL, Crist HS,
Yang Z, Yamashita H, Taylor JA III, Smith JA Jr, Chang SS, Cookson
MS, et al: Loss of FOXA1 drives sexually dimorphic changes in
urothelial differentiation and is an independent predictor of poor
prognosis in bladder cancer. Am J Pathol. 185:1385–1395. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McMullin RP, Dobi A, Mutton LN, Orosz A,
Maheshwari S, Shashikant CS and Bieberich CJ: A FOXA1-binding
enhancer regulates Hoxb13 expression in the prostate gland. Proc
Natl Acad Sci USA. 107:98–103. 2010. View Article : Google Scholar :
|
|
33
|
Song Y, Washington MK and Crawford HC:
Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal
transition in pancreatic cancer. Cancer Res. 70:2115–2125. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Y, Guo Z, Zhang D, Zhang W, Yan Q, Shi
X, Zhang M, Zhao Y, Zhang Y, Jiang B, et al: A novel colon cancer
gene therapy using rAAV-mediated expression of human shRNA-FHL2.
Int J Oncol. 43:1618–1626. 2013.PubMed/NCBI
|
|
35
|
Li X, Kong X, Kong X, Wang Y, Yan S and
Yang Q: 53BP1 sensitizes breast cancer cells to 5-fluorouracil.
PLoS One. 8:e749282013. View Article : Google Scholar : PubMed/NCBI
|