Introduction
Lung cancer is a leading cause of cancer-related
deaths worldwide, and >80% of lung cancers are diagnosed as
non-small cell lung cancer (NSCLC) (1). Unfortunately, the morbidity and
mortality rates of lung cancer in China have remained high and are
even continuously increasing; as a consequence, this disease has
resulted in heavy economic burden (2,3).
Despite the advancements in treatment strategies, the overall
5-year survival rate of lung cancer remains low (1). Cancer treatment remains challenging
because of the malignant growth and distant metastasis of cancer
cells (4). However, underlying
mechanisms remain largely unknown. Therefore, innovative research
efforts should be performed to understand the mechanism of
carcinogenesis and to develop new and effective therapeutic
methods.
Sirtuins (SIRTs) are nicotinamide adenine
dinucleotide oxidized form-dependent deacetylases belonging to
class III histone deacetylase (5).
SIRTs are also called longevity proteins because of their distinct
roles in extending lifespan (5).
SIRTs regulate various biological processes, including stress
responses, DNA repair, inflammation, metabolism, apoptosis, cell
cycle, and senescence; they also play critical roles in
cardiovascular and neurodegenerative diseases and cancer (5–8). Thus
far, seven members of the SIRT family (SIRT1-7) have been
characterized in mammals (9). Among
the SIRTs, SIRT1 is the most widely explored; this SIRT is
implicated in various cellular and metabolic processes (10). The important role of SIRT2 has been
observed in neurodegenerative diseases (11). SIRT3 is another thoroughly
investigated SIRT that represents a major mitochondrial deacetylase
(12,13). SIRT4 and SIRT5 participate in
metabolic processes (14,15). SIRT6 possibly regulates DNA damage
and genome integrity (16,17). SIRT7 is the latest characterized
SIRT and its function study has just begun (18). SIRT7 exhibits nuclear localization
and regulates ribosomal RNA synthesis (19,20).
SIRT7 has been implicated in various cellular processes, such as
cellular survival, protein synthesis, chromatin remodeling, and
lipid metabolism (18). SIRT7 has
also been proposed as an oncogene in various cancer types,
including hepatocellular carcinoma (21) and colorectal cancer (22). Nevertheless, the role of SIRT7 in
lung cancer remains unclear.
MicroRNAs (miRNAs), a group of small and non-coding
RNAs consisting of ~22 nucleotides, have been extensively
investigated in terms of cancer pathogenesis and treatments because
of their negative regulatory effect on gene expression (23). miRNAs can directly target the
3′-untranslated region (UTR) of target messenger RNA (mRNA) via
complementary sequences; as a result, mRNA becomes unstable and
undergoes degradation; thus, translation is inhibited (24–26).
Therefore, miRNAs participate in various biological processes, such
as cell proliferation, apoptosis, migration, and invasion (24). A series of miRNAs has been
aberrantly expressed in cancer tissues, such as NSCLC, functioning
as oncogenes or tumor suppressive genes (27,28).
These miRNAs participate in cancer pathogenesis and thus can be
potentially used as new diagnostic biomarkers and therapeutic
targets (27,28).
In our study, to better understand the biological
role of SIRT7 in NSCLC, we analyzed the expression of SIRT7 in
several NSCLC cell lines. The results showed that SIRT7 was highly
expressed in NSCLC cell lines. SIRT7 knockdown by small interfering
RNA (siRNA) significantly inhibited the growth of NSCLC cells and
promoted their apoptosis. To develop a novel inhibitor for SIRT7,
we predicted novel and potential miRNAs that could target and
regulate SIRT7 through bioinformatics algorithms. A novel miRNA,
the miR-3666, contained the predicted binding sites for 3′-UTR of
SIRT7. The finding was confirmed through dual-luciferase reporter
assay. Further data revealed that miR-3666 markedly inhibited the
protein expression of SIRT7. The overexpression of miR-3666
significantly inhibited NSCLC cell growth directly through SIRT7.
Moreover, the loss of SIRT7 induced by siRNA or miR-3666 activated
the pro-apoptotic gene expression. Thus, miR-3666/SIRT7 could be
used as a novel and potential molecular target for NSCLC
treatment.
Materials and methods
Cell lines
Normal human keratinocyte cell line (HaCaT), human
NSCLC cell lines (A549, H1299, NCI-H157, and SK-MES-1), and 293T
cells were purchased from American Type Culture Collection (ATCC,
Manassas, VA, USA). These cells were grown in Dulbecco's modified
Eagle's medium (DMEM; Gibco, Rockville, MD, USA) supplemented with
10% fetal calf serum (Gibco) and penicillin (100 U/ml)/streptomycin
(100 µg/ml) and then placed in a humidified incubator
containing 5% CO2 at 37°C.
Real-time quantitative polymerase chain
reaction (RT-qPCR)
Total RNAs were isolated using an RNeasy kit and an
miRNeasy mini kit (Qiagen, Dusseldorf, Germany) according to the
manufacturer's instructions. cDNA was synthesized with M-MLV
reverse transcriptase (Clontech, Palo Alto, CA, USA) and a
Primescript RT reagent kit (Takara, Dalian, China). PCR
amplification was performed using a SYBR Premix Ex Taq GC
kit (Takara) on an ABI7500 real-time PCR detection system (Applied
Biosystems, Carlsbad, CA, USA). Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) or U6 was also used as an internal control.
Fold changes relative to the control were calculated via the
2−ΔΔCt method.
Western blot analysis
Protein extractions were separated through 12.5%
sodium dodecyl sulfate polyacrylamide gel electrophoresis and
subsequently transferred to a polyvinylidene difluoride membrane
(Millipore, Boston, MA, USA). The membrane was blocked with 3.5%
non-fat milk and probed with primary antibodies at 4°C overnight.
Afterward, the membrane was incubated with horseradish
peroxidase-conjugated secondary antibodies (1:2,000, Santa Cruz
Biotechnology, Santa Cruz, CA, USA) for 1 h. Protein bands were
developed through chemiluminescence (Amersham Biosciences, Little
Chalfont, UK). Band intensity was quantified using Image-Pro Plus
6.0 (Media Cybernetics, Inc., Rockville, MD, USA). The primary
antibodies (anti-SIRT7 and anti-GAPDH) used in this study were
purchased from Santa Cruz Biotechnology.
Cell transfection
SIRT7 siRNA (siSIRT7-1: 5′-CCUGCCGUGUGAGGCGGAA-3′
and siSIRT7-2: 5′-GCCGUGUGAGGCGGAAGCG-3′) and control siRNA
(siControl: 5′-CCUUGCGUGGGAGCGCGAA-3′) were synthesized by
Invitrogen (Carlsbad, CA, USA). miR-3666 mimics and negative
control miRNA mimics (NC miRNA) were purchased from OriGene
Technologies (Beijing, China). For SIRT7 overexpression, the open
reading frame of SIRT7 without 3′-UTR was cloned into pcDNA3.1
plasmid (BioVector, Beijing, China). The siRNA, miRNA mimics, or
plasmids were transfected by using Lipofectamine™ 2000 (Invitrogen)
according to the manufacturer's instructions.
3-(4,5-Dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell growth was detected through an MTT assay. In
brief, the cells were seeded into 96-well plates at a density of
1×104 cells/well and cultured overnight. After
transfection was performed for 48 h, 20 µl of MTT stock
solution (Sigma, St. Louis, MO, USA) was added to each well and
incubated for 4 h; afterward, 200 µl of dimethyl
sulfoxide/well (Sigma) was added. Absorbance was detected at 490 nm
with a microplate spectrophotometer (Bio-Tek Instruments, Winooski,
VT, USA).
Caspase-3 activity assay
Cell apoptosis was detected through caspase-3
activity assay by using a commercial kit (Roche Life Science,
Shanghai, China). The cells were lysed in ice-cold cell lysis
buffer. The supernatant was collected, and protein concentration
was measured. Subsequently, 100 µg of protein was incubated
with 50 µl of reaction buffer and 5 µl of 4 mM
DEVD-pNA substrate at 37°C for 2 h. Absorbance was determined at
405 nm by using a microplate spectrophotometer (Bio-Tek
Instruments).
Dual-luciferase reporter assay
The 3′-UTR of SIRT7 containing the predicted binding
site or a mutant at miR-3666 binding site was cloned into pmirGLO
dual-luciferase vector (Promega, Madison, WI, USA) downstream of
the luciferase gene. The recombinant dual-luciferase vector was
co-transfected with miR-3666 mimics into 293T cells by using
Lipofectamine™ 2000 (Invitrogen). After 48 h of incubation,
luciferase activity was determined by using a dual-Glo luciferase
assay system (Promega).
Statistical analysis
Data were expressed as mean ± standard deviation.
Statistical analyses were performed with Student's t-test or ANOVA
with a Bonferroni correction in SPSS version 11.5 software (SPSS
Inc., Chicago, IL, USA). A p-value of <0.05 was considered
statistically significant.
Results
SIRT7 is highly expressed in NSCLC
cells
To investigate the potential role of SIRT7 in NSCLC,
we detected the expression of SIRT7 in several NSCLC cell lines.
RT-qPCR analysis revealed that the mRNA expression level of SIRT7
in NSCLC cell lines was significantly higher than that in HaCaT
cells (Fig. 1A). The protein
expression level of SIRT7 in NSCLC cells was also higher than that
in the control HaCaT cells (Fig.
1B). These results suggested that SIRT7 plays an important role
in NSCLC.
SIRT7 knockdown inhibits the cell growth
of NSCLC cells
To investigate the biological role of SIRT7 in
NSCLC, we performed a loss-of-function experiment through cell
transfection by using siSIRT7. The expression of SIRT7 in H1299 and
SK-MES-1 cells was significantly depleted after transfection was
conducted with siSIRT7-1 and siSIRT7-2 (Fig. 2A and B). Subsequently, the effect of
SIRT7 depletion on the cell growth of NSCLC cells was detected. The
results showed that SIRT7 knockdown significantly inhibited the
growth of H1299 and SK-MES-1 cells, as detected by the MTT assay
(Fig. 2C). Furthermore, SIRT7
knockdown also markedly increased the caspase-3 activity, thereby
implying a high level of apoptosis induced (Fig. 2D). Taken together, the data
indicated that SIRT7 had an oncogenic role in NSCLC.
miR-3666 targets and inhibits SIRT7
protein expression
Considering the important role of SIRT7 in NSCLC, we
may assume that targeting SIRT7 is a promising strategy to prevent
NSCLC cancer. miRNAs are novel tools used to treat lung cancer by
negatively regulating oncogene or tumor suppressive gene (28,29).
We also predicted the novel and potential miRNAs that could target
and regulate SIRT7 through bioinformatics algorithms.
Interestingly, miR-3666, a novel tumor suppressive miRNA, contained
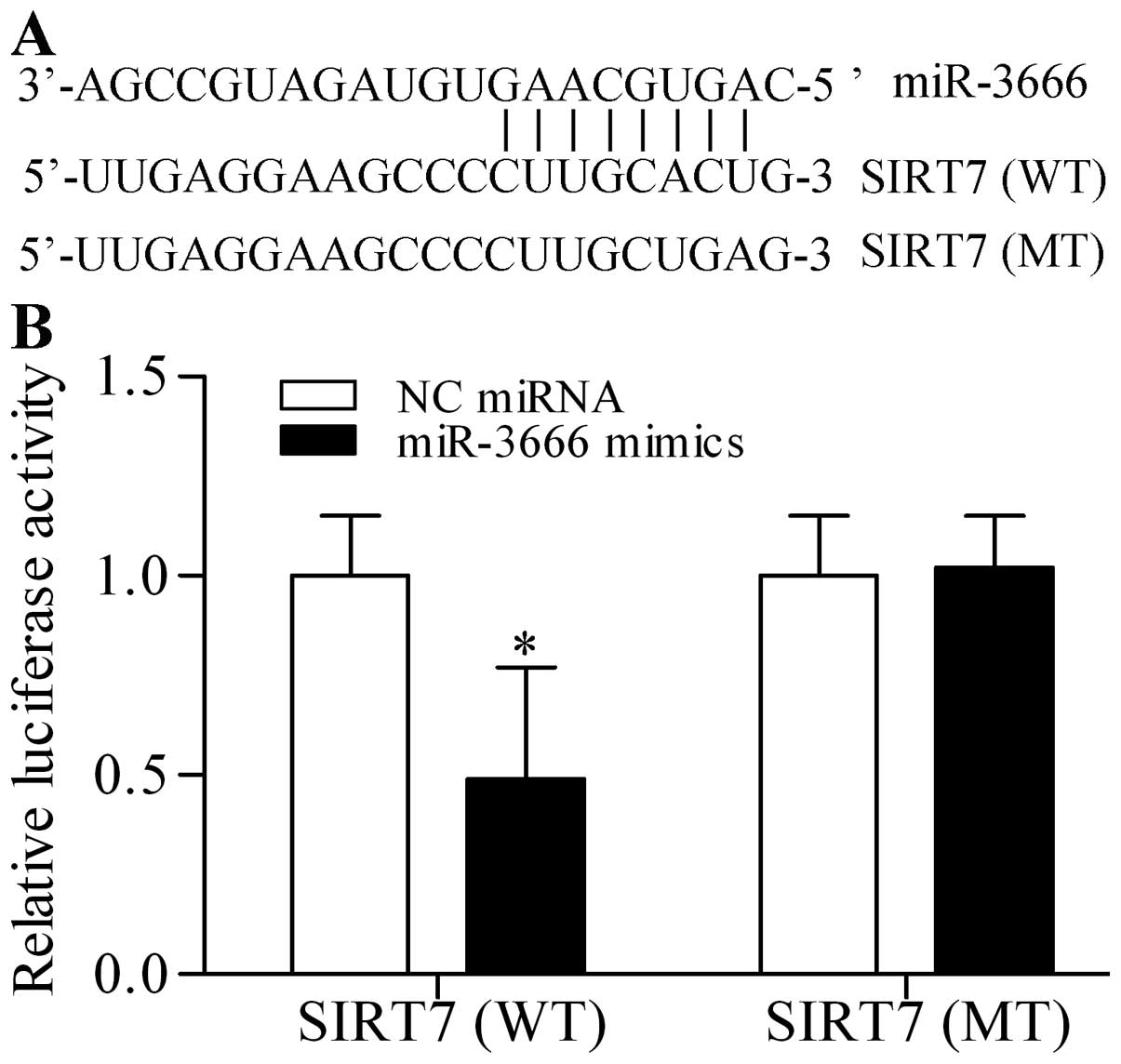
a theoretical seed region to target the 3′-UTR of SIRT7 (Fig. 3A). To confirm this prediction, we
cloned the 3′-UTR of SIRT7 (wild-type, WT) into pmirGLO
dual-luciferase reporter vector and co-transfected miR-3666 mimics
with this vector in 293T cells. Luciferase activities were
quantified in the transfected cells and the results showed that
miR-3666 mimics significantly decreased the luciferase activity
(Fig. 3B). By contrast, miR-3666
mimics did not evidently affect the mutant 3′-UTR constructs
(mutant type, MT) (Fig. 3B). To
investigate whether miR-3666 targeting the mRNA of SIRT7 regulates
the expression of SIRT7, we detected the effect of miR-3666 mimics
on SIRT7 expression in NSCLC cells. miR-3666 mimics did not
significantly affect the mRNA of SIRT7 (Fig. 4). By contrast, miR-3666 mimics
markedly decreased the protein expression of SIRT7 (Fig. 6A and B). Therefore, miR-3666
regulates the protein expression of SIRT7.
miR-3666 inhibits NSCLC cell growth
through targeting SIRT7
Considering the regulatory effect of miR-3666 on
SIRT7 expression, we speculated that miR-3666 could inhibit NSCLC
cell growth. To test this hypothesis, we transfected NSCLC cells
with miR-3666 mimics and detected its effect on cell growth and
apoptosis. The results showed that miR-3666 overexpression
significantly inhibited the growth (Fig. 5A) and apoptosis (Fig. 5B) of NSCLC cells. To validate
whether miR-3666 inhibited NSCLC cell growth through SIRT7, we
performed a rescue experiment. The cells were cotransfected with
miR-3666 mimics and SIRT7-overexpressing vector (without 3′-UTR).
The results showed that the decreased protein expression of SIRT7
induced by miR-3666 mimics was significantly restored by the
transfection of SIRT7-overexpressing vector (Fig. 6A and B). Furthermore, restoring
SIRT7 expression markedly decreased the inhibitory effect of
miR-3666 on NSCLC cell growth (Fig. 6C
and D). These data indicated that miR-3666 inhibited NSCLC cell
growth through targeting SIRT7.
Loss of SIRT7 promotes the activation of
pro-apoptotic gene expression
To further elucidate the underlying mechanism of
SIRT7 in regulating NSCLC cell growth and apoptosis, we detected
the effect of the loss of SIRT7 on pro-apoptotic gene expression.
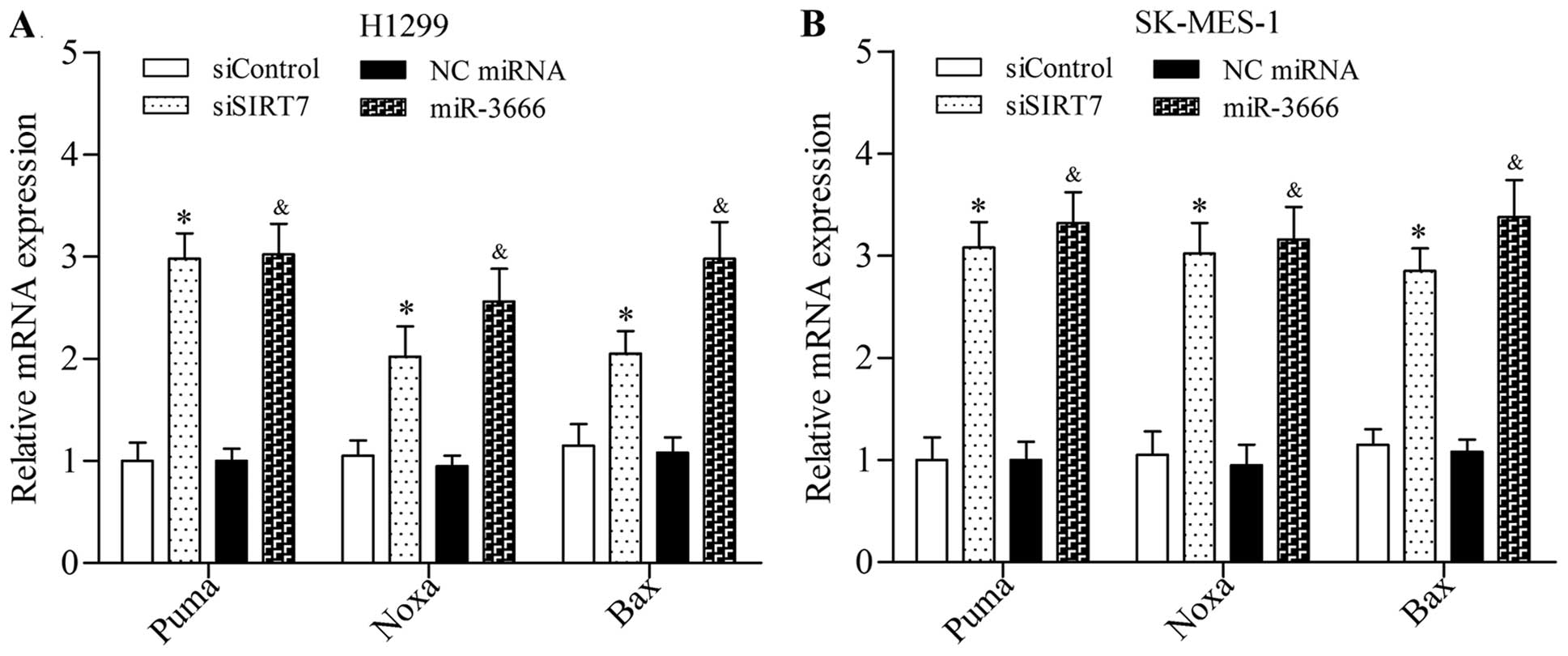
The results showed that SIRT7 depletion induced by siRNA or
miR-3666 mimics significantly increased the mRNA expression levels
of Puma, Noxa, and Bax in NSLCLC cells (Fig. 7). Taken together, these results
suggested that SIRT7 inhibition promoted the activation of
pro-apoptotic signaling pathway.
Discussion
In this study, we demonstrated a crucial role of
SIRT7 in NSCLC. SIRT7 was overexpressed in NSCLC cell lines, and
SIRT7 knockdown by siRNA significantly inhibited cell growth and
induced cell apoptosis of NSCLC. Intriguingly, miR-3666, a novel
tumor suppressive miRNA (30,31),
could target and inhibit SIRT7 protein expression through directly
interacting with the 3′-UTR of SIRT7 mRNA. The overexpression of
miR-3666 significantly inhibited NSCLC cell growth through
inhibiting SIRT7. Our data indicated an important role of
miR-3666/SIRT7 in regulating NSCLC cell growth.
SIRT7 regulates the ribosomal RNA synthesis
(19,20). SIRT7 is highly expressed in highly
proliferative tissues; SIRT7 is implicated in cell growth
regulation (9). This protein also
plays a critical role in cancers (18). Barber et al reported that
SIRT7 is essential for the maintenance of cancerous phenotype
because SIRT7 contributes to the maintenance of H3K18 deacetylation
leading to the suppression of many tumor suppressor genes (32). The depletion of SIRT7 in cancer
cells reduces the anchorage-independent growth and cell
proliferation (32). SIRT7 is
highly expressed in hepatocellular carcinoma tissues, and its
knockdown results in cell cycle arrest and inhibits cell grow in
vitro and in vivo (21).
SIRT7 is also gradually increased during cervical cancer
progression (33). In colorectal
cancer, SIRT7 protein expression is significantly correlated with
tumor stage, lymph node metastasis, and poor patient survival;
SIRT7 knockdown also suppresses the motility, proliferation, and
colony formation of colorectal cancer cells via the
mitogen-activated protein kinase pathway (22). Furthermore, SIRT7 is upregulated in
gastric cancer and is correlated with disease stage, metastasis,
and survival (34). SIRT7 knockdown
also inhibits gastric cancer cell growth in vitro and in
vivo (34). In addition, SIRT7
is highly expressed in ovarian cancer cell lines, and its loss
reduces cell growth and colony formation and promotes cell
apoptosis of ovarian cancer cells (35). High SIRT7 expression is also found
in breast cancer and is correlated with high histological grade and
poor survival rate (36,37). These findings indicate that SIRT7
functions as an oncogene. In line with these findings, our results
revealed that this protein functioned as an oncogene in lung
cancer. Our results further demonstrated that SIRT7 was
overexpressed in NSCLC cell lines, and the knockdown of this gene
impeded NSCLC cell growth. On the contrary, SIRT7 possesses
tumor-suppressor properties (38).
McGlynn et al demonstrated that decreased SIRT7 expression
is associated with an aggressive tumor phenotype and poor survival
(38). SIRT7 knockdown also induces
multidrug resistance in breast cancer cells (39).
miRNAs represent novel inhibitors for gene
expression (24–26). SIRT7 can be regulated by certain
miRNAs (21,40). Kim et al reported that tumor
suppressive miRNAs, miR-125a-5p, and miR-125b can target and
inhibit SIRT7 to suppress the growth of hepatocellular carcinoma
cells (21). Zhao and Wang found
that miR-125b inhibits the proliferation of hepatocellular
carcinoma cells by targeting SIRT7 (40). Another study reports that miR-125b
also inhibits bladder cancer development by targeting and
inhibiting SIRT7 (41). A recent
study revealed that SIRT7 is also a target gene of miR-93, which
regulates adiposity (42). In our
study, we found that the miR-3666 could directly target and inhibit
SIRT7 expression. miR-3666 has been suggested as an oncogene in
cervical cancer and can inhibit cancer cell invasion and metastases
by targeting zinc finger E-box binding homeobox 1 (30). Low miR-3666 expression is detected
in thyroid carcinoma, but the miR-3666 overexpression significantly
inhibits the growth of thyroid carcinoma cancer cells by targeting
the met proto-oncogene (31). In
this study, we found that miR-3666 also functioned as a potential
tumor suppressor in NSCLC. The miR-3666 overexpression could
inhibit NSCLC cell growth by suppressing SIRT7. Our study further
documented that SIRT7 could be targeted and inhibited by specific
miRNAs. Therefore, miR-3666 could be used as a novel inhibitor of
SIRT7 to inhibit SIRT7-related cancers, such as NSCLC.
SIRT7 plays an important role in regulating cell
apoptosis (43). The depletion of
SIRT7 triggers apoptosis in mammalian cells (19). SIRT7 can also prevent cardiomyocyte
apoptosis through deacetylation of p53 and inhibition of cell
apoptosis activation (43).
Moreover, SIRT7 knockdown increases pro-apoptotic proteins and
inhibits anti-apoptotic proteins in gastric cancer cells (34). In osteosarcoma cells, SIRT7 induces
resistance to doxorubicin-induced apoptosis by inhibiting the
activation of the pro-apoptotic signaling pathway (44). In line with these findings, our
results demonstrated that the loss of SIRT7 significantly promoted
the expression of pro-apoptotic genes, including Puma, Noxa, and
Bax. Our results further confirmed the role of SIRT7 in regulating
cell apoptosis in cancer cells.
Our results indicated that SIRT7 functioned as an
oncogene in NSCLC. miR-3666-induced SIRT7 inhibition could suppress
cell growth and trigger apoptosis of NSCLC cells. Therefore,
miR-3666/SIRT7 may be considered novel and potential molecular
targets for NSCLC treatment.
Acknowledgments
This study was supported by the Fundamental Research
Funds for the Central Universities (no. XJJ2012057) and Natural
Science Basic Research Project of Shaanxi Province (no.
2016JQ8054).
Abbreviations:
|
SIRTs
|
sirtuins
|
|
NSCLC
|
non-small cell lung cancer
|
|
RT-qPCR
|
real-time quantitative polymerase
chain reaction
|
|
siRNA
|
small interfering RNA
|
|
UTR
|
untranslated region
|
|
mRNA
|
messenger RNA
|
|
miRNAs
|
microRNAs
|
|
miR-3666
|
microRNA-3666
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qiu ZX, Sun RF, Mo XM and Li WM: The
p70S6K specific inhibitor PF-4708671 impedes non-small cell lung
cancer growth. PLoS One. 11:e01471852016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang C, Zhang H, Shi J, Wang D, Zhang X,
Yang J, Zhai Q and Ma A: Trial-based cost-utility analysis of
icotinib versus gefitinib as second-line therapy for advanced
non-small cell lung cancer in China. PLoS One. 11:e01518462016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Steeg PS: Metastasis suppressors alter the
signal transduction of cancer cells. Nat Rev Cancer. 3:55–63. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Houtkooper RH, Pirinen E and Auwerx J:
Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol
Cell Biol. 13:225–238. 2012.PubMed/NCBI
|
|
6
|
Bosch-Presegué L and Vaquero A: The dual
role of sirtuins in cancer. Genes Cancer. 2:648–662. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
North BJ and Verdin E: Sirtuins:
Sir2-related NAD-dependent protein deacetylases. Genome Biol.
5:2242004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsushima S and Sadoshima J: The role of
sirtuins in cardiac disease. Am J Physiol Heart Circ Physiol.
309:H1375–H1389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Michishita E, Park JY, Burneskis JM,
Barrett JC and Horikawa I: Evolutionarily conserved and
nonconserved cellular localizations and functions of human SIRT
proteins. Mol Biol Cell. 16:4623–4635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng X, Tan J, Li M, Song S, Miao Y and
Zhang Q: Sirt1: Role under the condition of ischemia/hypoxia. Cell
Mol Neurobiol. Mar 14–2016.Epub ahead of print. View Article : Google Scholar
|
|
11
|
Donmez G and Outeiro TF: SIRT1 and SIRT2:
Emerging targets in neurodegeneration. EMBO Mol Med. 5:344–352.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lombard DB, Alt FW, Cheng HL, Bunkenborg
J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D,
Murphy A, et al: Mammalian Sir2 homolog SIRT3 regulates global
mitochondrial lysine acetylation. Mol Cell Biol. 27:8807–8814.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Onyango P, Celic I, McCaffery JM, Boeke JD
and Feinberg AP: SIRT3, a human SIR2 homologue, is an NAD-dependent
deacetylase localized to mitochondria. Proc Natl Acad Sci USA.
99:13653–13658. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakagawa T, Lomb DJ, Haigis MC and
Guarente L: SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and
regulates the urea cycle. Cell. 137:560–570. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haigis MC, Mostoslavsky R, Haigis KM,
Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos
GD, Karow M, Blander G, et al: SIRT4 inhibits glutamate
dehydrogenase and opposes the effects of calorie restriction in
pancreatic beta cells. Cell. 126:941–954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mao Z, Hine C, Tian X, Van Meter M, Au M,
Vaidya A, Seluanov A and Gorbunova V: SIRT6 promotes DNA repair
under stress by activating PARP1. Science. 332:1443–1446. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McCord RA, Michishita E, Hong T, Berber E,
Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, et
al: SIRT6 stabilizes DNA-dependent protein kinase at chromatin for
DNA double-strand break repair. Aging (Albany, NY). 1:109–121.
2009. View Article : Google Scholar
|
|
18
|
Kiran S, Anwar T, Kiran M and Ramakrishna
G: Sirtuin 7 in cell proliferation, stress and disease: Rise of the
seventh sirtuin! Cell Signal. 27:673–682. 2015. View Article : Google Scholar
|
|
19
|
Ford E, Voit R, Liszt G, Magin C, Grummt I
and Guarente L: Mammalian Sir2 homolog SIRT7 is an activator of RNA
polymerase I transcription. Genes Dev. 20:1075–1080. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grob A, Roussel P, Wright JE, McStay B,
Hernandez-Verdun D and Sirri V: Involvement of SIRT7 in resumption
of rDNA transcription at the exit from mitosis. J Cell Sci.
122:489–498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ,
Kim MG, Chang YG, Shen Q, Park WS, Lee JY, et al: Sirtuin7
oncogenic potential in human hepatocellular carcinoma and its
regulation by the tumor suppressors MiR-125a-5p and MiR-125b.
Hepatology. 57:1055–1067. 2013. View Article : Google Scholar
|
|
22
|
Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X,
Zhou Y, Wang H, Pan C and Huang W: Overexpression of sirt7 exhibits
oncogenic property and serves as a prognostic factor in colorectal
cancer. Clin Cancer Res. 20:3434–3445. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ebrahimi A and Sadroddiny E: MicroRNAs in
lung diseases: Recent findings and their pathophysiological
implications. Pulm Pharmacol Ther. 34:55–63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cortinovis D, Monica V, Pietrantonio F,
Ceresoli GL, La Spina CM and Wannesson L: MicroRNAs in non-small
cell lung cancer: Current status and future therapeutic promises.
Curr Pharm Des. 20:3982–3990. 2014. View Article : Google Scholar
|
|
28
|
Feng B, Zhang K, Wang R and Chen L:
Non-small-cell lung cancer and miRNAs: Novel biomarkers and
promising tools for treatment. Clin Sci (Lond). 128:619–634. 2015.
View Article : Google Scholar
|
|
29
|
Barger JF and Nana-Sinkam SP: MicroRNA as
tools and therapeutics in lung cancer. Respir Med. 109:803–812.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Han LY, Yu M, Zhou Q, Xu JC and Li
P: Pituitary tumor-transforming gene 1 enhances metastases of
cervical cancer cells through miR-3666-regulated ZEB1. Tumour Biol.
2015:Sep 17–2015.Epub ahead of print.
|
|
31
|
Wang G, Cai C and Chen L: MicroRNA-3666
regulates thyroid carcinoma cell proliferation via MET. Cell
Physiol Biochem. 38:1030–1039. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barber MF, Michishita-Kioi E, Xi Y,
Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL,
Chen K, et al: SIRT7 links H3K18 deacetylation to maintenance of
oncogenic transformation. Nature. 487:114–118. 2012.PubMed/NCBI
|
|
33
|
Singh S, Kumar PU, Thakur S, Kiran S, Sen
B, Sharma S, Rao VV, Poongothai AR and Ramakrishna G:
Expression/localization patterns of sirtuins (SIRT1, SIRT2, and
SIRT7) during progression of cervical cancer and effects of sirtuin
inhibitors on growth of cervical cancer cells. Tumour Biol.
36:6159–6171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu
S, Hu Y and Cai T: Sirt7 promotes gastric cancer growth and
inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep.
5:97872015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang HL, Lu RQ, Xie SH, Zheng H, Wen XM,
Gao X and Guo L: SIRT7 exhibits oncogenic potential in human
ovarian cancer cells. Asian Pac J Cancer Prev. 16:3573–3577. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Aljada A, Saleh AM, Alkathiri M, Shamsa
HB, Al-Bawab A and Nasr A: Altered sirtuin 7 expression is
associated with early stage breast cancer. Breast Cancer (Auckl).
9:3–8. 2015.
|
|
37
|
Geng Q, Peng H, Chen F, Luo R and Li R:
High expression of Sirt7 served as a predictor of adverse outcome
in breast cancer. Int J Clin Exp Pathol. 8:1938–1945.
2015.PubMed/NCBI
|
|
38
|
McGlynn LM, McCluney S, Jamieson NB,
Thomson J, MacDonald AI, Oien K, Dickson EJ, Carter CR, McKay CJ
and Shiels PG: SIRT3 & SIRT7: Potential novel biomarkers for
determining outcome in pancreatic cancer patients. PLoS One.
10:e01313442015. View Article : Google Scholar :
|
|
39
|
Aljada A, Saleh AM and Al Suwaidan S:
Modulation of insulin/IGFs pathways by sirtuin-7 inhibition in
drug-induced chemoreistance. Diagn Pathol. 9:942014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao L and Wang W: miR-125b suppresses the
proliferation of hepatocellular carcinoma cells by targeting
Sirtuin7. Int J Clin Exp Med. 8:18469–18475. 2015.
|
|
41
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar
|
|
42
|
Cioffi M, Vallespinos-Serrano M, Trabulo
SM, Fernandez-Marcos PJ, Firment AN, Vazquez BN, Vieira CR, Mulero
F, Camara JA, Cronin UP, et al: MiR-93 controls adiposity via
inhibition of Sirt7 and Tbx3. Cell Rep. 12:1594–1605. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vakhrusheva O, Smolka C, Gajawada P,
Kostin S, Boettger T, Kubin T, Braun T and Bober E: Sirt7 increases
stress resistance of cardiomyocytes and prevents apoptosis and
inflammatory cardiomyopathy in mice. Circ Res. 102:703–710. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kiran S, Oddi V and Ramakrishna G: Sirtuin
7 promotes cellular survival following genomic stress by
attenuation of DNA damage, SAPK activation and p53 response. Exp
Cell Res. 331:123–141. 2015. View Article : Google Scholar
|