Introduction
Esophageal cancer is the sixth leading cause of
cancer-related death, and in 2013, it was estimated that 442,000
new cases were diagnosed with 440,000 deaths due to the disease
worldwide, and the overall 5-year survival ranges from 15 to 25%
(1–3). Esophageal squamous cell carcinoma
(ESCC) is one of the major histological types of esophageal cancer,
particularly in the 'Asian Esophageal Cancer Belt̓, which extends
from Middle East countries to Eastern Asia, including Japan and is
highly aggressive due to its tendency to metastasize to the lymph
nodes and organs (4,5). To improve the poor prognosis, it is
necessary to reveal the mechanism of cancer progression and detect
prognostic biomarkers for the prognosis.
Exosomes are small membrane vesicles secreted from
most cell types. They contain proteins, lipids and nucleic acids
(e.g., mRNA and miRNA) and are thought to have some function,
particularly in signal transduction (6,7). Our
laboratory previously reported that exosomes from the serum and
proximal lymph nodes of patients with ESCC contained high miR-1246
levels (8). Thus, some information
may be delivered to the proximal lymph node via the exosome.
Indeed, in the field of cancer research, tumor-released exosomes
have been reported to influence tumor progression (9), resistance to chemotherapy (10) or immune evasion (11,12).
Exosomes released from the bone marrow or cancer-associated
fibroblasts (CAFs) also correlate with chemoresistance (13) or tumor dormancy (14,15).
Although exosome contents isolated from the patients peripheral
blood were reported to be useful as a diagnostic or prognostic
factor for cancers (16), it is
difficult to distinguish whether exosomes originate from the tumor
itself or as a result of the host response against the tumor.
Moreover, no precise quantification method for exosomes has to date
been developed.
In the present study, we report on the behavior of
tumor-derived exosomes in a mouse model with fluorescence imaging
and determine the relationship between the quantification of plasma
exosomes, measured by the acetylcholinesterase (AChE) activity,
which is known to be elevated within the exosome (17,18),
and the clinical characteristics in human ESCC (19,20).
Materials and methods
Cell culture
TE2 human ESCC cell line and SCCVII murine SCC cell
line were used in the present study. TE2 cell line was obtained
from Cell Resource Center for Biomedical Research Institute of
Development, Aging and Cancer, Tohoku University (Sendai, Japan).
SCCVII cell line was kindly provided by Professor Yuta Shibamoto
(Department of Quantum Radiology, Nagoya City University, Nagoya,
Japan).
The cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) (Life Technologies, Grand Island, NY, USA)
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
sodium and 100 µg/ml streptomycin in a humidified atmosphere
containing 5% CO2 at 37°C.
CD63-GFP expressing cells
A CD63-GFP fusion protein expression vector
(CYTO120-VA1; System Biosciences, Mountain View, CA, USA) was used
to transfect TE2 cells. After selection by puromycin-containing
medium and the limiting dilution technique, a single cell-derived
cell line, CD63-GFP stably expressing TE2 (TE2-CD63-GFP), was
established.
Extraction of exosomes from culture media
of ESCC cell line
Exosomes were isolated from culture media using an
isolation reagent (Exosome Isolation kit from culture media;
Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
protocol. The cells were seeded onto 10 cm dishes at a
concentration of 1×106 cells/dish with DMEM containing
10% of exosome-free FBS (System Biosciences). After a 48-h
incubation, conditioned media were harvested for exosome
extraction. The culture medium was centrifuged at 2,000 × g for 30
min to remove cells and debris. Then, these supernatants were
passed through a 220 nm filter and were transferred to new tubes,
and the reagents were added. After incubation, the samples were
centrifuged at 10,000 × g for 1 h and the supernatants were
discarded. The exosomes were pelleted at the bottom of the tubes
and the pellets were resuspended in phosphate-buffered saline (PBS)
for fluorescence imaging.
Animal care and mouse model of human
ESCC
Six-week-old female nude mice (BALB/c Slc-nu/nu)
were used for the present study. The 8 mice were divided into two
groups. TE2 (1×107 cells in 50 µl PBS) were
injected subcutaneously (s.c.) into the backs of mice in one group
(n=4), and the same amount of TE2-CD63-GFP cells were injected s.c.
into the mice in another group (n=4). Eight weeks after injection,
the mice were sacrificed and the tumor and plasma were harvested.
The exosomes were isolated from plasma, and used for fluorescence
imaging and quantification as hereinafter described.
The tumors were assessed with hematoxylin and eosin
(H&E) stain, immunohistochemistry (IHC) staining of CD63, and
fluorescence imaging. Anti-human CD63 rabbit polyclonal antibody
(cat. #15363; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was
used as a primary antibody for IHC staining.
Six-week-old female C3H/He mice were used to assess
the exosome amount within the early and advanced cancer models.
SCCVII (5×105 cells in 50 µl PBS) were injected
s.c. into the backs of mice. The plasma were harvested 1 or 6 weeks
after injection from each 4 mice as the models for early and
advanced cancer. The plasma were also harvested from 4 mice without
tumor, and each plasma was used for exosome isolation and
quantification as hereinafter described. The tumor volume in
mm3 was calculated by the formula: Volume =
(width)2 × length/2.
The present study was performed according to the
guidelines on animal experiments, and approved by the animal
experiment and welfare committee at Chiba University.
Patient samples
The plasma samples were collected from
histologically proven ESCC patients at their first visit to Chiba
University Hospital (Chiba, Japan) between December 2011 to
December 2012. Patients with active malignancy in any other organ
were excluded, and none of the patients had been treated at the
time of sample collection. A total of 66 samples from cancer
patients were collected, and 20 samples from patients with no
malignancy (e.g., esophageal achalasia and reflux esophagitis) were
collected during the same period.
The clinical data including blood testing and
clinical characteristics of the cancer were obtained from the
clinical database. The neutrophil to lymphocyte ratio (NLR) was
calculated as neutrophils/lymphocytes (21). The Glasgow Prognostic Score (GPS)
was calculated according to the cut-off values of 1.0 mg/dl for
C-reactive protein and 3.5 g/dl for albumin, as previously reported
(22). The staging classification
was confirmed by the classification of the union for International
Cancer Control (UICC) TNM staging system 7th edition. The present
study was approved by the Ethics Committee of Graduate School of
Medicine, Chiba University. A written informed consent was obtained
from all the patients.
Extraction of exosomes from the
plasma
Each plasma sample was centrifuged at 2,000 × g for
20 min at room temperature to remove cells and debris. The
supernatant containing the partially clarified plasma was
transferred to a new tube, and then centrifuged at 10,000 × g for
20 min at room temperature to remove debris. Then, the supernatant
containing the clarified plasma was transferred to a new tube, 100
µl of plasma was transferred to a new tube and exosomes were
isolated using an exosome isolation reagent (Total Exosome
Isolation kit from plasma; Invitrogen) according to the
manufacturer's protocol.
Exosome quantification
Each obtained exosome preparation from culture media
or plasma was dissolved in 100 µl of PBS and quantified
according to the AChE activity (EXOCET Exosome Quantification kit;
System Biosciences) according to the manufacturer's protocol. To
confirm the reliability of this protocol, differing concentrations
of exosome were isolated from 50, 100 and 200 µl plasma,
collected from healthy controls and quantified. In addition, 10
ESCC patients samples were quantified using both AChE activity and
nanoparticle tracking analysis, and the correlation was assessed.
Nanoparticle tracking analysis was kindly performed by Mr. Rii
Morimura (Toppan Printing, Japan) using NanoSight NS500 (Malvern
Instruments, Malvern, UK).
Fluorescence imaging
The fluorescent and bright field images of the mouse
tumor tissue sections and isolated exosomes were acquired using an
Axio Observer Z1 microscope and an AxioCam MRm camera (Carl Zeiss,
Oberkochen, Germany).
Electron microscope
The exosome sample was absorbed to formvar film
coated copper grids (400 mesh) and was stained with 2%
phosphotungstic acid solution (pH 7.0) for 30 sec. The sample was
observed by a transmission electron microscope (JEM-1400Plus; JEOL
Ltd., Tokyo, Japan) at an acceleration voltage of 80 kV. Digital
images were captured by a CCD camera (Veleta; Olympus Soft Imaging
Solutions GmbH, Münster, Germany).
Statistical analysis
Data of the exosome number are expressed as the
average ± SD (standard deviation). Differences in the relationships
between the number of exosomes and categorical clinicopathological
features were assessed using the Mann-Whitney U test or Student's
t-test. Comparisons among more than two groups were assessed using
a one-way factorial ANOVA and the Kruskal-Wallis test. The
Kaplan-Meier method was used for the survival analysis and the
log-rank test was used to determine the statistical significance of
the difference between the two groups. Prognostic factors were
determined by univariate and multivariate analyses (Cox
proportional hazard regression model). Statistical significance was
considered to exist at P-values <0.05. All data were
statistically analyzed using JMP version 12 (SAS Institute Inc.,
Cary, NC, USA).
Results
CD63-GFP expressing cells release
GFP-tagged exosomes
Stable CD63-GFP expressing TE2 (TE-CD63-GFP) cells
were established as described above (Fig. 1A and B). The exosomes were isolated
from the culture medium of TE2-CD63-GFP and TE2 cells using an
exosome isolation kit. The exosomes were imaged by electron
microscope and fluorescence. The small round particles, some 50 nm
in diameter, were recognized and the presence of GFP-positive, but
RFP-negative, small vesicles were confirmed only in exosome
isolated from TE2-CD63-GFP (Fig. 1C and
D).
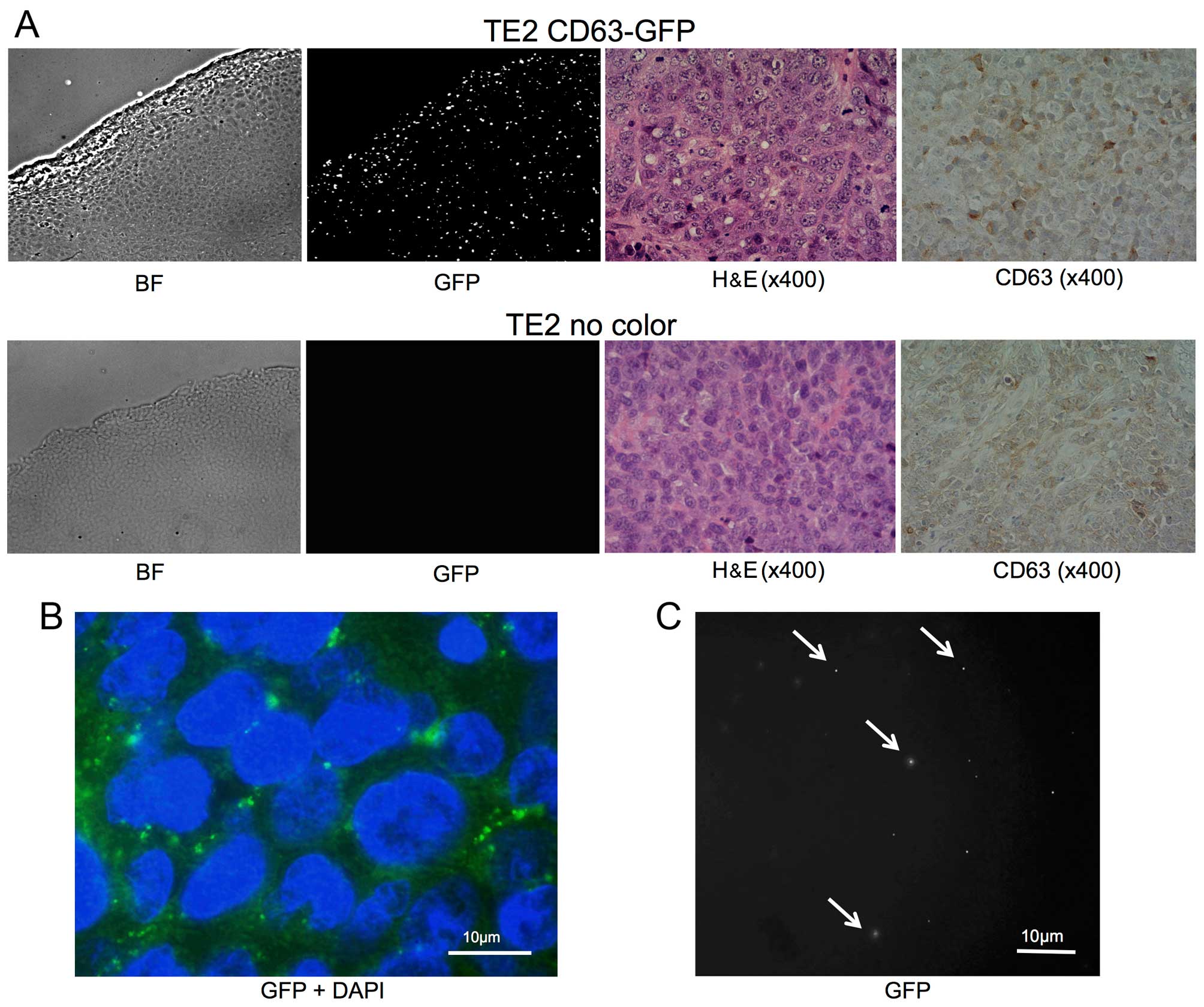
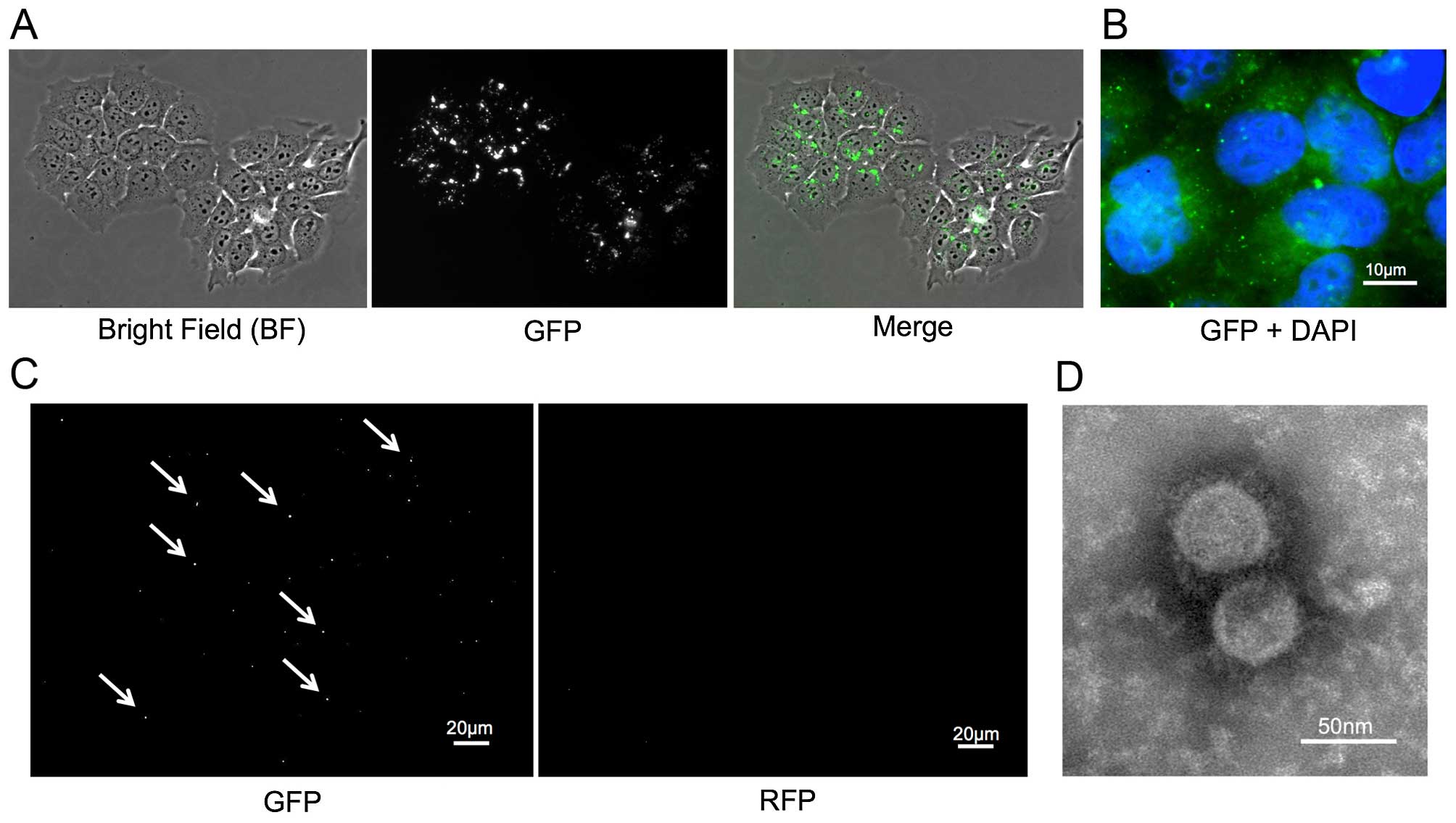 | Figure 1(A) A stable TE2 cell line expressing
CD63-GFP (TE2-CD63-GFP) was established by transduction with a
CD63-GFP expression vector. Left, bright-field (BF); middle, GFP;
right, merge (BF + GFP). (B) The enlarged fluorescence image of
TE2-CD63-GFP cells. GFP-expressing foci were confirmed in the
cytoplasm and the matrix of the cells (GFP + DAPI, bar, 10
µm). (C) GFP and RFP images of isolated exosome from
TE2-CD63-GFP culture medium. GFP positive, but RFP negative, small
foci were confirmed (bar, 20 µm). (D) An electron microscope
image of exosome isolated from TE2-CD63-GFP culture medium. Small
round particles, some 50 nm in diameter were confirmed (bar, 50
nm). |
Tumor-derived exosomes are released
outside the tumor and circulated in the blood flow
Subcutaneous tumors comprising TE2-CD63-GFP or TE2
cells were successfully formed following s.c. injection in each 4
mice. Eight weeks after s.c. injection of the cells, the tumor and
blood samples were harvested. There was no difference between the
two groups in the appearance of H&E stain and IHC staining of
CD63 (Fig. 2A).
Fluorescent imaging with GFP revealed intracellular
foci existed mainly in the cytoplasm among the TE2-CD63-GFP tumor,
and the appearance was similar to that of cultured cells (Fig. 2A and B). In addition, the presence
of GFP-positive, but RFP-negative, small vesicles were confirmed in
isolated exosomes from the plasma of TE2-CD63-GFP tumor-bearing
mice (3/4), but no GFP-positive particle was confirmed in the group
of TE2 tumor (0/4) (Fig. 2C).
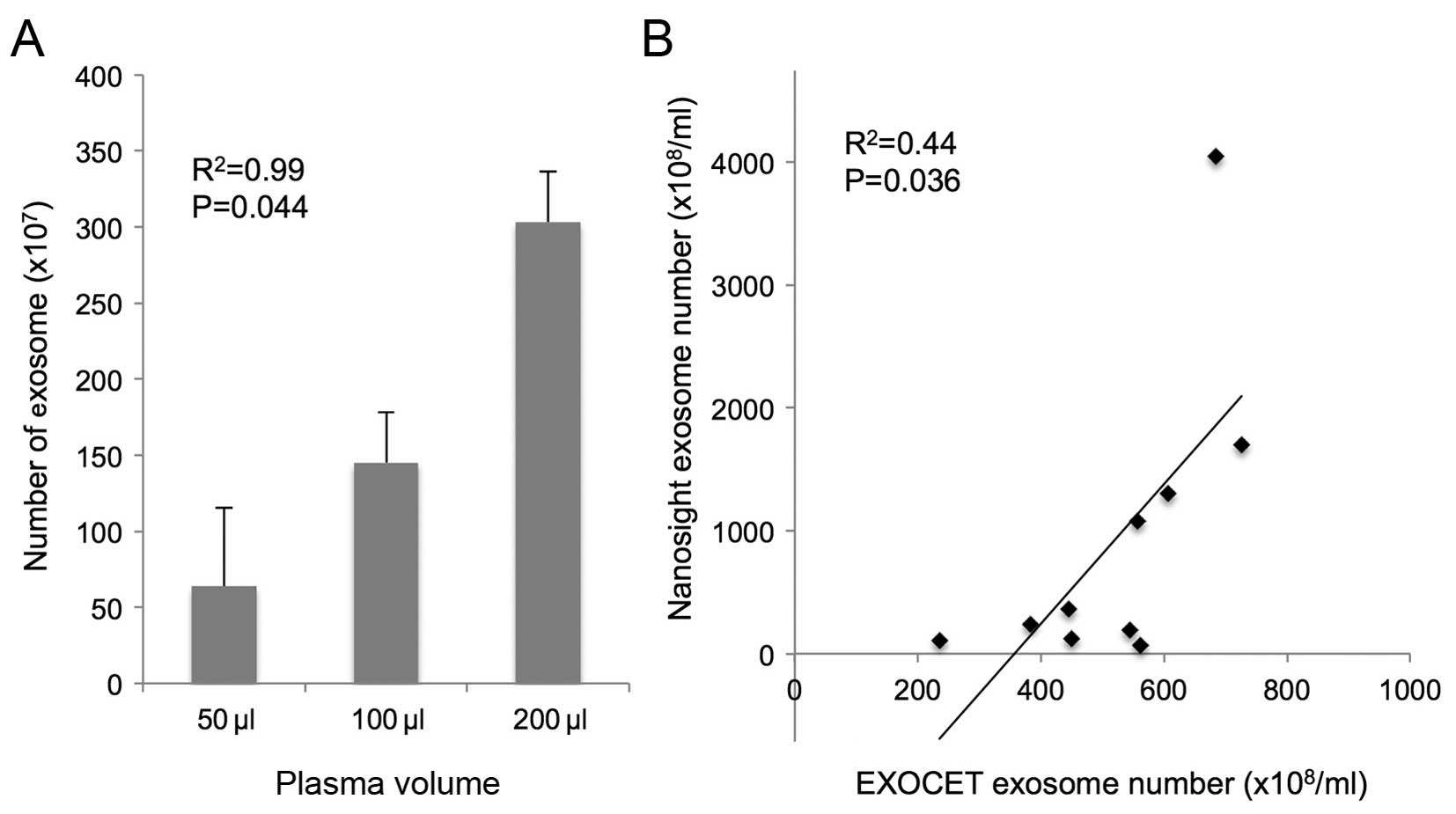
Exosome can be quantified by AChE
activity
The number of exosomes isolated from 50, 100 and 200
µl of plasma from healthy controls was 63.9±51.2
(×107), 145.2±33.2 (×107) and 303.2±33.2
(×107), respectively. The exosomes were isolated in a
dose-dependent manner (R2=0.99; P=0.044; Fig. 3A). There were positive correlation
between the exosome number quantified by AChE activity and
nanoparticle tracking analysis (R2=0.44, P=0.036;
Fig. 3B).
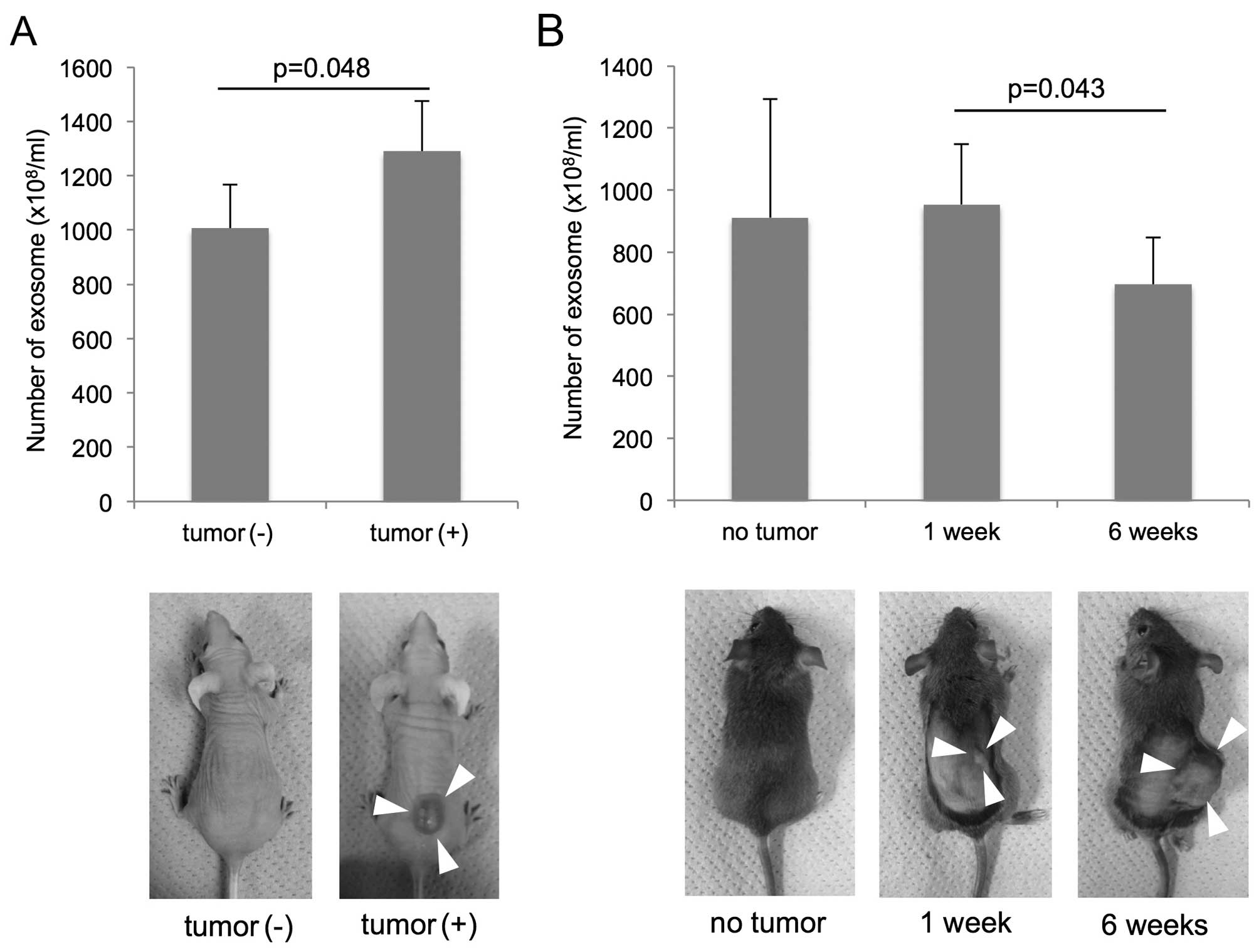
The quantification of plasma exosome
isolates from murine SCC models
The group of nude mice (BALB/c Slc-nu-nu) with
subcutaneous tumor composed of TE2-CD63-GFP (tumor volume, 1042±293
mm3) had higher amount of plasma exosome
(1290±186×108/ml) than the group of mice without tumor
(1007±162×108/ml) (P=0.048; t-test; Fig. 4A).
Concerning the C3H/He mouse groups, the exosome
amounts were 911±382×108/ml in the group without tumor,
979±194×108/ml in the group with early cancer (1 week
after injection), 695±170×108/ml in the group with
advanced cancer (6 weeks after injection). There was a significant
difference in the exosome number between the groups of early and
advanced cancer (P=0.043; t-test; Fig.
4B). The tumor volume was 1.2±1.2 mm3 in the mice 1
week after injection, 2712±1339 mm3 in the mice 6 weeks
after injection, and there was no correlation between the tumor
volume and the exosome number.
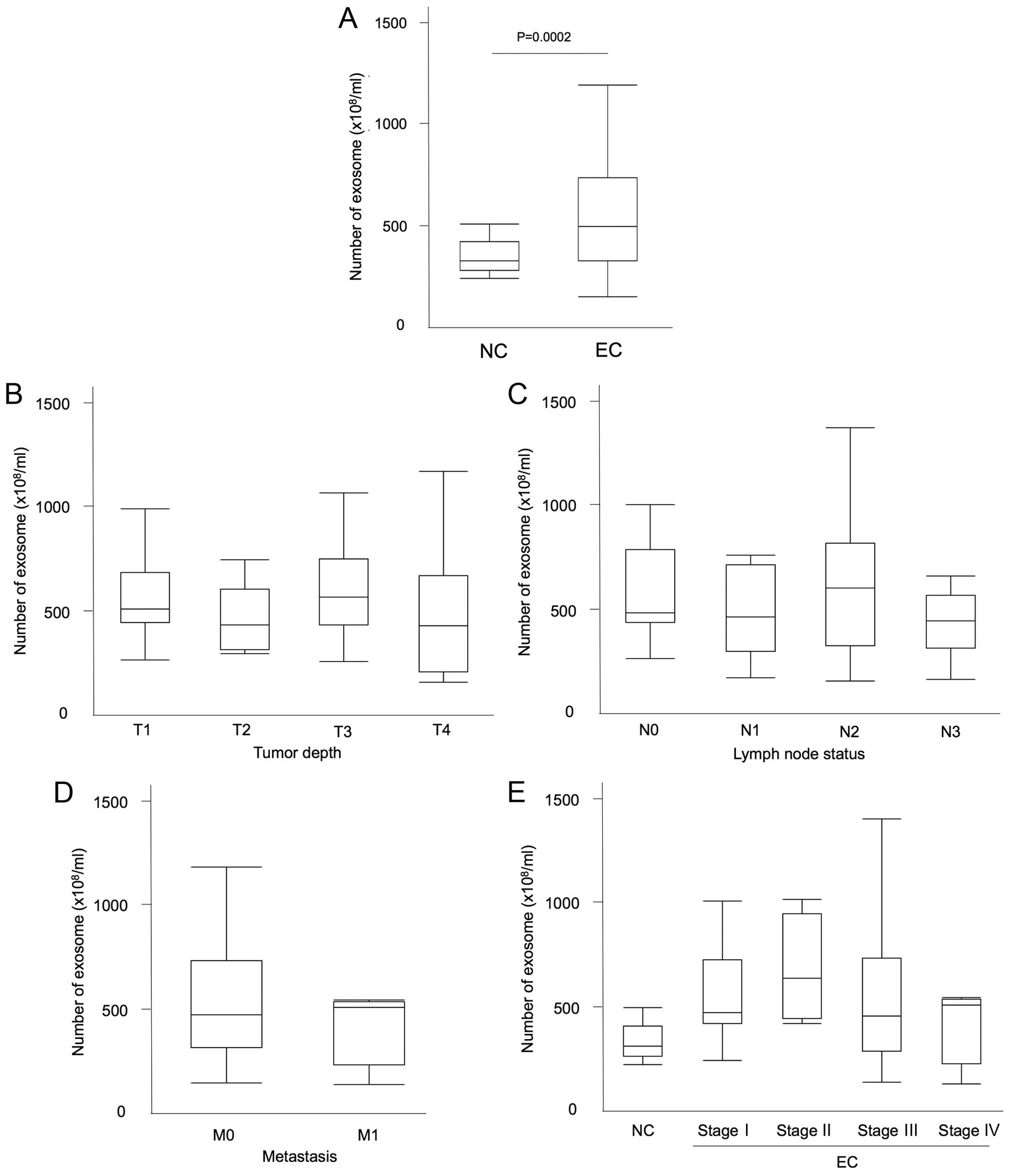
Exosome quantification in patient plasma
is higher in cancer patients
The quantification of exosomes isolated from 100
µl of the patients plasma was similarly measured. The
clinical characteristics of the patients are shown in Table I. The median number of plasma
exosomes was 493.9×108/ml in esophageal cancer patients
and 325.9×108/ml in non-malignant patients (P=0.0002;
Mann-Whitney U test; Fig. 5A).
There was no relationship between the exosome amount and several
factors, such as the white blood cell count (WBC), hemoglobin (Hb),
total protein (TP), albumin, C-reactive protein (CRP), CEA, CYFRA,
SCC-Ag level, as well as NLR (R2=0.014, 0.006, 0.123,
0.055, 0.014, 0.009, 0.003, 0.01 and 0.02, respectively).
Furthermore, there was also no difference in exosome amount within
the groups of GPS score 0, 1 and 2.
 | Table IClinical features of patients with
esophageal squamous cell carcinoma (n=66). |
Table I
Clinical features of patients with
esophageal squamous cell carcinoma (n=66).
| Clinicopathological
characteristics | |
|---|
| Mean age ± SD | 69.0±9.3 |
| Gender | |
| Male | 57 |
| Female | 9 |
| Clinical T
factor | |
| T1 | 20 |
| T2 | 5 |
| T3 | 21 |
| T4 | 20 |
| Clinical N
factor | |
| N0 | 19 |
| N1 | 12 |
| N2 | 20 |
| N3 | 15 |
| Clinical M
factor | |
| M0 | 62 |
| M1 | 4 |
| Stage (UICC
7th) | |
| I | 18 |
| II | 4 |
| III | 40 |
| IV | 4 |
| Treatment
procedure | |
| ESD | 11 |
| Surgery | 22 |
| Non-surgical | 25 |
| CRT | 18 |
| CT | 5 |
| RT | 2 |
| Palliative | 8 |
The exosome amount did not correlate with
the tumor progression
The number of exosomes and tumor characteristics
were then assessed. Regarding the tumor depth, the number of
exosomes was 565.1±210.6 in T1, 451.0±149.0 in T2, 627.7±283.0 in
T3, and 510.8±340.2 in T4 with median values of 508.0, 428.9, 564.6
and 425.2 (×108/ml), respectively (Fig. 5B). There was no significant
difference between the groups (P= 0.45; ANOVA). In relation to the
lymph node status, the number of exosomes was 585.9±237.6 in N0,
481.6±202.0 in N1, 632.4±331.4 in N2 and 492.7±288.5 in N3 with
median values of 482.4, 462.1, 598.9 and 440.5
(×108/ml), respectively (Fig. 5C; P=0.34; ANOVA). The number of
exosomes was 585.9±237.6 in the negative lymph node metastasis
group and 549.3±292.7 in the positive group with median values of
482.4 and 505.4 (×108/ml), respectively (P=0.48;
Mann-Whitney U test). Regarding the distant metastatic status, the
number of exosomes was 568.1±280.6 in M0 and 433.9±187.2 in M1 with
median values of 482.0 and 516.4 (×108/ml), respectively
(Fig. 5D; P=0.54; Mann-Whitney U
test). Regarding the tumor stage, the number of exosomes was
563.0±221.8 in stage I, 676.9±252.0 in stage II, 559.4±3090.1 in
stage III, and 433.9±187.2 in stage IV, with median values of
482.0, 640.1, 464.2 and 516.4 (×108/ml), respectively
(Fig. 5E). There was no statistical
difference between the clinical stages (P=0.68; ANOVA).
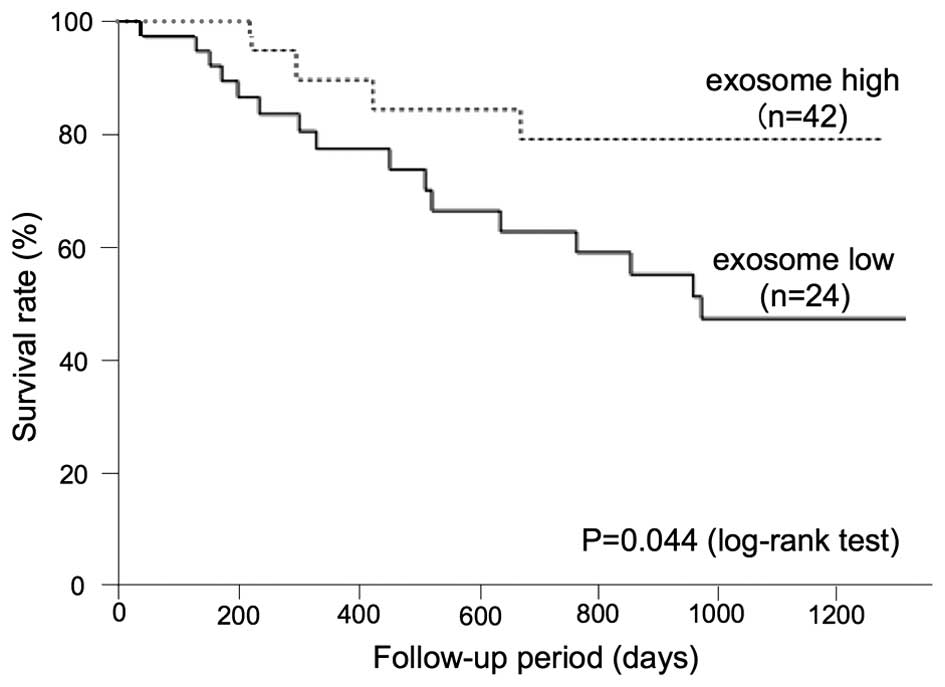
A low exosome amount predicts a poor
prognosis in ESCC patients
To determine the correlation between the plasma
exosome amount and the prognosis of ESCC patients, an analysis of
the overall survival was performed. The Kaplan-Meier approach, with
a statistical analysis using the log-rank test, was performed for
various clinicopathological factors, as presented in Table II. Regarding the exosome level, a
cut-off value was set as 600×108/ml.
 | Table IIUnivariate analyses of the survival
(n=66). |
Table II
Univariate analyses of the survival
(n=66).
| Variables | Categories | No. of cases | 3-year survival
rate (%) | P-value |
|---|
| Age (years) | <65 | 21 | 47.6 | 0.357 |
| ≥65 | 45 | 63.9 | |
| Gender | Male | 57 | 57.1 | 0.410 |
| Female | 9 | 74.1 | |
| Tumor depth | T1, T2 | 25 | 75.0 | 0.011 |
| T3, T4 | 41 | 48.2 | |
| Lymph node
status | Negative | 19 | 85.7 | 0.008 |
| Positive | 47 | 46.6 | |
| Metastasis
status | Negative | 64 | 59.5 | 0.002 |
| Positive | 4 | N/A | |
| Stage | I,II | 21 | 81.3 | 0.009 |
| III,IV | 45 | 47.2 | |
| NLR | <3.7 | 49 | 67.2 | 0.001 |
| ≥3.7 | 17 | 33.7 | |
| GPS | 0 | 55 | 63.1 | 0.164 |
| 1,2 | 11 | 37.5 | |
| Exosome | High | 24 | 79.2 | 0.044 |
| Low | 42 | 47.2 | |
The log-rank test showed that there was significant
difference in the tumor depth, lymph node status, distant
metastasis, clinical stage, the NLR and exosome number (P=0.011,
P=0.008, P=0.002, P=0.009, P=0.009 and P=0.044, respectively), but
age, gender, the serum level of SCC-Ag, CEA, and CYFRA and the GPS
did not significantly affect the survival (P=0.357, P=0.410,
P=0.225, P=0.306, P=0.333 and P=0.164, respectively). The 3-year
overall survival rate in patients with high plasma exosome amounts
was 79.2%, whereas that of the patients with lower plasma exosome
amounts was 47.2% (Fig. 6).
We next analyzed the importance of various factors
that may be able to predict a poor prognosis for the survival.
According to the univariate analysis, the clinical stage, the NLR
and the exosome number were selected. We subsequently performed a
multivariate analysis using a Cox proportional hazards regression
analysis, and a lower exosome number in the plasma was found to be
an independent risk factor for a poor survival with a hazard ratio
of 3.152 (95% confidence interval, 1.107–11.416; P=0.03; Table III).
 | Table IIIMultivariate analyses of the survival
(n=66). |
Table III
Multivariate analyses of the survival
(n=66).
| Categories | Odds ratio | 95% CI | P-value |
|---|
| Stage III,IV | 4.85 | 1.62–21.01 | 0.004 |
| NLR ≥3.7 | 4.10 | 1.49–11.12 | 0.007 |
| Low exosome | 3.15 | 1.11–11.41 | 0.030 |
Discussion
Exosome analyses have been previously performed in
cancer research. The previous studies mainly concerned the contents
of exosomes, such as proteins or microRNAs. Numerous types of
proteins and microRNAs have been reported to be useful as potential
biomarkers, particularly miR-21 for ESCC (23), however, the origin of exosomes
remains unclear and is difficult to discriminate. Although
cancer-derived exosomes were considered to be released outside the
tumor and circulate throughout the whole body (24), only a few studies have confirmed the
exosome behavior in vivo. In the present study,
cancer-derived exosomes were labeled by transfection with a
CD63-GFP expression vector, and the circulation of ESCC
tumor-derived exosomes in the blood flow was confirmed using mouse
models as previously reported (25). Notably, a recent study reported that
tumor-derived exosomes may integrate into specific organs and
affect organotropic metastasis (26).
Various studies confirmed that the number of
exosomes or extracellular vesicles (EVs) in the peripheral blood
are useful as a biomarker (27–29).
The quantification of EVs has not yet been standardized. Therefore,
in the present study, we quantified the exosome number according to
the AChE activity, which is known to be rich in exosomes. Although
quite a few proteins outside the exosome can be contaminated in the
isolated exosome, our data showed there was no correlation between
the quantified exosome number and the protein, included total
protein, albumin and C-reactive protein. Although the AChE
inhibitors, used for Alzheimer's disease, may influence the value,
the patients did not take these medicines in the present study. The
number of exosomes quantified by AChE activity was correlated with
the number quantified by nanoparticle tracking analysis, and the
plasma exosome number of non-malignant patients existed within a
certain range. Furthermore, this procedure takes ~1 h for exosome
isolation and quantification, and needs only 100 µl of
plasma samples, thus, it is reasonable to quantify the exosomes
according to the AChE activity.
Our data, including clinical analysis and in
vivo mouse model analysis, indicated that tumor-derived
exosomes can circulate in the blood and the plasma exosome amount
of cancer patients was significantly higher than that of patients
without cancer, however, the amount of plasma exosomes was not
increased according to the tumor progression, such as lymph node
metastasis or distant organ metastasis. Thus, we thought the
majority of plasma exosomes were host derived or the amount of
exosome released from the tumor decreased as the tumor grows.
Concerning host factors, a biochemical examination
of the blood identify potential relationships with the exosome
amount, however, there were no significant correlations. Immune
cells including T cells and dendritic cells were thought to release
exosomes (30), and exosomes also
regulate the immune system (31).
Therefore, we assessed the correlation between the exosome quantity
and the NLR or the GPS. The NLR and exosome number are both
independent prognostic factors, however, there was no correlation
between these factors. A further assessment is necessary concerning
the relationship between tumor-specific immune responses or immune
evasion and the exosome level.
Whether the release of exosomes from the tumor
changes as the tumor progresses was not revealed, however, some
studies have indicated that the state of hypoxia enhances exosome
release from the tumor (32), as
well as a low pH (33,34). According to this theory, it may be
reasonable that cancer-derived exosomes change or decrease when the
tumor becomes far advanced and rich in vascular formation.
Taken together, although the precise factors remain
unknown, it is possible that the exosome quantity reflects the
state of cancer-specific immune responses or the tumor
microenvironment and may predict the prognosis.
In conclusion, tumor-derived exosomes can be
detected in the blood from the plasma in our established mouse
model. The exosome quantity in the patient plasma did not correlate
with the respective tumor progression in each ESCC patient,
however, the amount of exosomes in the plasma, as measured using
the AChE activity, was an independent prognostic factor for ESCC
patients.
Acknowledgments
The present study was supported by a Grant-in-Aid
for Scientific research (grant nos. 15K19872 and 26670597) from the
Japan Society for the Promotion of Science.
References
|
1
|
Global Burden of Disease Cancer
Collaboration; Fitzmaurice C, Dicker D, Pain A, Hamavid H,
Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe
C, et al: The Global Burden of Cancer 2013. JAMA Oncol. 1:505–527.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tachimori Y, Ozawa S, Numasaki H,
Fujishiro M, Matsubara H, Oyama T, Shinoda M, Toh Y, Udagawa H and
Uno T; The Registration Committee for Esophageal Cancer of the
Japan Esophageal Society: Comprehensive Registry of Esophageal
Cancer in Japan, 2008. Esophagus. 12:130–157. 2015. View Article : Google Scholar
|
|
5
|
Akutsu Y, Uesato M, Shuto K, Kono T,
Hoshino I, Horibe D, Sazuka T, Takeshita N, Maruyama T, Isozaki Y,
et al: The overall prevalence of metastasis in T1 esophageal
squamous cell carcinoma: A retrospective analysis of 295 patients.
Ann Surg. 257:1032–1038. 2013. View Article : Google Scholar
|
|
6
|
Denzer K, Kleijmeer MJ, Heijnen HF,
Stoorvogel W and Geuze HJ: Exosome: From internal vesicle of the
multivesicular body to intercellular signaling device. J Cell Sci.
113:3365–3374. 2000.PubMed/NCBI
|
|
7
|
van den Boorn JG, Dassler J, Coch C,
Schlee M and Hartmann G: Exosomes as nucleic acid nanocarriers. Adv
Drug Deliv Rev. 65:331–335. 2013. View Article : Google Scholar
|
|
8
|
Takeshita N, Hoshino I, Mori M, Akutsu Y,
Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuman N,
et al: Serum microRNA expression profile: miR-1246 as a novel
diagnostic and prognostic biomarker for oesophageal squamous cell
carcinoma. Br J Cancer. 108:644–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen WX, Liu XM, Lv MM, Chen L, Zhao JH,
Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, et al: Exosomes from
drug-resistant breast cancer cells transmit chemoresistance by a
horizontal transfer of microRNAs. PLoS One. 9:e952402014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ichim TE, Zhong Z, Kaushal S, Zheng X, Ren
X, Hao X, Joyce JA, Hanley HH, Riordan NH, Koropatnick J, et al:
Exosomes as a tumor immune escape mechanism: Possible therapeutic
implications. J Transl Med. 6:372008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lundholm M, Schröder M, Nagaeva O, Baranov
V, Widmark A, Mincheva-Nilsson L and Wikström P: Prostate
tumor-derived exosomes down-regulate NKG2D expression on natural
killer cells and CD8+ T cells: Mechanism of immune
evasion. PLoS One. 9:e1089252014. View Article : Google Scholar
|
|
13
|
Hu Y, Yan C, Mu L, Huang K, Li X, Tao D,
Wu Y and Qin J: Fibroblast-derived exosomes contribute to
chemoresistance through priming cancer stem cells in colorectal
cancer. PLoS One. 10:e01256252015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ono M, Kosaka N, Tominaga N, Yoshioka Y,
Takeshita F, Takahashi RU, Yoshida M, Tsuda H, Tamura K and Ochiya
T: Exosomes from bone marrow mesenchymal stem cells contain a
microRNA that promotes dormancy in metastatic breast cancer cells.
Sci Signal. 7:ra632014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Walker ND, Patel J, Munoz JL, Hu M, Guiro
K, Sinha G, et al: The bone marrow niche in support of breast
cancer dormancy. Cancer Lett. Nov 3–2015.(Epub ahead of print).
pii: S0304–3835(15)00664-3. View Article : Google Scholar
|
|
16
|
An T, Qin S, Xu Y, Tang Y, Huang Y, Situ
B, Inal JM and Zheng L: Exosomes serve as tumour markers for
personalized diagnostics owing to their important role in cancer
metastasis. J Extracell Vesicles. 4:2752242015. View Article : Google Scholar
|
|
17
|
Savina A, Vidal M and Colombo MI: The
exosome pathway in K562 cells is regulated by Rab11. J Cell Sci.
115:2505–2515. 2002.PubMed/NCBI
|
|
18
|
Gupta S and Knowlton AA: HSP60 trafficking
in adult cardiac myocytes: Role of the exosomal pathway. Am J
Physiol Heart Circ Physiol. 292:H3052–H3056. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Usui A, Hoshino I, Akutsu Y, Sakata H,
Nishimori T, Murakami K, Kano M, Shuto K and Matsubara H: The
molecular role of Fra-1 and its prognostic significance in human
esophageal squamous cell carcinoma. Cancer. 118:3387–3396. 2012.
View Article : Google Scholar
|
|
20
|
Akanuma N, Hoshino I, Akutsu Y, Murakami
K, Isozaki Y, Maruyama T, Yusup G, Qin W, Toyozumi T, Takahashi M,
et al: MicroRNA-133a regulates the mRNAs of two invadopodia-related
proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer.
110:189–198. 2014. View Article : Google Scholar :
|
|
21
|
Yodying H, Matsuda A, Miyashita M,
Matsumoto S, Sakurazawa N and Uchida E: Prognostic significance of
neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio in
oncologic outcomes of esophageal cancer: A systematic review and
meta-analysis. Ann Surg Oncol. 23:646–654. 2016. View Article : Google Scholar
|
|
22
|
McMillan DC: The systemic
inflammation-based Glasgow Prognostic Score: A decade of experience
in patients with cancer. Cancer Treat Rev. 39:534–540. 2013.
View Article : Google Scholar
|
|
23
|
Tanaka Y, Kamohara H, Kinoshita K,
Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M and Baba H:
Clinical impact of serum exosomal microRNA-21 as a clinical
biomarker in human esophageal squamous cell carcinoma. Cancer.
119:1159–1167. 2013. View Article : Google Scholar
|
|
24
|
Yoshioka Y, Kosaka N, Konishi Y, Ohta H,
Okamoto H, Sonoda H, Nonaka R, Yamamoto H, Ishii H, Mori M, et al:
Ultra-sensitive liquid biopsy of circulating extracellular vesicles
using ExoScreen. Nat Commun. 5:35912014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suetsugu A, Honma K, Saji S, Moriwaki H,
Ochiya T and Hoffman RM: Imaging exosome transfer from breast
cancer cells to stroma at metastatic sites in orthotopic nude-mouse
models. Adv Drug Deliv Rev. 65:383–390. 2013. View Article : Google Scholar
|
|
26
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fleitas T, Martínez-Sales V, Vila V,
Reganon E, Mesado D, Martín M, Gómez-Codina J, Montalar J and
Reynés G: Circulating endothelial cells and microparticles as
prognostic markers in advanced non-small cell lung cancer. PLoS
One. 7:e473652012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tavoosidana G, Ronquist G, Darmanis S, Yan
J, Carlsson L, Wu D, Conze T, Ek P, Semjonow A, Eltze E, et al:
Multiple recognition assay reveals prostasomes as promising plasma
biomarkers for prostate cancer. Proc Natl Acad Sci USA.
108:8809–8814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Silva J, Garcia V, Rodriguez M, Compte M,
Cisneros E, Veguillas P, Garcia JM, Dominguez G, Campos-Martin Y,
Cuevas J, et al: Analysis of exosome release and its prognostic
value in human colorectal cancer. Genes Chromosomes Cancer.
51:409–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Quah B and O'Neill HC: Review: The
application of dendritic cell-derived exosomes in tumour
immunotherapy. Cancer Biother Radiopharm. 15:185–194. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Robbins PD and Morelli AE: Regulation of
immune responses by extracellular vesicles. Nat Rev Immunol.
14:195–208. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
King HW, Michael MZ and Gleadle JM:
Hypoxic enhancement of exosome release by breast cancer cells. BMC
Cancer. 12:4212012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Parolini I, Federici C, Raggi C, Lugini L,
Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A,
et al: Microenvironmental pH is a key factor for exosome traffic in
tumor cells. J Biol Chem. 284:34211–34222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ban JJ, Lee M, Im W and Kim M: Low pH
increases the yield of exosome isolation. Biochem Biophys Res
Commun. 461:76–79. 2015. View Article : Google Scholar : PubMed/NCBI
|