Introduction
Vulvar squamous cell carcinoma (VSCC) is a
relatively rare malignant tumor in women which accounts for 80–90%
of female genital tract cancers (1). Early stage VSCC is primarily treated
with surgery, however, postoperative complications can occur.
Several of these complications, such as lower limb lymphatic
obstruction and vulvar morphologic changes, can have a deep impact
on the quality of life of patients. The treatment for advanced
vulvar cancer includes radiotherapy. The survival rate for patients
with advanced VSCC is only ~30% due to the reduced tolerance of
vulvar tissues to radiation. Thus, it is important to better
understand the pathogenesis of VSCC and to identify possible
effective biomarkers to aid diagnosis.
Over the past decade, research into the non-coding
RNA (ncRNA) domain has attracted significant attention. Long
non-coding RNAs (lncRNAs) >200 nucleotides in length do not
encode any proteins (2,3) but are involved in regulating gene
functions at transcriptional and post-transcriptional levels
(4). These gene functions include
epigenetic regulation, chromatin modification, cell cycle
regulation, nuclear trafficking, transcription and splicing
(5–8). The dysregulation of lncRNAs is linked
to various human diseases, particularly cancer (9–11).
In the present study, an lncRNA expression profile
was established from samples obtained from four paired VSCC
subjects and their adjacent non-tumor (NT) tissues through a
microarray platform. The aim of the present study was to identify
dysregulated lncRNAs and mRNAs in VSCC patients. Six of these
lncRNAs were evaluated by real-time reverse
transcription-polymerase chain reaction (RT-PCR) in 35 cases with
benign vulvar diseases, VSCC and their pericarcinoma tissues
according to the top 30 upregulated and top 30 down-regulated
lncRNAs.
Materials and methods
Patients and sample collection
The present study protocol was approved by the
Institutional Review Board of Shengjing Hospital Affiliated to
China Medical University. Written informed consent was obtained
from all patients. A total of 16 VSCC patients, diagnosed with VSCC
between January 2013 and June 2015, were enrolled in the present
study. Adjacent tissues in 4 VSCC patients, 7 cases of vulvar
lichen sclerosus, 6 cases of vulvar leukoplakia and 2 cases of
vulvar intraepithelial neoplasia (VIN) grade I were obtained as the
control group. The clinical characteristics of the patients and
their tumors are summarized in Table
I. None of the patients had received prior radiotherapy or
other anticancer treatment. All histopathological samples were
obtained at surgery and were confirmed as VSCC. All samples were
stored at −80°C until use.
 | Table IRelationship between the expression of
NEAT1, LINC00478 or MIR31HG and the clinicopathological features of
the patients with VSCC. |
Table I
Relationship between the expression of
NEAT1, LINC00478 or MIR31HG and the clinicopathological features of
the patients with VSCC.
| No. of patients
(%) | NEAT1 | LINC00478 | MIR31HG |
|---|
| Age (years) |
| ≤63 | 8 (50) | 2.59 (1.23,
2.90) | 8.44 (5.06,
7.09) | 9.09 (6.61,
13.67) |
| >63 | 8 (50) | 4.09 (1.86,
7.68) | 8.93 (8.35,
10.45) | 8.91 (7.67,
10.04) |
| P-value | | 0.27 | 0.012 | 1.000 |
| FIGO Stage |
| I | 7 (43.8) | 3.40 (1.49,
8.28) | 8.33 (5.55,
9.32) | 10.08 (6.46,
14.49) |
| II | 1 (6.3) | | | |
| III | 8 (50) | 2.59 (1.23,
2.90) | 6.68 (5.26,
10.02) | 8.91 (7.67,
10.04) |
| IV | 0 (0) | | | |
| P-value | | 0.08 | 0.252 | 1.000 |
| Tumor
differentitation |
| Well | 9 (56.3) | 4.09 (1.86,
7.68) | 9.32 (8.36,
11.43) | 10.08 (9.62,
15.58) |
| Moderate | 5 (31.3) | 2.71 (1.91,
3.00) | 5.55 (5.28,
6.68) | 7.08 (6.26,
7.90) |
| Poor | 2 (12.5) | 1.41 (0.23) | 4.74 (4.47) | 7.64 (6.74) |
| P-value | | 0.214 | 0.003 | 0.007 |
| Lymphatic
metastasis |
| N0 | 10 (60) | 3.22 (2.90,
6.46) | 8.93 (8.33,
10.22) | 8.91 (6.92,
12.02) |
| N1, N2, N3 | 6 (37.5) | 1.34 (0.97,
1.77) | 5.71 (5.17,
9.32) | 9.14 (7.28,
11.72) |
| P-value | | 0.002 | 0.065 | 1.000 |
Total RNA extraction
Total RNA was isolated from the frozen samples using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer's protocols. All samples were quantified using a
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific,
Waltham, MA. USA). The purity of total RNA was examined by its
absorbance ratio at 260–280 nm. The absorbance ratios at A260/280
were between 1.8 and 2.0.
lncRNA and mRNA microarray analysis
Four paired samples were chosen for the experiments
performed at CapitalBio Corporation Laboratories (CapitalBio Corp.,
Beijing, China). Generally, total RNA which was extracted using
TRIzol reagent for microarray was reverse-transcripted to
double-stranded complementary DNA (cDNA). cDNA synthesis,
purification, labeling and hybridization were carried out according
to the manufacturer's instructions. lncRNA expression profiling was
performed using lncRNAs and mRNA 4×180K Human Gene Expression
Microarray V4.0 (CapitalBio Corp.). The data extracted were
analyzed by Agilent Feature Extraction version 10.7 (Agilent
Technologies, Santa Clara, CA, USA), and were summarized and
normalized using Agilent GeneSpring software version 11.5.
Differentially expressed lncRNAs and mRNAs were identified through
an absolute fold-change >2 at a p<0.05.
Functional analysis
Gene ontology (GO) and pathway analyses were
performed in order to better understand the functions of
differentially expressed lncRNAs and mRNAs in VSCC. GO terms
consisted of biological processes, cellular components, and
molecular functions that were used to annotate and classify gene
function. Pathway analysis was based on the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database that place differentially
expressed mRNAs. In brief, the Fisher's exact test and t-test were
used. A p<0.05 was considered to indicate a statistically
significant result.
lncRNA-mRNA co-expression network
analysis
A correlation analysis was constructed between
differentially expressed lncRNAs and mRNA by calculating the
Pearson correlation coefficients for each dysregulated lncRNA and
mRNA. A significant correlation was defined as a correlation
>0.99 or ≤0.99 at a p<0.05.
Real-time PCR
Single-stranded cDNA was reverse transcribed using
the PrimeScript RT reagent kit with gDNA Eraser Perfect Real-Time
(Takara Bio, Kyoto, Japan), according to the manufacturer's
instructions. Real-time PCR was performed using SYBR Premix Ex Taq
Tli RNase H Plus (Takara Bio) and a Roche LightCycler 480 II system
(Roche, Basel, Switzerland). The 20 μl real-time PCR
reaction mixture contained 3 μl cDNA, 10 μl SYBR
Premix Ex Taq, 4 μl RNase-Free Water, and 1.5 μl,
each, of forward and reverse primers. The PCR conditions were as
follows: 30 sec at 95°C, followed by 40 cycles at 95°C for 5 sec
and at 60°C for 30 sec. All samples were run in triplicate for
analysis. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used
as an endogenous control. The relative abundance of lncRNA
expression was calculated using the threshold cycle (Ct) method
with relative quantitation (12).
The primer sequences were designed in the laboratory and were
synthesized by Sangon Biotech (Sangon, Shanghai, China), and are
shown in Table II.
 | Table IIRT-PCR primers for lncRNA expression
analysis. |
Table II
RT-PCR primers for lncRNA expression
analysis.
| Target name | Primer sequence
(5′-3′) |
|---|
|
MIR31HG-Forward |
CAGGTCTCCAGGTGTTCCAG |
|
MIR31HG-Reverse |
CCCAGGCTATGTCTTTCCTCT |
|
LINC00478-Forward |
AAGATGACAAGAGCACCTCAAAG |
|
LINC00478-Reverse |
GACCTCAGCCTCCTCCATTA |
| MEG3-Forward |
GCTGCCCATCTACACCTCA |
| MEG3-1-Reverse |
CCTCTTCATCCTTTGCCATC |
| NEAT1-Forward |
GATGCGCGCCTGGGTGTAGTT |
| NEAT1-Reverse |
CATGCAGCCTGCCCCACTGT |
| MALAT1-Forward |
CCGAGCTGTGCGGTAGGCATT |
| MALAT1-Reverse |
CGGTTTCCTCAAGCTCCGCCT |
| HOTAIR-Forward |
GGTCCTGCTCCGCTTCGCAG |
| HOTAIR-Reverse |
ACGCCCCTCCTTCCTCTCGC |
| GAPDH-Forward |
ACCCACTCCTCCACCTTTGAC |
| GAPDH-Reverse |
TGTTGCTGTAGCCAAATTCGT |
Statistical analysis
Statistical analysis was performed using Social
Sciences (SPSS) 19.0 software (SPSS, Inc., Chicago, IL, USA). The
independent-sample t-test was used to evaluate the expression
levels of lncRNAs. Association between lncRNAs and
clinicopathological factors was analyzed with the Mann-Whitney U
test which was used for comparison between the two groups and the
Kruskal-Wallis test which was used for comparison among the three
groups. A p<0.05 (two-tailed) was regarded as statistically
significant.
Results
Expression profiles of lncRNAs in VSCC
tissues
From the lncRNA gene expression, dysregulated
lncRNAs were identified in four paired vulvar squamous cell tumor
tissues and the adjacent normal vulvar tissue samples. Among the
15,840 lncRNA transcripts analyzed, 312 lncRNAs were observed as
upregulated (fold-change, >2; p<0.05) whereas 1,469 lncRNAs
were detected as downregulated (fold-change >2; p<0.05) in
the VSCC samples. Of these, AI769947 (fold-change, 34.2) was the
most highly upregulated lncRNA in the VSCC tissues. FER1L4
(fold-change, 48.9) was the most extensively downregulated lncRNA.
Hierarchical clustering analysis was performed to determine whether
the expression patterns in the VSCC tissues were significantly
different from these patterns in the adjacent normal tissues
(Fig. 1A).
Overview of the mRNA profile in VSCC
Up to 33,045 coding transcripts were detected in the
VSCC tissues and the NTs, of which 21,788 showed significantly
differential expression (fold-change, >2; p<0.05). Among
them, 1,521 were upregulated and 4,694 were downregulated in the
VSCC tissues. Their distinct expression patterns were evaluated by
hierarchical clustering analysis (Fig.
1B).
GO and pathway analysis
Go analysis (comprised of biological processes,
cellular components and molecular functions) was performed in order
to determine the genes and gene product enrichment. Through GO
analysis, the significant differentially expressed mRNAs were
principally associated with extracellular organelles (Fig. 2), but none were linked with
molecular function on GO analysis. Pathway analysis indicated that
38 pathways corresponded to the most significant differential
pathways.
Overview of the co-expression
network
In order to verify the correlation between
differentially expressed lncRNAs and mRNAs, a coding/non-coding
gene co-expression network was constructed in VSCC patients
compared with the control group. The co-network showed that one
lncRNA correlated with multiple mRNAs and reciprocally. According
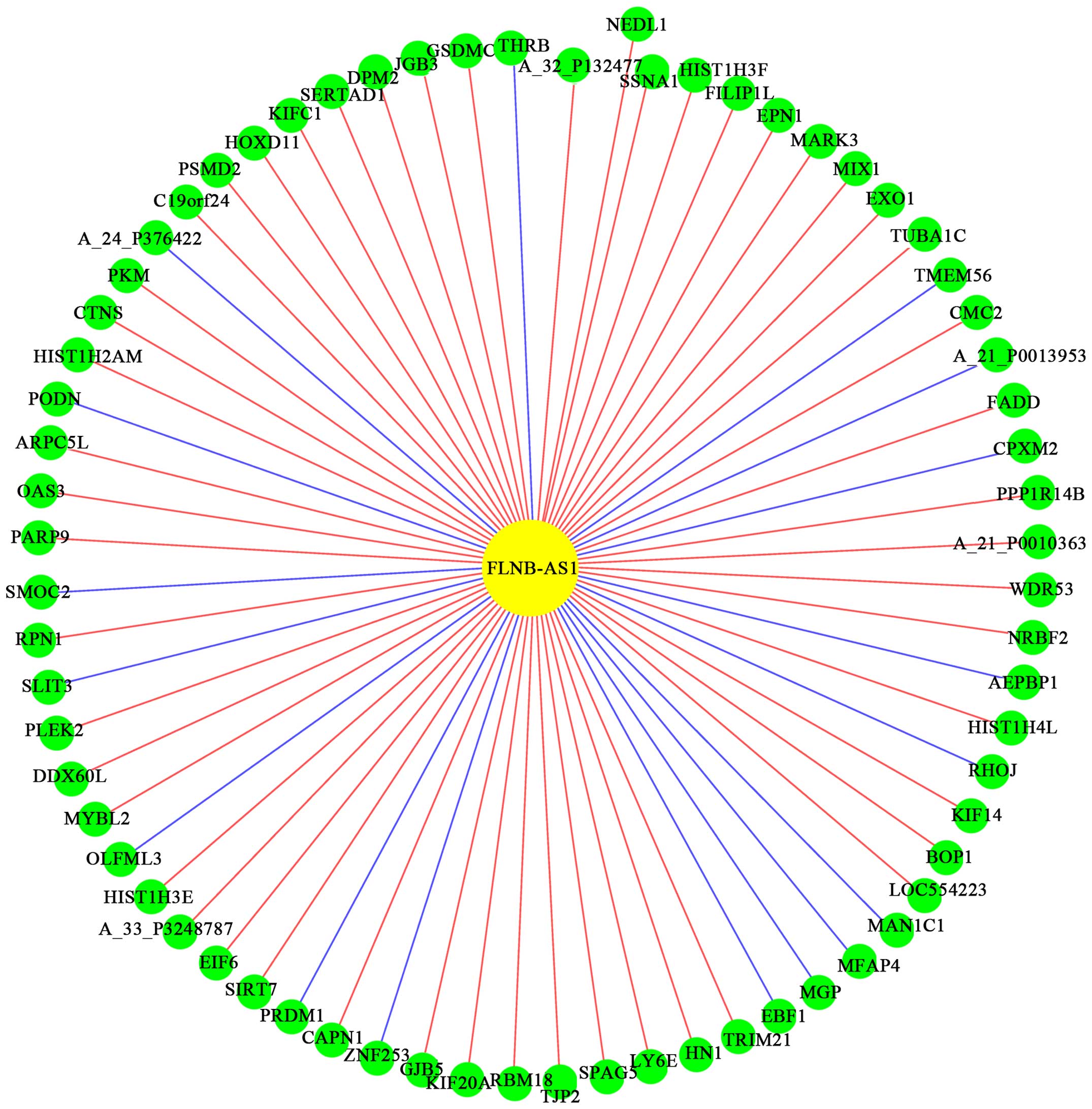
to the network, we found that lncRNA, FLNB-AS1, had the most
correlative mRNAs such as HOXD11 and PKM (Fig. 3). Therefore, it was proposed that
the expression profile of lncRNAs and mRNAs was significantly
correlated.
Real-time PCR validation of several
differentially expressed lncRNAs
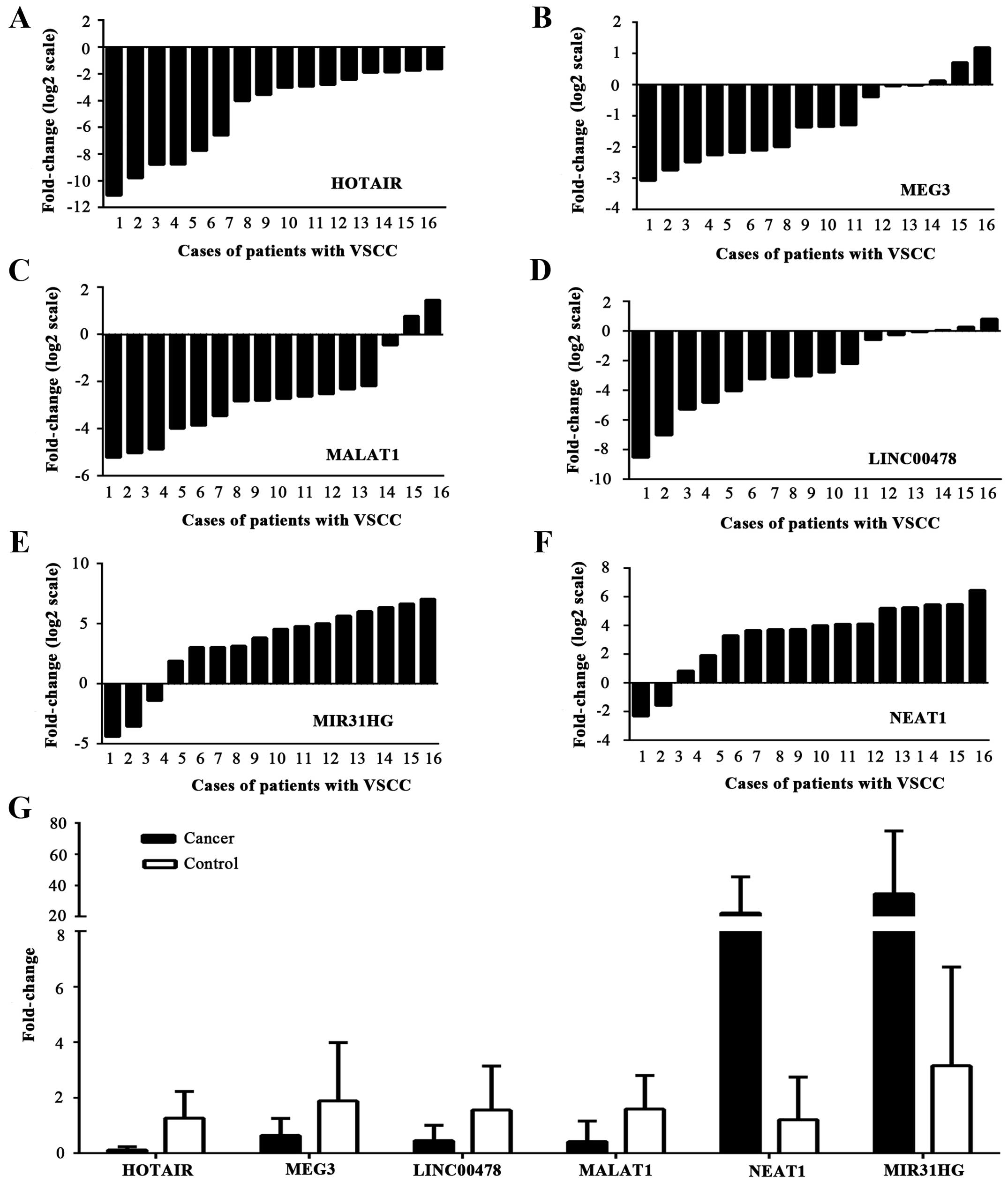
According to fold difference, one upregulated lncRNA
(NEAT1) was initially selected and five downregulated lncRNAs
(MIR31HG, MALAT1 HOTAIR, LINC00478 and MEG3) were also selected for
verification of expression in two sets of tissue samples. A general
consistency between the micro-array and real-time PCR showed that
the six selected lncRNAs were dysregulated between VSCC tissues and
the control group (Fig. 4). The
levels of HOTAIR, MALAT1, MEG3 and LINC00478 (p=0.000, p=0.001,
p=0.023 and p=0.009, respectively) expression were decreased and
NEAT1 was upregulated as indicated by the microarray analysis
(p=0.002); whereas high expression of MIR31HG in the 35 samples,
was detected, in opposition to the microarray data (p=0.007).
Evaluation of MEG3 and MALAT1 as
diagnostic biomarkers for VSCC
We constructed a receiver operating characteristic
(ROC) curve to evaluate the diagnostic value of our findings. Both
MEG3 and MALAT1 were downregulated in the VSCC samples. Among them,
MEG3 was only detected in 19% (3/16) of the tumor samples,
meanwhile, MALAT1 was found downregulated in 87.5% (14/16) of the
VSCC samples. Furthermore, the areas under the ROC curve (AUCs)
were 0.707 and 0.839, respectively (p<0.05) (Fig. 5).
Correlation between NEAT1, LINC00478 and
MIR31HG expression and clinicopathological features in VSCC
patients
Lymphatic metastasis is the important measure of
prognosis. As shown in Table I, we
found that the level of NEAT1 expression in VSCC tissues was
associated with lymph node metastasis (p=0.002). We also observed
that there was a statistical relationship between LINC00478 and
MIR31HG expression and tumor differentiation (p=0.003, p=0.007
respectively). However, no significant correlations were found
between lncRNAs levels and other clinicopathological factors
studied, including the International Federation of Gynecology and
Obstetrics (FIGO) stage.
Discussion
Recent research has demonstrated the key role of
lncRNAs in regulating embryogenesis and gene expression, and
lncRNAs have emerged as drivers of tumor suppressive and oncogenic
functions in various human solid tumors (13–16).
Our results revealed that aberrantly expressed lncRNAs may be a
factor in VSCC pathogenesis, potentially providing new biomarkers
and therapeutic targets for VSCC.
VSCC is a rare malignant tumor, however, its
morbidity and mortality rates have risen in the past few decades.
It is generally believed that VSCC has two etiological pathways,
i.e., a high-risk human papillomavirus (HPV)-dependent route and
genetic alterations such as p53 mutations and silencing of p16INK4a
(17,18).
Homeobox transcript antisense RNA (HOTAIR) is a
well-known lncRNA transcribed from the HOXC cluster located on
chromosome 12q13.3 (19). It has
been proven that HOTAIR is involved in cancer progression and
prognosis and may be an important target for cancer diagnosis and
therapy (20–22). According to microarray analysis, the
present study revealed that the level of HOTAIR expression was
decreased in 75% (3/4) of the paired samples of tumor tissues.
According to Sharma et al, there is a significant linear
trend towards progressive HOTAIR downregulation among HPV-negative
controls, HPV-positive non-malignant tumors and cervical carcinoma
samples. They speculated that HOTAIR could be a potential target of
E7, in HPV16-related cervical cancers (23).
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1) is another long lncRNA correlated with HPV. Jiang et
al found that MALAT1 was expressed in HPV-positive cervical
squamous cells, but not in HPV-negative normal cervical squamous
cells, which suggests that HPV correlates with MALAT1 deregulation
in cervical cancer (24). Various
previous studies have indicated that HOTAIR and MALAT1 are
overexpressed in tumor tissues compared with the normal tissues
(25–28). It was thought that HOTAIR and MALAT1
contribute to carcinogenesis in most tumors whereas other studies
proved the role of HOTAIR and MALAT1 as tumor suppressors. HOTAIR
expression was found to be increased in breast cancer patients;
nonetheless, knockdown of HOTAIR could inhibit invasion (15). Lu et al found that breast
cancer patients with HOTAIR high-expression had lower risk of
recurrence and poor prognosis (29). It has also been demonstrated that
the level of expression of MALAT1 in gliomas was lower than that in
normal tissues (30). These
discrepant results suggest that lncRNAs can differentially express
depending on the cancer types and that lncRNAs have active roles in
tumorigenesis. Moreover, our data revealed the opposite results
when compared to many other studies, namely that HOTAIR and MALAT1
were down-regulated in VSCC tissues. Firstly, we inferred that HPV
infection may be involved. Both HOTAIR and MALAT1 are correlated
with HPV. Unfortunately, some of the patients we evaluated refused
HPV examination and we were unable to determine the number of
patients originally infected with HPV. HPV infection is not the
most crucial factor in the level of expression of lncRNAs, since it
is irrelevant in other types of tumors. However, HPV examination is
still necessary for VSCC patients. In addition, it is meaningful
that HPV infection be considered in lncRNA expression analysis as
it is a useful factor. We hypothesized that lncRNAs such as HOTAIR
and MALAT1 are expressed differentially in HPV-positive and
HPV-negative VSCC samples and that these two lncRNAs may be
involved in VSCC tumorigenesis by targeting HPV. Secondly, the
number of cases is one limitation in our research. Thus,
larger-scale studies are needed to validate our findings for
further research.
The TP53-associated route and other genetic changes,
such as CDKN2A (p16), also play a key role in the development of
VSCC. Genetic mutations in TP53 have been detected in HPV-negative
VSCCs. They may represent early changes in HPV-independent vulvar
carcinogenesis (31). Expression of
p53 was significantly increased in VSCC patient samples compared
with the control group (32).
Maternally expressed gene 3 (MEG3) is a lncRNA which
has been recently associated with p53. Lu et al found that
p53 protein levels are affected by MEG3 overexpression in
vitro. MEG3 was significantly downregulated in non-small cell
lung cancer (NSCLC) tissues that may be affected by DNA
methylation, and it partially regulated NSCLC cell proliferation
and apoptosis via the activation of p53 (33). Zhu et al found that the level
of MEG3 expression was reduced in hepatoma samples compared with
adjacent non-tumor samples and interacts with the p53 DNA binding
domain. Moreover, various p53 target genes were found to be
deregulated after overexpression of MEG3 in hepatoma cells
(34). Our data showed that MEG3
was downregulated in the VSCC patient samples. We speculate that
MEG3 may operate as a tumor-suppressor in VSCC, whose mechanism may
involve activation of p53.
Epigenetic silencing of p16 is also an early and
important event in vulvar neoplasia (19). Montes et al showed that the
lncRNA, MIR31HG, is upregulated in oncogene-induced senescence
(OIS) which is considered an important mechanism of tumor
suppression. They also found that knockdown of MIR31HG promoted a
strong p16INK4A-dependent senescence phenotype. These results
suggest that MIR31HG may repress p16INK4A expression in human
cancer (35). MIR31HG is
underactive in most cancers, but in lung and breast cancers, it
appears to be upregulated by an, as yet, unknown mechanism
(36,37). According to the microarray analysis,
the present study revealed that the level of MIR31HG expression was
decreased in 75% (3/4) of the paired samples of tumor tissues while
it was upregulated in VSCC samples by RT-PCR. These findings imply
that the microarray profiling results that are based on a small
sample size may not entirely be reliable and need to be further
verified by RT-PCR analysis using a greater number of samples.
The lncRNA MIR31HG is also correlated with poor
prognosis and may be an important target for cancer diagnosis and
therapy in gastric cancer (38).
According to the present study, we found that the increased level
of MIR31HG expression in VSCC was significantly associated with
tumor differentitation (Table I),
suggesting that MIR31HG may play a critical role, not only in the
tumorigenesis of VSCC, but also in its prognosis. Patients with
higher expression levels of MIR31HG may have poorer prognosis.
However, not all patients who were diagnosed with VSCC within the
last three years at our institution underwent surgery, and only two
patients died of VSCC, therefore, we were unable to obtain reliable
survival analysis.
Nuclear paraspeckle assembly transcript 1 (NEAT1) is
a prognostic biomarker as well as MIR31HG (39). NEAT1 was upregulated in 75% (3/4) of
VSCC tissues in accordance with the results of the microarray
profile (Fig. 4). According to the
study of Li et al, NEAT1 may play an oncogenic role in
colorectal cancer which is involved in differentiation, invasion
and metastasis (40). Our results
revealed a greater association between NEAT1 expression and lymph
node metastasis (Table I).
Lymphatic metastasis usually indicates poor prognosis, suggesting
that NEAT1 may play an important role in the prognosis of VSCC.
lncRNAs are emerging as biomarkers based upon ncRNA
biology. Similar to microRNAs, various lncRNAs have been found in
both urine and plasma, which can be easily obtained and provide for
readily-available and stable diagnostics in order to detect cancers
and cancer subtypes (41,42).
lncRNAs, such as MEG3 and MALAT1, have been shown to
be potential biomarkers in other types of cancers. However, there
are some critical issues that still need to be resolved before they
can be applied clinically. Most recent studies have been performed
using only small-scale samples. Therefore, further large-scale
studies are needed to validate their findings. In addition, since
conventional methods such as RNA extraction take too long and the
quality of lncRNAs can be influenced by multiple factors during the
analytical process, more stable platforms and more rapid analytical
methods are required. Finally, the expression of lncRNAs could be
confounded by multiple factors in tumorigenesis procedures such as
pathology. Thus, our results must be validated in larger
independent cohorts to confirm that lncRNAs can be used as reliable
biomarkers for cancer detection.
The present study had several limitations including
its small sample size. Although our results suggested that several
lncRNAs were potentially important biomarkers in VSCC, we collected
only 16 samples from VSCC patients. Thus, our results require
validation using larger prospective studies involving a larger
number of samples. Furthermore, not all patients who were diagnosed
with VSCC within the last three years underwent surgery; some were
lost in the follow-up and only two patients died from VSCC. Thus,
it was difficult to arrive at a reliable survival curve due to the
small number of cases available. In addition, there were no stage
IV patients in our cases since most patients with stage IV chose
not to undergo surgery. Therefore, future experiments targeting
these aspects are needed to verify our results.
In conclusion, the present study was the first to
determine global aberrant lncRNA expression in VSCC tissues
compared with the adjacent non-tumorous tissues using microarray
analysis. We also identified a panel of dysregulated lncRNAs that
may be potential biomarkers as they were also correlated with VSCC
carcinogenesis. These dysregulated lncRNAs may be VSCC-specific
lncRNAs and may form the basis for future diagnostic, therapeutic
and functional research on VSCC.
References
|
1
|
Woelber L, Kock L, Gieseking F, Petersen
C, Trillsch F, Choschzick M, Jaenicke F and Mahner S: Clinical
management of primary vulvar cancer. Eur J Cancer. 47:2315–2321.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bertone P, Stolc V, Royce TE, Rozowsky JS,
Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et
al: Global identification of human transcribed sequences with
genome tiling arrays. Science. 306:2242–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roberts TC, Morris KV and Weinberg MS:
Perspectives on the mechanism of transcriptional regulation by long
non-coding RNAs. Epigenetics. 9:13–20. 2014. View Article : Google Scholar :
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi K, Yan I, Haga H and Patel T:
Long noncoding RNA in liver diseases. Hepatology. 60:744–753. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song H, Sun W, Ye G, Ding X, Liu Z, Zhang
S, Xia T, Xiao B, Xi Y and Guo J: Long non-coding RNA expression
profile in human gastric cancer and its clinical significances. J
Transl Med. 11:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔC T method. Methods. 25:402–408.
2001. View Article : Google Scholar
|
|
13
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trietsch MD, Nooij LS, Gaarenstroom KN and
van Poelgeest MI: Genetic and epigenetic changes in vulvar squamous
cell carcinoma and its precursor lesions: A review of the current
literature. Gynecol Oncol. 136:143–157. 2015. View Article : Google Scholar
|
|
18
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gasco M, Sullivan A, Repellin C, Brooks L,
Farrell PJ, Tidy JA, Dunne B, Gusterson B, Evans DJ and Crook T:
Coincident inactivation of 14-3-3σ and p16INK4a is an
early event in vulval squamous neoplasia. Oncogene. 21:1876–1881.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lv XB, Lian GY, Wang HR, Song E, Yao H and
Wang MH: Long noncoding RNA HOTAIR is a prognostic marker for
esophageal squamous cell carcinoma progression and survival. PLoS
One. 8:e635162013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen FJ, Sun M, Li SQL, Wu QQ, Ji L, Liu
ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al: Upregulation of the long
non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma
metastasis and poor prognosis. Mol Carcinog. 52:908–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY,
Xue WQ, Chen YB, Zhang Y and Jia WH: HOTAIR, a prognostic factor in
esophageal squamous cell carcinoma, inhibits WIF-1 expression and
activates Wnt pathway. Cancer Sci. 104:1675–1682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sharma S, Mandal P, Sadhukhan T, Roy
Chowdhury R, Ranjan Mondal N, Chakravarty B, Chatterjee T, Roy S
and Sengupta S: Bridging links between long noncoding RNA HOTAIR
and HPV oncoprotein E7 in cervical cancer pathogenesis. Sci Rep.
5:117242015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie
P, Zhou G and Li G: The role of MALAT1 correlates with HPV in
cervical cancer. Oncol Lett. 7:2135–2141. 2014.PubMed/NCBI
|
|
25
|
Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J
and Liu M: Long inter-genic noncoding RNA HOTAIR is overexpressed
and regulates PTEN methylation in laryngeal squamous cell
carcinoma. Am J Pathol. 182:64–70. 2013. View Article : Google Scholar
|
|
26
|
Wu ZH, Wang XL, Tang HM, Jiang T, Chen J,
Lu S, Qiu GQ, Peng ZH and Yan DW: Long non-coding RNA HOTAIR is a
powerful predictor of metastasis and poor prognosis and is
associated with epithelial-mesenchymal transition in colon cancer.
Oncol Rep. 32:395–402. 2014.PubMed/NCBI
|
|
27
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013.PubMed/NCBI
|
|
28
|
Li X, Wu Z, Mei Q, Li X, Guo M, Fu X and
Han W: Long non-coding RNA HOTAIR, a driver of malignancy, predicts
negative prognosis and exhibits oncogenic activity in oesophageal
squamous cell carcinoma. Br J Cancer. 109:2266–2278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu L, Zhu G, Zhang C, Deng Q, Katsaros D,
Mayne ST, Risch HA, Mu L, Canuto EM, Gregori G, et al: Association
of large noncoding RNA HOTAIR expression and its downstream
intergenic CpG island methylation with survival in breast cancer.
Breast Cancer Res Treat. 136:875–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X,
Sun T, Xie X, Zhou Y and Du Z: Tumor-suppressive function of long
noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and
inactivation of ERK/MAPK signaling. Cell Death Dis. 7:e21232016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Del Pino M, Rodriguez-Carunchio L and Ordi
J: Pathways of vulvar intraepithelial neoplasia and squamous cell
carcinoma. Histopathology. 62:161–175. 2013. View Article : Google Scholar
|
|
32
|
Sadalla JC, Lourenço SV, Sotto MN, Baracat
EC and Carvalho JP: Claudin and p53 expression in vulvar lichen
sclerosus and squamous-cell carcinoma. J Clin Pathol. 64:853–857.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu KH and Li W, Liu XH, Sun M, Zhang ML,
Wu WQ, Xie WP, Hou YY, Lu KH and Li W: Long non-coding RNA MEG3
inhibits NSCLC cells proliferation and induces apoptosis by
affecting p53 expression. BMC Cancer. 13:4612013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Wei L, Jin Y, Fu H, Wu Y, et al: Long noncoding RNA MEG3 interacts
with p53 protein and regulates partial p53 target genes in hepatoma
cells. PLoS One. 10:e01397902015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Montes M, Nielsen MM, Maglieri G, Jacobsen
A, Højfeldt J, Agrawal-Singh S, Hansen K, Helin K, van de Werken
HJ, Pedersen JS, et al: The lncRNA MIR31HG regulates
p16INK4A expression to modulate senescence. Nat Commun.
6:69672015. View Article : Google Scholar
|
|
36
|
Shi Y, Lu J, Zhou J, Tan X, He Y, Ding J,
Tian Y, Wang L and Wang K: Long non-coding RNA Loc554202 regulates
proliferation and migration in breast cancer cells. Biochem Biophys
Res Commun. 446:448–453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xi S, Yang M, Tao Y, Xu H, Shan J,
Inchauste S, Zhang M, Mercedes L, Hong JA, Rao M, et al: Cigarette
smoke induces C/EBP-β-mediated activation of miR-31 in normal human
respiratory epithelia and lung cancer cells. PLoS One.
5:e137642010. View Article : Google Scholar
|
|
38
|
Nie FQ, Ma S, Xie M, Liu YW, De W and Liu
XH: Decreased long noncoding RNA MIR31HG is correlated with poor
prognosis and contributes to cell proliferation in gastric cancer.
Tumour Biol. Dec 21–2015.Epub ahead of print. PubMed/NCBI
|
|
39
|
Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F,
Zhuo C, Zheng Y, Li B, Wang Z, et al: Nuclear-enriched abundant
transcript 1 as a diagnostic and prognostic biomarker in colorectal
cancer. Mol Cancer. 14:1912015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Y, Li Y, Chen W, He F, Tan Z, Zheng J,
Wang W, Zhao Q and Li J: NEAT expression is associated with tumor
recurrence and unfavorable prognosis in colorectal cancer.
Oncotarget. 6:27641–27650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang X, Yuan T, Tschannen M, Sun Z, Jacob
H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, et al:
Characterization of human plasma-derived exosomal RNAs by deep
sequencing. BMC Genomics. 14:3192013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee GL, Dobi A and Srivastava S: Prostate
cancer: Diagnostic performance of the PCA3 urine test. Nat Rev
Urol. 8:123–124. 2011. View Article : Google Scholar : PubMed/NCBI
|