Introduction
The molecular pathogenesis of the development of
non-small cell lung cancer (NSCLC) is very complex (1). Understanding the molecular basis of
the development of this malignant tumor, especially lung squamous
cell carcinoma (LSCC), may enable the use of targeted therapy,
which may result in a greater efficiency in the treatment of these
patients. It is therefore important to search for new and more
effective therapeutic strategies as well as new proteins that may
be used as potential targets (1,2). In
this sense, the SOX protein family appears to be an auspicious
element in anticancer therapy (3).
SRY-related HMG-box (SOX) family genes were isolated
in mammals in 1990 on the basis of the presence of the conservative
high mobility group (HMG) box protein domain, primarily occurring
in the sex-determining region Y (SRY) (4). Varying expression levels of SOX
proteins have been attested depending on the type of cancer in
which they occur. This may indicate that the same protein can serve
opposing functions in different tumors (5). Approximately 20 proteins belong to the
SOX family, and they are divided into 8 main groups denoted from A
to H (6,7). Group F comprises the proteins SOX7
(8), SOX17 (9) and SRY-related HMG-box 18 (SOX18)
(10), which are involved in the
same pathways as the vascular endothelial growth factor (VEGF). The
SOX18 protein is one of the most important proteins involved in the
development of blood and lymphatic vessels during embryogenesis
(11–17). Recent studies have also shown that
the SOX18 protein may play a significant role in the progression of
malignant diseases (6,17–21).
Based on the results of our previous research
(3), we observed a differential
SOX18 expression both at the mRNA and protein level in NSCLC and
non-malignant lung tissues (NMLTs). We noted significantly lower
mRNA expression levels of this transcription factor in paired
tissues and in all of the studied NSCLC tissues as compared to
NMLTs. In contrast, increased SOX18 protein levels were observed in
NSCLC cases compared to that noted in the NMLTs (3). Interestingly, the level of mRNA did
not reflect in any way the level of protein, determined by western
blot analysis. This allowed us to hypothesize that the SOX18
transcript level could be controlled by microRNA (miRNA) molecules,
since similar mechanisms are observed in many other types of tumors
in relation to different types of proteins, as for example
miRNA-34b in prostate cancer (22).
miRNAs are involved in many important biological
processes, such as the regulation of cell proliferation, cell
differentiation, apoptosis, embryogenesis and organogenesis
(23–26). An increasingly visible role of
miRNAs in the regulation of cell proliferation processes, cell
differentiation and apoptosis has drawn the attention of scientists
to the relationship between miRNAs and carcinogenesis (26). As evidenced, miRNAs not only
regulate the expression of multiple oncogenes and tumor-suppressor
genes, but may also act themselves as oncogenes and tumor
suppressors. Those miRNAs with pro-apoptotic activity can function
as tumor suppressors, inhibiting proliferation. The correlation
between miRNAs and patient survival indicates the possibility to
use miRNAs as potential tumor prognostic markers (24,27–30). A
relationship between the expression level of 8 miRNAs and the
survival of patients with lung adenocarcinoma (AC) has been shown
(29). Patients with increased
expression of miR-155, miR-17-3p, miR-106a, miR-93 or miR-21, or
reduced expression of miR-7a-2, miR-7b or miR-145 exhibited a
significantly lower survival rate (31). The prospects for the use of miRNAs
in cancer therapy appear promising as well. It has been shown that
inhibition of miRNAs may lead to a reduction in tumor cell
proliferation in vitro (31).
The role of SOX18 expression in LSCC and other types
of lung cancer is not fully understood. Yet, considering previous
reports, this protein may be a significant factor in the
development and progression of NSCLC. The determination of the role
of specific miRNAs may be used in the future in cancer diagnosis,
prognostic assessment and NSCLC-targeted therapy.
Materials and methods
Patients and clinical samples
The present study was carried out using paraffin
blocks of LSCC and pairs of LSCC and NMLTs resected adjacent to the
primary tumor. All samples were obtained during surgical resection
from 2007–2014 at the Lower Silesian Centre of Lung Diseases in
Wroclaw. Paraffin sections of the obtained LSCC samples were
stained with hematoxylin and eosin (H&E) to verify the utility
for immunohistochemical (IHC) analysis. The study group consisted
of 25 formalin-fixed paraffin-embedded (FFPE) samples of LSCC used
for further IHC analysis and 25 pairs of LSCC and NMLT which were
collected in RNAlater solution (Qiagen, Hilden, Germany), and
stored at −20°C for RT-qPCR and droplet digital PCR (ddPCR)
experiments. Additionally, the same samples were collected, frozen
in liquid nitrogen and stored at −80°C for western blot analysis.
Clinical data were derived from hospital archives and are
summarized in Table I.
 | Table IPatient and tumor
characteristics. |
Table I
Patient and tumor
characteristics.
| Parameters | Data |
|---|
| Total no. of LSCC
cases | 25 |
| Age (years) | |
| Mean | 67.52±8.82 |
| Range | 57-81 |
| Gender, n (%) | |
| Male | 19 (76.0) |
| Female | 6 (24.0) |
| Tumor size, n
(%) | |
| T1 | 9 (36.0) |
| T2 | 13 (52.0) |
| T3 | 3 (12.0) |
| T4 | 0 (0.0) |
| Lymph nodes, n
(%) | |
| N0 | 20 (80.0) |
| N1, N2, N3 | 5 (20.0) |
| pTNM, n (%) | |
| 1A | 8 (32.0) |
| 1B | 6 (24.0) |
| 2A | 6 (24.0) |
| 2B | 4 (16.0) |
| 3A | 0 (0.0) |
| 3B | 0 (0.0) |
| 4 | 1 (4.0) |
| Grade, n (%) | |
| G1 | 0 (0.0) |
| G2 | 21 (87.5) |
| G3 | 4 (12.5) |
Immunohistochemistry
LSCC samples fixed in 10% buffered formalin and
embedded in paraffin were used for the IHC reactions. In order to
determine the SOX18 expression, the murine monoclonal mouse
antibody directed against SOX18 (D-8, Sc-166025; Santa Cruz
Biotechnology, Santa Cruz, CA, USA) was used in a dilution of 1:100
according to a previously established protocol (3). The IHC procedure was performed using
the Autostainer Link 48 (DakoCytomation, Glostrup, Denmark) to
provide reliable and repeatable conditions.
RNA extraction, cDNA synthesis and
real-time PCR reactions
Total RNA was isolated from the RNAlater-fixed
samples of LSCC and the corresponding NMLT samples with the use of
the RNeasy Mini Kit (Qiagen). This total RNA was transcribed to
cDNA with the High-Capacity cDNA Reverse Transcription Kit (Applied
Biosystems, Foster City, CA, USA) according to the manufacturer's
instructions. RT-qPCR was carried out in 20 μl volumes using
the TaqMan Universal PCR Master Mix on a 7900HT Fast Real-Time PCR
System (Applied Biosystems). The TaqMan-specific probes used in the
experiment (Hs00746079_s1 for SOX18 and Hs00188166_m1 for
SDHA as a reference gene) were also obtained from Applied
Biosystems. All reactions were performed in triplicates under the
following conditions: activation of polymerase at 50°C for 2 min,
initial denaturation at 94°C for 10 min followed by 40 cycles of
denaturation at 94°C for 15 sec and annealing and elongation at
60°C for 1 min. The relative mRNA expression of the studied markers
was calculated with the ΔΔCq method.
miRNA quantification using ddPCR
Small RNA fractions containing miRNAs from the
RNAlater-fixed samples of LSCC and NMLT were isolated with the use
of the mirVana miRNA Isolation kit (Ambion, Waltham, MA, USA)
according to the manufacturer's instructions. For reverse
transcription (RT-PCR), the TaqMan MicroRNA Reverse Transcription
kit was used, as well as miRNA-specific stem-loop primers (both
from Applied Biosystems), 20 primers for SOX18 (Table II) and 2 as a reference for miRNA
genes (Table III). An input of 30
ng of RNA from each sample was reversely transcribed using a C1000
Touch Thermal Cycler (Bio-Rad, Hercules, CA, USA). The miRNAs that
most probably interact with the SOX18 transcript were
selected from miRNA libraries and repositories available online:
miRBase, TargetScanHuman 6.2, miRanda and RepTar database (date of
access, 15 May 2015). The thermocycler parameters were as follows:
hold for 30 min at 16°C, for 30 min at 42°C, and for 5 min at
85°C.
 | Table IIList of TaqMan® microRNAs
used in this study. |
Table II
List of TaqMan® microRNAs
used in this study.
| miRNA
transcript | Assay ID | Mature miRNA
sequence |
|---|
| hsa-miR-7-5p | 000268 |
UGGAAGACUAGUGAUUUUGUUGU |
| hsa-miR-20a-3p | 002437 |
ACUGCAUUAUGAGCACUUAAAG |
| hsa-miR-24-3p | 000402 |
UGGCUCAGUUCAGCAGGAACAG |
| hsa-miR-202 | 002362 |
UUCCUAUGCAUAUACUUCUUUG |
| hsa-miR-335-5p | 000546 |
UCAAGAGCAAUAACGAAAAAUGU |
| hsa-miR-374a | 000563 |
UUAUAAUACAACCUGAUAAGUG |
| hsa-miR-374b | 001319 |
AUAUAAUACAACCUGCUAAGUG |
| hsa-miR-488 | 001106 |
CCCAGAUAAUGGCACUCUCAA |
| hsa-miR-499-5p | 001352 |
UUAAGACUUGCAGUGAUGUUU |
| hsa-miR-548ab | 463573_mat |
AAAAGUAAUUGUGGAUUUUGCU |
| hsa-miR-548ak | 463366_mat |
AAAAGUAACUGCGGUUUUUGA |
| hsa-miR-548i | 002909 |
AAAAGUAAUUGCGGAUUUUGCC |
| hsa-miR-764 | 241115_mat |
GCAGGUGCUCACUUGUCCUCCU |
| hsa-miR-1205 | 002778 |
UCUGCAGGGUUUGCUUUGAG |
| hsa-miR-1909 | 121123_mat |
UGAGUGCCGGUGCCUGCCCUG |
| hsa-miR-3128 | 244506_mat |
UCUGGCAAGUAAAAAACUCUCAU |
| hsa-miR-3973 | 464008_mat |
ACAAAGUACAGCAUUAGCCUUAG |
|
hsa-miR-4645-5p | 463591_mat |
ACCAGGCAAGAAAUAUUGU |
|
hsa-miR-4716-3p | 462953_mat |
AAGGGGGAAGGAAACAUGGAGA |
|
hsa-miR-4802-3p | 462011_mat |
UACAUGGAUGGAAACCUUCAAGC |
 | Table IIITaqMan® probes used as
reference genes in this study. |
Table III
TaqMan® probes used as
reference genes in this study.
| miRNA
transcript | Assay ID | Mature miRNA
sequence |
|---|
| hsa-miR-103 | 000439 |
AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-191 | 000490 |
CAACGGAAUCCCAAAAGCAGCU |
The ddPCR reaction mixtures contained: 1.33
μl of RT product, 1 μl of TaqMan miRNA-specific probe
(Life Technologies), 7.67 μl of molecular biology-grade
water and 10 μl of 2x ddPCR™ Master Mix for Probes
(Bio-Rad). A total of 20 μl of the reaction mixtures was
loaded into a plastic cartridge with 70 μl of Droplet
Generation Oil for Probes in the QX100 Droplet Generator (all from
Bio-Rad). The droplets obtained from each sample were then
transferred to a 96-well PCR plate (Eppendorf, Hamburg, Germany).
PCR amplifications were carried out in the C1000 Touch Thermal
Cycler at 95°C for 10 min, followed by 40 cycles at 95°C for 3 sec
and 60°C for 1 min, and 1 cycle at 98°C for 10 min ending at room
temperature (RT). Finally, the plate was loaded on a Droplet Reader
(Bio-Rad) and read automatically. Absolute quantification (AQ) of
each miRNA was calculated from the number of positive counts per
panel using Poisson distribution. The quantification of the target
miRNAs is presented as the number of copies/μl of the PCR
reaction mixture.
SDS-PAGE and western blot analysis
Whole cell lysates of LSCC and NMLT samples were
obtained by using the T-PER Tissue Protein Extraction kit (Thermo
Fisher Scientific, Walthman, MA, USA) with the addition of a
cocktail of inhibitors (Sigma, St. Louis, MO, USA), 250 U of
Benzonase (Merck Millipore, Bedford, MA, USA) and 2 mM
phenylmethylsulfonyl fluoride (PMSF). The lysates were mixed with
4X SDS-PAGE gel loading buffer (200 mM Tris-HCl - pH 6.8, 400 mM
DTT, 8% SDS, 0.4% bromophenol blue, 40% glycerol), loaded on 10%
acrylamide gel and separated by SDS-PAGE under reducing conditions,
and then transferred onto a PVDF membrane in the XCell SureLock™
Mini-Cell Electrophoresis System (Thermo Fisher Scientific, Santa
Clara, CA, USA). After protein transfer, the membrane was incubated
in blocker solution (4% BSA in TBST buffer) for 1 h at RT followed
by overnight incubation at 4°C with the anti-SOX18 monoclonal mouse
antibody, diluted at 1:100 (D-8, Sc-166025; Santa Cruz
Biotechnology). Next, the membrane was washed with TBST buffer and
incubated for 1 h at RT with the secondary donkey anti-mouse
antibody conjugated with HRP, diluted at 1:3,000 (709-035-149;
Jackson ImmunoResearch, Mill Valley, CA, USA), then rinsed and
treated with the Immun-Star HRP Chemiluminescent kit (Bio-Rad).
Rabbit anti-human β-actin monoclonal antibody (#4970; Cell
Signaling Technology, Inc., Danvers, MA, USA) diluted at 1:1,000
was used as an internal control. The western blotting results were
analyzed in the ChemiDoc MP System (Bio-Rad).
Statistical analysis
The Shapiro-Wilk test was used for the evaluation of
the normality assumption of the groups examined. In order to
compare the differences between the LSCC and NMLT groups, the
Wilcoxon signed-rank test was used. Additionally, the Spearman's
correlation test was carried out to analyze the existing
correlations. All the statistical analyses were performed using
Prism 5.0 (GraphPad, La Jolla, CA, USA). The results were
considered statistically significant at p<0.05.
Results
Immunohistochemistry
In total, 25 cases of LSCC were tested in this
study. There were 19 men (76%) and 6 women (24%), and the mean ± SD
age at surgery was 67.52±8.82.
In the presented results, SOX18 expression was
observed mostly in the nuclei of both cancer and endothelial cells
(Fig. 1). The nuclear localization
of the SOX18 protein was observed in 23 cases (92%), and
cytoplasmic expression was noted in 1 case (4%). The quantitation
of the IHC analysis was based on scoring for the number of
positively stained nuclei. In the case of SOX18 (nSOX18) expression
in LSCC cancer cells, a semi-quantitative scale based on tumor cell
positivity in the whole tissue section was employed. This scale is
encoded as: 0 (0% cells stained), 1 (1–10% cells stained), 2
(11–25% cells stained), 3 (26–50% cells stained) and 4 (51–100%
cells stained). We were not able to observe SOX18 protein
expression in the fibroblastic-like cells of the tumor stroma and
healthy lung tissue. By using the Spearman's correlation test,
significant correlations were observed between IRS SOX18 and
RQ-values of SOX18 in LSCC samples (r=0.43, p=0.041), IRS SOX18 and
AQ-values of miR-24-3p (r=−0.48, p=0.02), and nSOX18 and miR-7a in
the NMLT cases (r=−0.49, p=0.018).
SOX18 mRNA expression levels in LSCC and
NMLT - RT-qPCR
SOX18 mRNA expression level was determined in
21/25 cases (84%) of LSCC and in all 25 cases (100%) of NMLT. We
observed a lower expression of SOX18 in LSCC as compared to
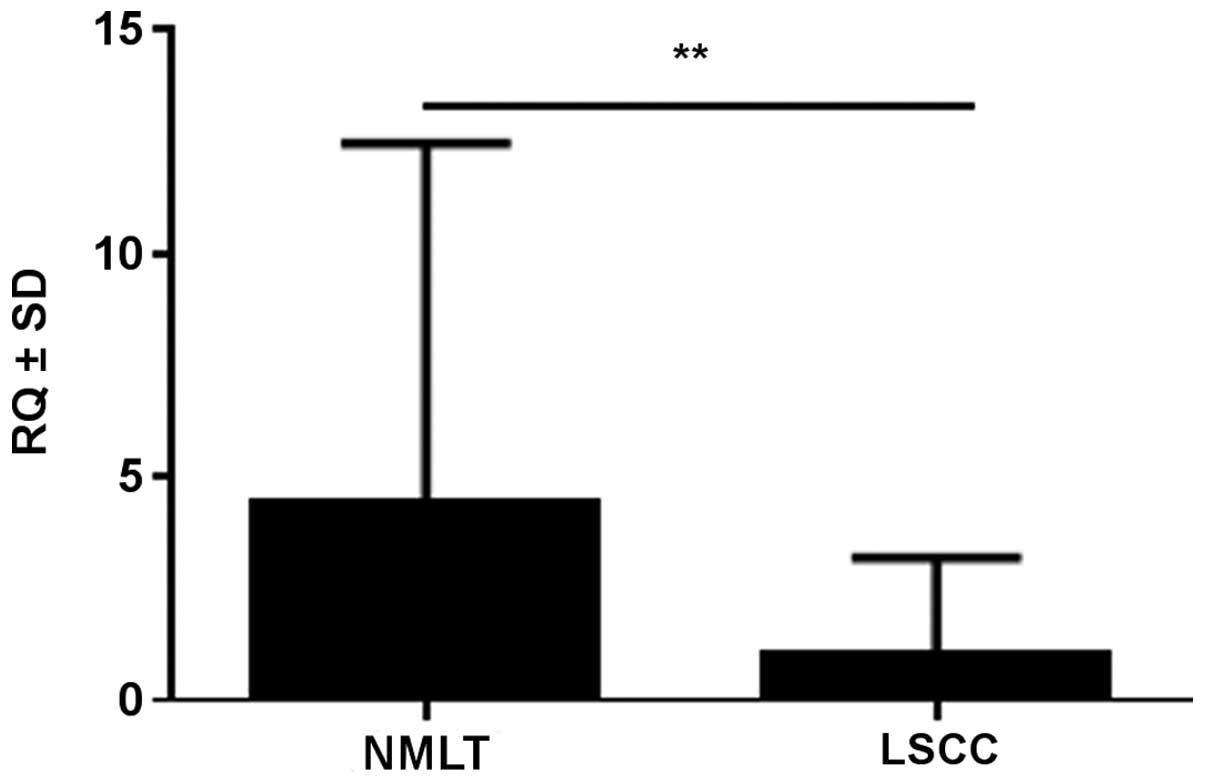
NMLT in 22 cases (88%) (mean RQ ± SD, 1.05±1.26 vs. 4.44±4.99,
respectively). The difference was statistically significant for the
analyzed pairs (p<0.01, Wilcoxon signed-rank test) (Fig. 2). SOX18 expression in NMLT
was positively correlated with SOX18 expression in the LSCC
samples (r=0.48, p=0.019; Spearman's correlation test).
SOX18 protein level - western blot
analysis
The bands of SOX18 protein were observed at 41 kDa
in the whole cell fractions of all 25 cases (100%) of LSCC and in
only 3 cases (12%) of NMLT. The expression of SOX18 protein was
significantly higher in all of the analyzed cases of LSCC compared
to that noted in the NMLT (mean OD ± SD, 9.97±6.24 vs. 0.32±1.20,
respectively; p<0.0001, Wilcoxon signed-rank test) (Fig. 3).
miRNAs expression levels – ddPCR
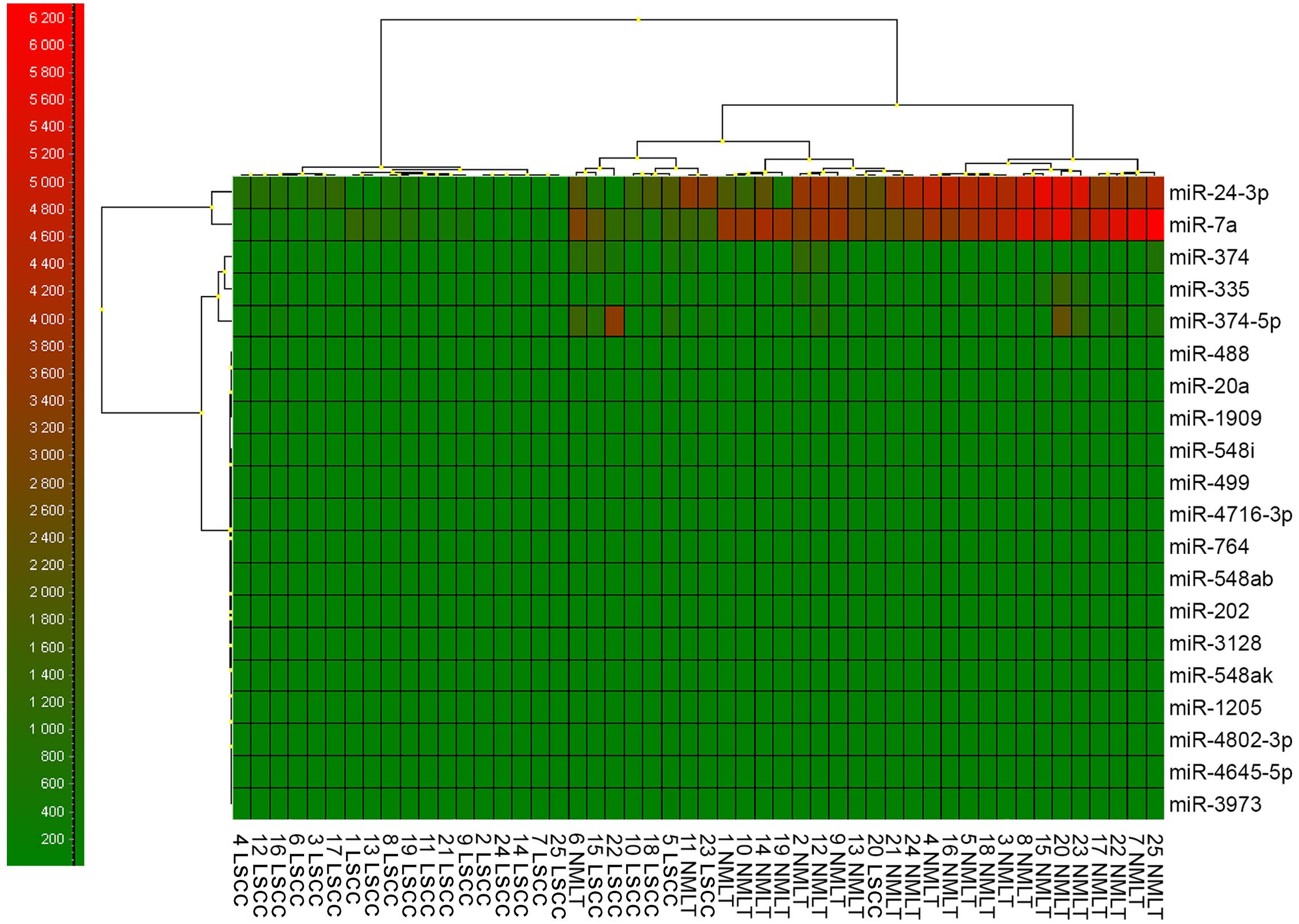
From all the 20 potentially miRNAs that could
interact with the SOX18 transcript, we could justify a closer
examination of only two of them that were variably expressed:
miR-7a and miR-24-3p (Fig. 4).
According to the ddPCR AQ method, miR-7a was significantly more
highly expressed in 23 cases (92%) of NMLT compared to LSCC (mean
AQ ± SD, 4026±1,158 vs. 658.1±670, respectively; p<0.0001,
Wilcoxon signed-rank test). The same observation was made for
miR-24-3p: there was a higher expression in 18 cases (72%) of NMLT
compared to LSCC (mean AQ ± SD, 3,674±1,304 vs. 735.9±835.7,
respectively; p<0.0001, Wilcoxon signed-rank test) (Fig. 5).
However, only one of the miRNAs used as a reference
gene showed a relatively constant and invariant expression in all
examined samples - miR-191 (5,654±764 copies/μl), as
previously described (28,32). The reference genes were not required
for analysis purposes, but helped us to ensure the quality and
relevance of the chosen samples.
Overall, both miR-7a and miR-24-3p had a
significantly higher copy number in the lung tissue samples (NMLTs)
compared to the cancer samples (LSCC). Statistically higher copy
numbers per μl of miR-7a were observed in the NMLTs rather
than in the LSCC, both for all the analyzed samples and the paired
cases. By using the Spearman's correlation test, positive
correlations were observed between AQ-values of miR-7a and
miR-24-3p in the NMLT cases (r=0.4, p=0.057), AQ-values of miR-7a
and miR-24-3p in the LSCC samples (r=0.51, p=0.012), and AQ-values
of miR-24-3p in the NMLTs and miR-24-3p in the LSCC samples (r=0.4,
p=0.017).
Discussion
The proteins encoded by SOX genes act as
transcription factors in cells mostly at the embryonic stage of
development. SOX proteins can be found in many tissues at different
stages of development, fulfilling important functions in a variety
of processes occurring in the body, such as embryonic development
and disease processes - atherosclerosis or carcinogenesis (33). In recent years, their role in tumors
has been intensively studied, as a result of which it has been
possible to demonstrate the participation of these transcription
factors in the pathogenesis of many malignant tumors (34).
In previous study we demonstrated that the
cytoplasmic expression of the SOX18 protein could be a new
prognostic marker in NSCLC patients and that it plays a possible
role in the regulation of lung cancer cell proliferation (3). The molecular mechanisms that explain
the observed disparity between the mRNA and the protein levels of
SOX18 have not been fully discovered yet. Previous observations by
Azhikina et al and Dammann et al considered the
methylation of promotors as a mechanism of regulation of variable
genes in NSCLC (35,36). Although the hypermethylation of
promotors in lung carcinomas can be observed quite often, there is
also some evidence for the role of miRNAs in lung cancer pathology
(23,24,27,28).
Balakrishnan et al identified two miRNA molecules that
interact with the mRNA of the SOX18 gene (37). In the present study, we aimed to
identify the miRNAs that are responsible for the observed disparity
between SOX18 mRNA and the protein level in NMLT and LSCC
cells.
The results obtained with the RT-qPCR technique
showed a statistically higher expression level of SOX18 mRNA
in NMLTs compared to the corresponding LSCC samples. Moreover, we
observed a statistically higher expression level of the SOX18
protein in LSCC samples compared to the NMLTs.
In addition to the involvement of the SOX18
transcription factor in a series of embryonic development
processes, its expression has also been shown in cells of many
organs, such as the heart, the lungs, the skeletal muscles, the
stomach or the jejunum, in mature organisms (10,38).
In this study, the RT-qPCR data showed that the SOX18 mRNA
level was at an approximately average level in the mature lung
tissues, but that in almost all cases of NMLT (88%) there was a
lack of its protein. We postulated that miR-24-3p together with
miR-7a could play a role in the mechanism of SOX18
transcript inhibition. It is very probable that after the embryonic
development of lung tissue, the SOX18 gene product could be
inhibited or even degraded via miRNAs. Their role in blood vessel
development, vascular adaptations and arterial occlusions in normal
and tumor tissues has already been firmly confirmed (39,40).
To date, many studies have demonstrated that members of the
SOX gene family play important roles in the development and
maintenance of the lung (5).
Moreover, the expression of SOX2 in neural progenitor cells (NPCs)
is proven to be controlled by miRNAs (41), as well as SOX4 and SOX15 in cancers
(42). It has been also confirmed
that miR-124 downregulates SOX8 expression and suppresses
cell proliferation in NSCLC (43).
Up until now, there have been many reports that
strengthen the role of a variety of miRNAs in lung tissue and the
epigenetics of lung cancers (28,29,31,44–46).
Li et al showed that miR-7a was suppressed in NSCLC cells,
and B-cell lymphoma 2 (BCL-2) protein was identified as a possible
target (47). Furthermore,
miR-24-3p was also found to be significantly downregulated in NSCLC
samples, where it regulates the autophagy process. Since miRNAs
interact with many different mRNA targets, it is not surprising
that miR-7a and miR-24-3p could also bind to the SOX18
transcript. Therefore, we propose that miR-7a together with
miR-24-3p can act as major factors that control SOX18
expression in LSCC.
The results presented in our study correspond to
those of Balakrishnan et al, where miR-7a and miR-24-3p were
confirmed to interact with the SOX18 transcript (37). In their study, they used
immunoprecipitation as a technique to confirm the binding
properties of those miRNAs with the SOX18 mRNA.
It has been firmly confirmed that miRNAs can be
downregulated or upregulated during the development of lung
cancers. On the one hand, most of the miRNAs that are
down-regulated are essential to inhibit the growth and survival of
tumor cells (23). On the other
hand, the genes that are upregulated by miRNAs are essential for
cell adhesion, mobility and development. Tavazoie et al
demonstrated that miR-335, also examined in our study, regulated
metastasis and invasion through the suppression of the SOX4 gene in
the breast cancer cell line MDA-MB-231 (48). Our data do not confirm these
properties for miR-335 in the case of NMLT and LSCC, but
SOX18 suppression is most probably caused by miR-7a and
miR-24-3p molecules.
In the present study, we have, most probably, a
situation where the cancer cells successfully downregulate miR-7a
and miR-24-3p, which leads to higher expression of the SOX18
protein. The mechanism involved in these modulations is not fully
understood and requires further analysis, but we postulate that the
downregulation of these miRNAs is due to two different models:
chromatin remodeling or natural antagomiRs (anti-miRs). Although
the chromatin remodeling process modulates the expression of miRNAs
in dendritic cells, it can only be hypothesized that miRNAs, in
particular miR-7a, are also downregulated via this mechanism in
LSCC (49). We believe that miR-7a
and miR-24-3p expression in LSCC and other types of lung cancer is
effectively modulated via natural anti-miRs. Until recently,
anti-miRs were considered as artificial particles that could be
used as a new and highly specific weapon against pro-oncogenic
miRNAs in many diseases (50–55),
but the latest studies discovered natural antisense transcripts
(NATs), which are natural endogenous RNA molecules transcribed from
the opposite strand of other protein- or non-protein-coding genes
(56). The mechanisms by which NATs
regulate gene expression are highly incomprehensible. Faghihi et
al proved that in Alzheimer's disease the β-secretase 1 (BACE1)
protein expression is modulated via the competition of miR-485-5p
and BACE1-AS (NATs) for a binding site in the exon region of BACE1
mRNA (56). The opposing effects of
BACE1-antisense and miR-485-5p on BACE1 protein were proven in
vitro. They also demonstrated that the expression of both
BACE1-antisense and miR-485-5p are dysregulated in RNA samples from
Alzheimer's disease subjects compared to control individuals.
It has not been verified yet whether the same
suppression model takes place in lung cancer pathology, especially
in regards to SOX18 and its role in cancer angiogenesis. Yet,
results from ddPCR (data not shown) indicate that, in LSCC samples,
we can observe populations of unspecific products that compete with
miR-7a and miR-24-3p for SOX18 mRNA binding sites. Those
ʻunspecific productsʼ could be similar to those NATs that were
discovered by Faghihi et al, but further analyses are
required to prove this theory (56).
In conclusion, our data, along with our previous
findings, indicate that the disparity between the mRNA and protein
levels of the SOX18 transcription factor in NSCLC and NMLT could be
caused by the abilities of miR-7a and/or miR-24-3p to bind to the
transcript. The proper mechanism via which it is carried out
remains unknown and will be our next research goal. Yet, it is most
probable that NATs are involved in the suppression of these miRNAs
allowing cancer cells to express SOX18 protein. The presence of
SOX18 is highly desirable for cancer cells mostly due to its role
in angiogenesis and the intensification of the metastasis process.
However, further studies are required in order to fully understand
the role of SOX18 in cancer development and progression.
Acknowledgments
The present study was supported by a research grant
from the WROVASC-Integrated Cardiovascular Centre project, and
co-financed by the European Regional Development Fund within the
Innovative Economy Operational Program, 2007–2013, realized at the
Research and Development Centre of the Provincial Specialist
Hospital in Wroclaw, Poland. Additionally, the authors would like
to thank Dr Adam Rzechonek and Dr Maciej Majchrzak for their
support.
References
|
1
|
Brandao GD, Brega EF and Spatz A: The role
of molecular pathology in non-small-cell lung carcinoma-now and in
the future. Curr Oncol. 19(Suppl 1): S24–S32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Malvezzi M, Bertuccio P, Levi F, La
Vecchia C and Negri E: European cancer mortality predictions for
the year 2014. Ann Oncol. 25:1650–1656. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jethon A, Pula B, Olbromski M, Werynska B,
Muszczynska- Bernhard B, Witkiewicz W, Dziegiel P and
Podhorska-Okolow M: Prognostic significance of SOX18 expression in
non-small cell lung cancer. Int J Oncol. 46:123–132. 2015.
|
|
4
|
Gubbay J, Collignon J, Koopman P, Capel B,
Economou A, Münsterberg A, Vivian N, Goodfellow P and Lovell-Badge
R: A gene mapping to the sex-determining region of the mouse Y
chromosome is a member of a novel family of embryonically expressed
genes. Nature. 346:245–250. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu Y, Li Y, Jun Wei JW and Liu X: The
role of Sox genes in lung morphogenesis and cancer. Int J Mol Sci.
13:15767–15783. 2012. View Article : Google Scholar
|
|
6
|
Wegner M: From head to toes: The multiple
facets of Sox proteins. Nucleic Acids Res. 27:1409–1420. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bowles J, Schepers G and Koopman P:
Phylogeny of the SOX family of developmental transcription factors
based on sequence and structural indicators. Dev Biol. 227:239–255.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taniguchi K, Hiraoka Y, Ogawa M, Sakai Y,
Kido S and Aiso S: Isolation and characterization of a mouse
SRY-related cDNA, mSox7. Biochim Biophys Acta. 1445:225–231. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanai Y, Kanai-Azuma M, Noce T, Saido TC,
Shiroishi T, Hayashi Y and Yazaki K: Identification of two Sox17
messenger RNA isoforms, with and without the high mobility group
box region, and their differential expression in mouse
spermato-genesis. J Cell Biol. 133:667–681. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dunn TL, Mynett-Johnson L, Wright EM,
Hosking BM, Koopman PA and Muscat GE: Sequence and expression of
Sox-18 encoding a new HMG-box transcription factor. Gene.
161:223–225. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cermenati S, Moleri S, Cimbro S, Corti P,
Del Giacco L, Amodeo R, Dejana E, Koopman P, Cotelli F and Beltrame
M: Sox18 and Sox7 play redundant roles in vascular development.
Blood. 111:2657–2666. 2008. View Article : Google Scholar
|
|
12
|
Cermenati S, Moleri S, Neyt C, Bresciani
E, Carra S, Grassini DR, Omini A, Goi M, Cotelli F, François M, et
al: Sox18 genetically interacts with VegfC to regulate
lymphangiogenesis in zebrafish. Arterioscler Thromb Vasc Biol.
33:1238–1247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Downes M, François M, Ferguson C, Parton
RG and Koopman P: Vascular defects in a mouse model of
hypotrichosis-lymphedema-telangiectasia syndrome indicate a role
for SOX18 in blood vessel maturation. Hum Mol Genet. 18:2839–2850.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
François M, Caprini A, Hosking B, Orsenigo
F, Wilhelm D, Browne C, Paavonen K, Karnezis T, Shayan R, Downes M,
et al: Sox18 induces development of the lymphatic vasculature in
mice. Nature. 456:643–647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pendeville H, Winandy M, Manfroid I,
Nivelles O, Motte P, Pasque V, Peers B, Struman I, Martial JA and
Voz ML: Zebrafish Sox7 and Sox18 function together to control
arterial-venous identity. Dev Biol. 317:405–416. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pula B, Olbromski M, Wojnar A,
Gomulkiewicz A, Witkiewicz W, Ugorski M, Dziegiel P and
Podhorska-Okolow M: Impact of SOX18 expression in cancer cells and
vessels on the outcome of invasive ductal breast carcinoma. Cell
Oncol (Dordr). 36:469–483. 2013. View Article : Google Scholar
|
|
17
|
Young N, Hahn CN, Poh A, Dong C, Wilhelm
D, Olsson J, Muscat GE, Parsons P, Gamble JR and Koopman P: Effect
of disrupted SOX18 transcription factor function on tumor growth,
vascularization, and endothelial development. J Natl Cancer Inst.
98:1060–1067. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Duong T, Proulx ST, Luciani P, Leroux JC,
Detmar M, Koopman P and Francois M: Genetic ablation of SOX18
function suppresses tumor lymphangiogenesis and metastasis of
melanoma in mice. Cancer Res. 72:3105–3114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang G, Wei Z, Jia H, Zhao W, Yang G and
Zhao H: Knockdown of SOX18 inhibits the proliferation, migration
and invasion of hepatocellular carcinoma cells. Oncol Rep.
34:1121–1128. 2015.PubMed/NCBI
|
|
20
|
Petrovic I, Milivojevic M, Popovic J,
Schwirtlich M, Rankovic B and Stevanovic M: SOX18 is a novel target
gene of Hedgehog signaling in cervical carcinoma cell line. PLoS
One. 10:e01435912015. View Article : Google Scholar
|
|
21
|
Zhang J, Ma Y, Wang S, Chen F and Gu Y:
Suppression of SOX18 by siRNA inhibits cell growth and invasion of
breast cancer cells. Oncol Rep. 35:3721–3727. 2016.PubMed/NCBI
|
|
22
|
Majid S, Dar AA, Saini S, Shahryari V,
Arora S, Zaman MS, Chang I, Yamamura S, Tanaka Y, Chiyomaru T, et
al: miRNA-34b inhibits prostate cancer through demethylation,
active chromatin modifications, and AKT pathways. Clin Cancer Res.
19:73–84. 2013. View Article : Google Scholar
|
|
23
|
Devaraj S and Natarajan J: miRNA-mRNA
network detects hub mRNAs and cancer specific miRNAs in lung
cancer. In Silico Biol. 11:281–295. 2012.PubMed/NCBI
|
|
24
|
Lin PY and Yang PC: Circulating miRNA
signature for early diagnosis of lung cancer. EMBO Mol Med.
3:436–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Markou A, Sourvinou I, Vorkas PA, Yousef
GM and Lianidou E: Clinical evaluation of microRNA expression
profiling in non small cell lung cancer. Lung Cancer. 81:388–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang C, Hu X, Alattar M and Zhao H: miRNA
expression profiles associated with diagnosis and prognosis in lung
cancer. Expert Rev Anticancer Ther. 14:453–461. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lv J and Xu L, Xu Y, Qiu M, Yang X, Wang
J, Yin R and Xu L: Expression of miRNA-221 in non-small cell lung
cancer tissues and correlation with prognosis. Zhongguo Fei Ai Za
Zhi. 17:221–225. 2014.In Chinese. PubMed/NCBI
|
|
29
|
Mairinger FD, Ting S, Werner R, Walter RF,
Hager T, Vollbrecht C, Christoph D, Worm K, Mairinger T,
Sheu-Grabellus SY, et al: Different micro-RNA expression profiles
distinguish subtypes of neuroendocrine tumors of the lung: Results
of a profiling study. Mod Pathol. 27:1632–1640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Salim H, Arvanitis A, de Petris L, Kanter
L, Hååg P, Zovko A, Özata DM, Lui WO, Lundholm L, Zhivotovsky B, et
al: miRNA-214 is related to invasiveness of human non-small cell
lung cancer and directly regulates alpha protein kinase 2
expression. Genes Chromosomes Cancer. 52:895–911. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lang Y, Xu S, Ma J, Wu J, Jin S, Cao S and
Yu Y: MicroRNA-429 induces tumorigenesis of human non-small cell
lung cancer cells and targets multiple tumor suppressor genes.
Biochem Biophys Res Commun. 450:154–159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lovell-Badge R: The early history of the
Sox genes. Int J Biochem Cell Biol. 42:378–380. 2010. View Article : Google Scholar
|
|
34
|
Castillo SD and Sanchez-Cespedes M: The
SOX family of genes in cancer development: Biological relevance and
opportunities for therapy. Expert Opin Ther Targets. 16:903–919.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Azhikina T, Kozlova A, Skvortsov T and
Sverdlov E: Heterogeneity and degree of TIMP4, GATA4, SOX18, and
EGFL7 gene promoter methylation in non-small cell lung cancer and
surrounding tissues. Cancer Genet. 204:492–500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dammann R, Strunnikova M, Schagdarsurengin
U, Rastetter M, Papritz M, Hattenhorst UE, Hofmann HS, Silber RE,
Burdach S and Hansen G: CpG island methylation and expression of
tumour-associated genes in lung carcinoma. Eur J Cancer.
41:1223–1236. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Balakrishnan I, Yang X, Brown J,
Ramakrishnan A, Torok-Storb B, Kabos P, Hesselberth JR and Pillai
MM: Genome-wide analysis of miRNA-mRNA interactions in marrow
stromal cells. Stem Cells. 32:662–673. 2014. View Article : Google Scholar :
|
|
38
|
Crémazy F, Berta P and Girard F:
Genome-wide analysis of Sox genes in Drosophila melanogaster. Mech
Dev. 109:371–375. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu D, Krueger J and Le Noble F: The role
of blood flow and microRNAs in blood vessel development. Int J Dev
Biol. 55:419–429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sen CK, Gordillo GM, Khanna S and Roy S:
Micromanaging vascular biology: Tiny microRNAs play big band. J
Vasc Res. 46:527–540. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sarkar A and Hochedlinger K: The sox
family of transcription factors: Versatile regulators of stem and
progenitor cell fate. Cell Stem Cell. 12:15–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thu KL, Becker-Santos DD, Radulovich N,
Pikor LA, Lam WL and Tsao MS: SOX15 and other SOX family members
are important mediators of tumorigenesis in multiple cancer types.
Oncoscience. 1:326–335. 2014. View Article : Google Scholar
|
|
43
|
Xie C, Han Y, Liu Y, Han L and Liu J:
miRNA-124 down-regulates SOX8 expression and suppresses cell
proliferation in non-small cell lung cancer. Int J Clin Exp Pathol.
7:7518–7526. 2014.
|
|
44
|
Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F,
Bao G, Kong H, Ge C, Zhang F, et al: miRNA-200c inhibits invasion
and metastasis of human non-small cell lung cancer by directly
targeting ubiquitin specific peptidase 25. Mol Cancer. 13:1662014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Salim H, Akbar NS, Zong D, Vaculova AH,
Lewensohn R, Moshfegh A, Viktorsson K and Zhivotovsky B: miRNA-214
modulates radiotherapy response of non-small cell lung cancer cells
through regulation of p38MAPK, apoptosis and senescence. Br J
Cancer. 107:1361–1373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang C, Ge S, Hu C, Yang N and Zhang J:
MiRNA-218, a new regulator of HMGB1, suppresses cell migration and
invasion in non-small cell lung cancer. Acta Biochim Biophys Sin
(Shanghai). 45:1055–1061. 2013. View Article : Google Scholar
|
|
47
|
Li J, Zheng Y, Sun G and Xiong S:
Restoration of miR-7 expression suppresses the growth of Lewis lung
cancer cells by modulating epidermal growth factor receptor
signaling. Oncol Rep. 32:2511–2516. 2014.PubMed/NCBI
|
|
48
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mei S, Liu Y, Bao Y, Zhang Y, Min S, Liu
Y, Huang Y, Yuan X, Feng Y, Shi J, et al: Dendritic cell-associated
miRNAs are modulated via chromatin remodeling in response to
different environments. PLoS One. 9:e902312014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Brock M, Samillan VJ, Trenkmann M,
Schwarzwald C, Ulrich S, Gay RE, Gassmann M, Ostergaard L, Gay S,
Speich R, et al: AntagomiR directed against miR-20a restores
functional BMPR2 signalling and prevents vascular remodelling in
hypoxia-induced pulmonary hypertension. Eur Heart J. 35:3203–3211.
2014. View Article : Google Scholar
|
|
51
|
Li J, Bai H, Zhu Y, Wang XY, Wang F, Zhang
JW, Lavker RM and Yu J: Antagomir dependent microRNA-205 reduction
enhances adhesion ability of human corneal epithelial
keratinocytes. Chin Med Sci J. 25:65–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu D, Huang Y, Jia C, Li Y, Liang F and
Fu Q: Administration of antagomir-223 inhibits apoptosis, promotes
angiogenesis and functional recovery in rats with spinal cord
injury. Cell Mol Neurobiol. 35:483–491. 2015. View Article : Google Scholar
|
|
53
|
Selvamani A, Sathyan P, Miranda RC and
Sohrabji F: An antagomir to microRNA Let7f promotes neuroprotection
in an ischemic stroke model. PLoS One. 7:e326622012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Song MS and Rossi JJ: The anti-miR21
antagomir, a therapeutic tool for colorectal cancer, has a
potential synergistic effect by perturbing an
angiogenesis-associated miR30. Front Genet. 4:3012014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sun B, Yang N, Jiang Y, Zhang H, Hou C, Ji
C, Liu Y and Zuo P: Antagomir-1290 suppresses CD133+
cells in non-small cell lung cancer by targeting fyn-related Src
family tyrosine kinase. Tumour Biol. 36:6223–6230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Faghihi MA, Zhang M, Huang J, Modarresi F,
Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G III and
Wahlestedt C: Evidence for natural antisense transcript-mediated
inhibition of microRNA function. Genome Biol. 11:R562010.
View Article : Google Scholar : PubMed/NCBI
|