Introduction
Breast cancer is the leading cause of cancer-related
deaths in women worldwide, with ~1.38 million cases newly diagnosed
and 458,000 deaths in 2008 alone (1,2).
According to the histopathological classification of breast
carcinoma, 70–80% of the all breast cancers will eventually belong
to invasive ductal carcinomas (IDCs) or invasive lobular carcinoma
(ILC) (3). There have been great
advances in uncovering the biological mechanisms of breast cancer
progression. However, many details of the genetic causes have yet
to be determined.
Cyclins are a family of proteins that control cell
cycle progression by regulating the activity of various
cyclin-dependent kinases (CDKs) (4,5).
Cyclin A1 was discovered to be highly expressed in primary breast
cancer tissues and metastatic lesions, which leads to a significant
increase in cancer cell growth and angiogenesis (6). Co-expression of cyclin D1 and p21
proteins is required for the initial steps of breast cancer
development (7). Moreover,
inhibition of cyclin D1 or its binding partners CDK4/6 can increase
or decrease migration and stem-like cell activity in ER-negative
and ER-positive breast cancer, respectively, showing estrogen
receptor-dependent divergent functions (8). Cyclin D1 has also been reported to be
associated with a poor prognosis for breast cancer patients
(9,10). High cyclin E expression is common in
hormone receptor negative and high grade aggressive breast cancer
(11–13). A meta-analysis has shown that cyclin
E over-expression is associated with poor overall survival and
breast cancer-specific survival (14).
Cyclin Y is a PFTK1 interacting protein newly
identified using a yeast two-hybrid screen (15). Human PFTK1 protein has been
characterized as a Cdc2-related kinase that controls cell cycle
progression and is highly expressed in brain, pancreas and kidney
(16). Binding of cyclin Y to PFTK1
not only enhances the PFTK1 kinase activity but also changes its
intracellular location. Recently, cyclin Y was found to be
overexpressed in human non-small cell lung cancer (NSCLC) and is
associated with NSCLC proliferation and tumorigenesis (5). However, the role of cyclin Y in breast
cancer remains elusive. The present study found that cyclin Y is
highly expressed in human breast cancer specimens. To investigate
the physiological function of cyclin Y in breast cancer, we applied
gene knockdown using siRNA as an excellent tool to suppress cyclin
Y expression in two types of breast cancer cells, MDA-MB-231 and
MCF-7. The effects of cyclin Y on cell growth were also examined in
two breast cancer cell lines.
Materials and methods
Immunohistochemical staining
Eighty cases of breast cancer tissues including 65
cases of breast cancer and 15 cases of non-tumor tissues were
collected from the Department of Clinical Laboratory, Nanjing
Medical University Cancer Hospital and Jiangsu Cancer Hospital from
2008 to 2010. The clinical staging of breast cancer was evaluated
by the tumor-node-metastasis (TNM) staging systems. The samples
were used with the written informed consent from patient and
approval of the Ethics Committee of Nanjing Medical University
Cancer Hospital and Jiangsu Cancer Hospital. All tissue samples
were paraffin-embedded, dewaxed and rehydrated. The sections were
then microwaved for antigen rerieval. For immunohistochemical
staining, slides were treated with hydrogen peroxide
(H2O2) for 10 min, washed with water and
placed in phosphate-buffered saline (PBS) buffer. Anti-cyclin Y
(1:150; #ab114086; Abcam) was then applied for incubation at room
temperature. Biotinylated goat anti-rabbit IgG was used as the
secondary antibody. The immunoreactions were detected by staining
with 3,3′-diaminobenzidine (DAB). All stained slides were evaluated
under a light microscope. In each sample section, at least 5 visual
field areas were examined. The proportion of positive tumor cells
was recorded according to the following classification: 0, no cells
stained; 1, <30% of cells stained; 2, 30–60% of cells stained
and 3, >60% of cells stained. The intensity of the coloring was
recorded according to the following classification: 0, no coloring;
1, stramineous; 2, buffy; and 3, dark brown. The two scores were
combined to obtain the final one: scores equal to 0 indicate
negative (−), 1–2 indicate slightly positive (+), 3–4 indicate
moderately positive (++), and 5–6 indicate strongly positive
(+++).
Cell culture
Human breast cancer cell lines BT-474, MDA-MB-231,
T-47D and MCF-7 and human embryonic kidney cell line 293T were
obtained from the Cell Bank of Shanghai Institute of Cell Biology,
Chinese Academy of Sciences (Shanghai, China). BT-474 cells were
cultured in RPMI-1640 medium (HyClone) supplemented with 10% fetal
bovine serum (FBS) in 5% CO2 at 37°C. MDA-MB-231, T-47D
and 293T cells were grown in Dulbecco's modified Eagle's medium
(DMEM; HyClone) supplemented with 10% FBS in 5% CO2 at
37°C. MCF-7 cells were cultured in DMEM supplemented with 10% FBS,
1% sodium pyruvate and 0.01 mg/ml bovine insulin in 5%
CO2 at 37°C.
Constructions of lentiviruses
A short hairpin RNA (shRNA) sequence
(CCGGCAGGACAAATAGCAAGGAAATCTC GAGATTTCCTTGCTATTTGTCCTGTTTTTTG) was
designed for human cyclin Y gene (NM_145012.3). The non-silencing
siRNA sequence (TTCTCCGAACGTGTCACGT) was used as control. The
stem-loop-stem oligos (shRNAs) were ligated into the pFH-L vector
containing a GFP reporter (Shanghai Hollybio, China). 293T cells
were transfected with pFH-L-cyclin Y shRNA or control shRNA along
with two helper plasmids pVSVG-I and pCMV∆R8.92 (Shanghai Hollybio,
China) using Lipofectamine 2000 (Invitrogen) according to the
manufacturer's instructions. Lentiviruses were harvested 72 h after
transfection. The lentiviruses were purified using
ultracentrifugation, and the titer of the lentiviruses was
determined. MDA-MB-231 (5×104 cells/well) and MCF-7
(5×104 cells/well) cells were cultured in 6-well plates
and infected with the lentivirus constructs at a multiplicity of
infection (MOI) of 40 and 30, respectively. The transfection
efficiency was determined by fluorescence microscopy 96 h after
infection.
Quantitative real-time PCR
After infection for 96 h, MDA-MB-231 and MCF-7 cells
were collected for RNA extraction by TRIzol (Invitrogen, Carlsbad,
CA, USA). Total RNA (2 µg) was reverse transcribed using an
M-MLV reverse transcriptase kit (Promega) according to the
manufacturer's protocol. In quantitative real-time PCR, two sets of
primers were applied: cyclin Y sense, 5′-GTCAGTCAACCAAACCTCAAG-3′
and antisense, 5′-AACAGTGTCCGAACGAACC-3′; β-actin sense,
5′-GTGGACATCCGCAAAGAC-3′ and antisense, 5′-AAAGGGTGTAACGCAACTA-3′.
Relative expression levels of cyclin Y mRNA were calculated by
normalizing to the level of β-actin mRNA using comparative
threshold cycle method, in which the fold difference = 2 − (Δct of
target gene − Δct of reference). Each sample was performed in
triplicate. All results were analyzed with LightCycler software
version 3.5 (Roche Diagnostics).
MTT assay
After infection with recombined lentiviruses
(Lv-shCCNY and Lv-shCon), MDA-MB-231 (2×103 cells/well)
or MCF-7 (2×103 cells/well) cells were reseeded into
96-well plates, and were collected at 1-day intervals to perform
the methylthiazol tetrazolium (MTT) proliferation assay. In brief,
10 µl of MTT solution (5 mg/ml; Sigma) was added into each
well and incubated at 37°C for 4 h. Acidic isopropanol (10% SDS, 5%
isopropanol and 0.01 mol/l HCl) was then added to dissolve the
crystals. After 10 min, the absorbance of each sample was recorded
at 595 nm.
Colony formation assay
MDA-MB-231 (400 cells/well) or MCF-7 (200
cells/well) cells were reseeded in 6-well plates after lentivirus
infection. The medium was changed at three-day intervals. After 6
days of culture for MDA-MB-231 cells and 8 days of culture for
MCF-7 cells, the colonies formed were washed with PBS and fixed
with 4% paraformaldehyde for 30 min at room temperature. The fixed
cell samples were stained with crystals violet for 10 min. The
total number of colonies (>50 cells/colony) was counted.
Flow cytometric analysis
Fluorescence dye propidium iodide (PI) (Sigma) was
used to analyze the DNA contents in different cell cycle phases.
MDA-MB-231 (2×105 cells/well) cells infected with
Lv-shcyclin Y and Lv-shCon were reseeded into 6-cm dishes and
cultured for 40 h and harvested after trypsinization, washed with
PBS and fixed with 70% cold ethanol. The fixed cells were pelleted,
re-suspended in PBS containing PI (100 µg/ml) and RNase A
(10 µg/ml) for at least 30 min in the dark. The percentages
of cells in G0/G1, S and G2/M phases were determined by FACSCalibur
(BD Biosciences, USA).
Intracellular signaling assay
Phosphorylation and proteolysis are two widespread
covalent post-translational modifications. Detection of these
modifications on a set of cellular proteins that play a
well-understood role in cell biology can provide a broad snapshot
of intracellular signaling. The alteration of signaling molecules
in MDA-MB-231 cells was detected by PathScan®
intracellular signaling array kit (#7323; Cell Signaling
Technology) according to the protocol provided by CST.
Statistical analysis
The results of immunohistochemical staining were
evaluated by Pearson's χ2 test and the other data were
evaluated by Student's t-test and expressed as the mean ± SD of
three independent experiments. A p-value of <0.05 was considered
to indicate a statistically significant result.
Results
Cyclin Y expression in human breast
cancer and normal breast tissues
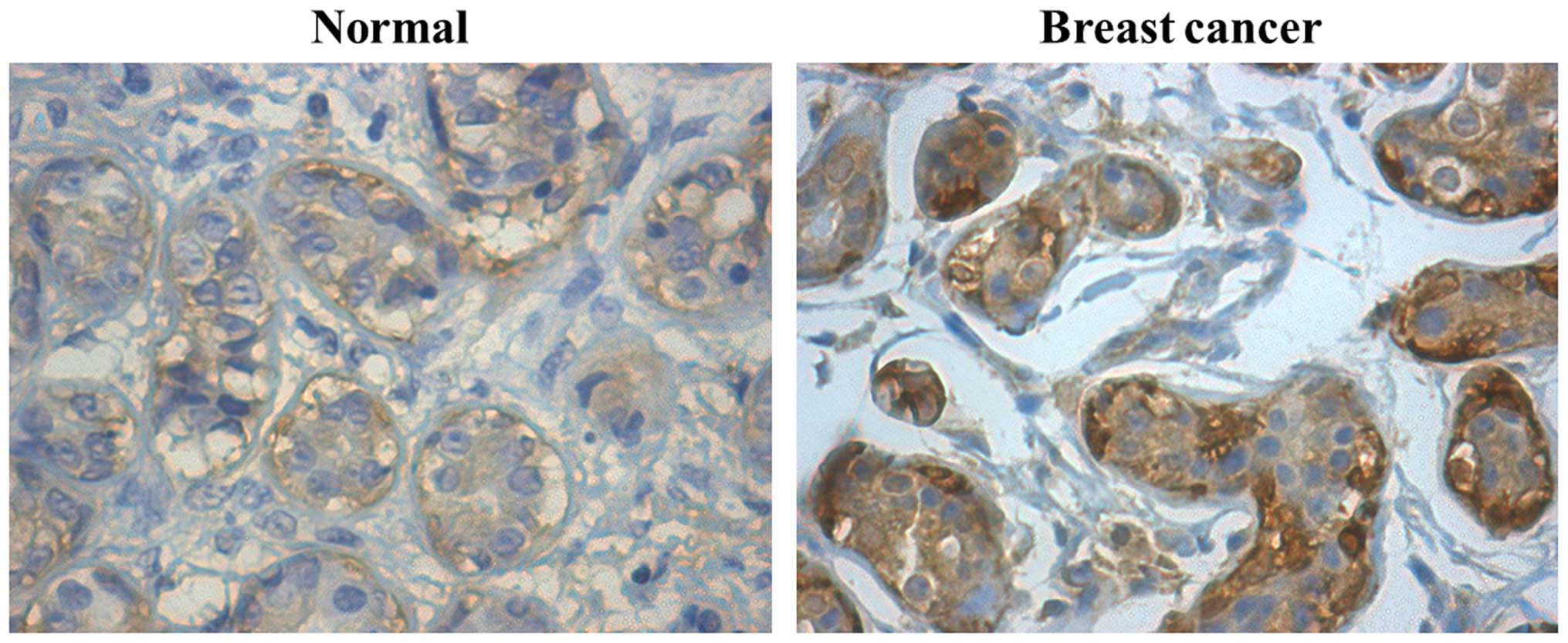
The expression patterns of cyclin Y protein in 65
stage I-III breast cancer and 15 normal breast tissues were
analyzed by immunohistochemistry. Representative
immunohistochemical staining is shown in Fig. 1. The rate of strong cyclin Y
expression (+++) in breast cancer tissues was significantly higher
than that in normal breast tissues (Table I; p<0.05; χ2 test).
Moreover, the expression of cyclin Y in breast cancer was
associated with lymph node metastasis (Table II; p<0.001; χ2 test).
These results suggest that the high cyclin Y expression may
contribute to breast cancer development and progression.
 | Table IExpression of cyclin Y in normal
breast and breast cancer tissues (n=80). |
Table I
Expression of cyclin Y in normal
breast and breast cancer tissues (n=80).
| Characteristics | Total | Cyclin Y
immunostaining
| P-valuea |
|---|
| − (%) | + (%) | ++ (%) | +++ (%) |
|---|
| Normal breast | 15 | 0 (0.0) | 4 (26.7) | 10 (66.7) | 1 (6.7) | 0.037 |
| Breast cancer | 65 | 0 (0.0) | 12 (18.5) | 26 (40.0) | 27 (41.5) | |
 | Table IIRelationship of cyclin Y expression
and clinicopathological parameters in breast cancer patients
(n=65). |
Table II
Relationship of cyclin Y expression
and clinicopathological parameters in breast cancer patients
(n=65).
| Cyclin Y
immunostaining
| |
|---|
| Characteristic | − | + | ++ | +++ | P-valuea |
|---|
| Stage | | | | | 0.458 |
| I | 0 | 0 | 1 | 2 | |
| II | 0 | 12 | 23 | 21 | |
| III | 0 | 0 | 2 | 4 | |
| Invasion depth | | | | | 0.139 |
| I | 0 | 0 | 1 | 0 | |
| II | 0 | 3 | 3 | 13 | |
| III | 0 | 9 | 8 | 11 | |
| Lymph node
metastasis | | | | | 0.001 |
| No | 0 | 1 | 17 | 20 | |
| Yes | 0 | 11 | 9 | 5 | |
Inhibition of cyclin Y expression by RNA
interference
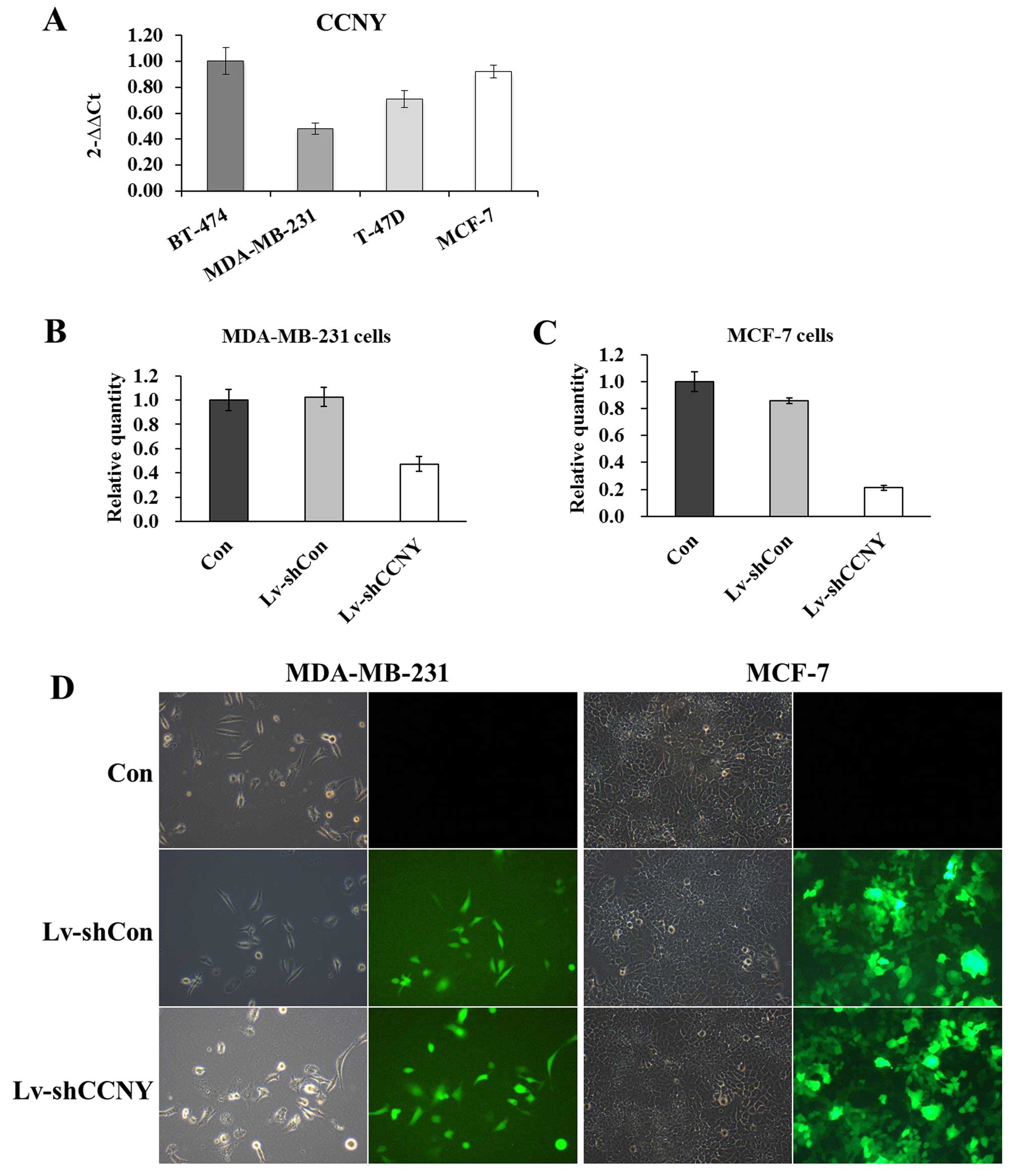
The expression levels of cyclin Y in four human
breast cancer cell lines were analyzed by RT-qPCR (Fig. 2A). The MCF-7 (ER-positive) cells
with high cyclin Y expression and MDA-MB-231 (ER-negative) cells
with low cyclin Y expression were chosen for further investigation.
The cells were treated with Lv-shcyclin Y and Lv-shCon. To
determine the transfection efficiency, fluorescent cells were
examined and photographed. As shown in Fig. 2D, >80% of cells were
GFP-positive, indicating that both MCF-7 and MDA-MB-231 cells were
successfully transfected. The mRNA levels of cyclin Y were then
measured to assess the knockdown efficiency of Lv-shcyclin Y. As
compared to Lv-shCon, the Lv-shcyclin Y conferred ~60% knockdown
efficiency in both MDA-MB-231 (Fig.
2B) and MCF-7 cells (Fig. 2C).
The results indicated that the lentivirus constructs were able to
efficiently suppress cyclin Y expression in both MDA-MB-231 and
MCF-7 cells.
Cyclin Y knockdown suppresses
proliferation of MDA-MB-231 and MCF-7 cells
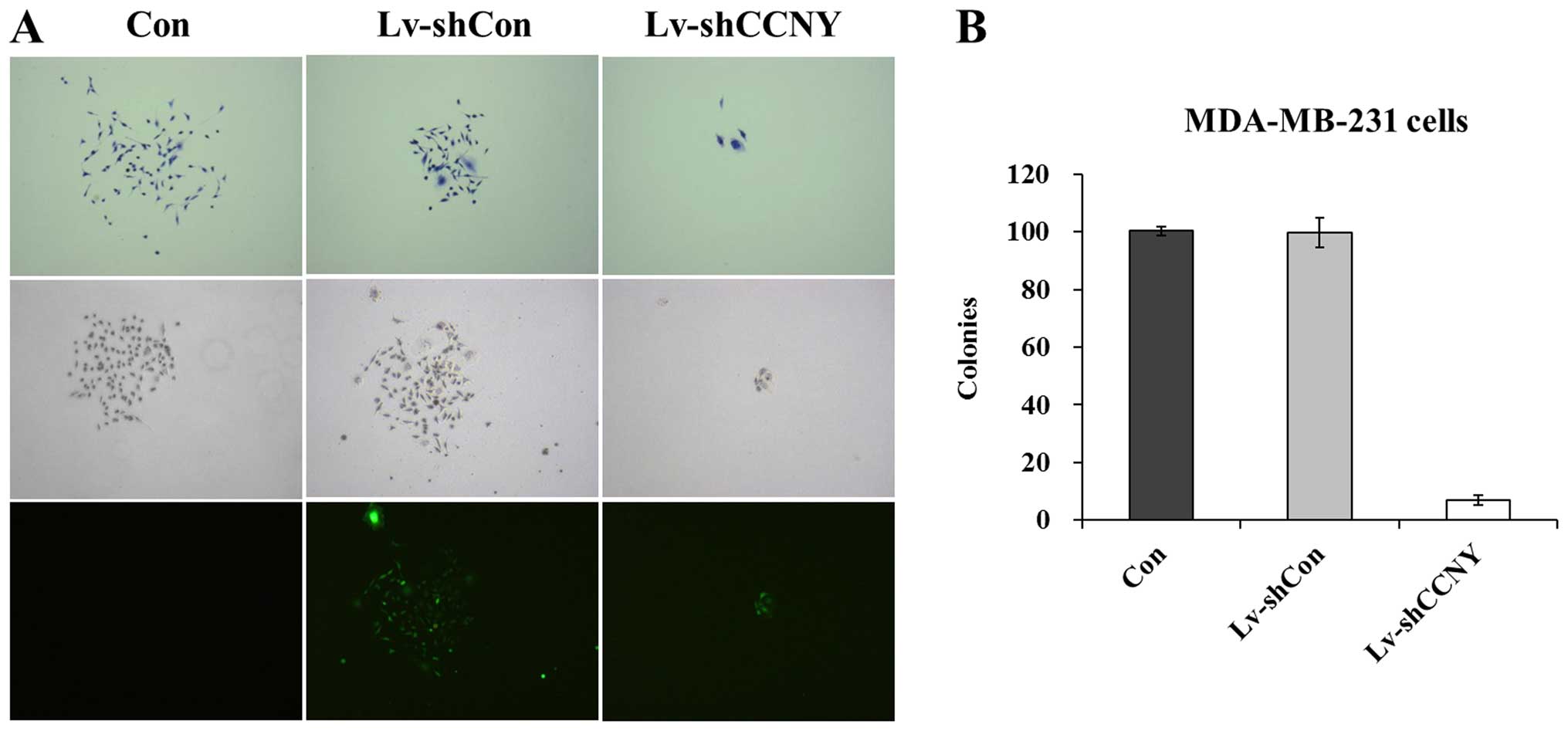
To evaluate the biological effect of cyclin Y
knockdown in regulating breast cancer cell proliferation, MTT and
colony formation assays were used. As shown in Fig. 3, the growth curves of Lv-shcyclin Y
groups were much lower than those of Lv-shCon and control groups in
both MDA-MB-231 and MCF-7 cells. The results showed that the
Lv-shcyclin Y had a short-term inhibitory effect on cell
proliferation. Moreover, we further determined its relative
long-term function on cell proliferation by colony formation assay.
As revealed in Fig. 4A, the size of
single colony in the Lv-shcyclin Y group was much smaller than that
in Lv-shCon and control groups. Also, the total number of colonies
formed in MDA-MB-231 cells was markedly reduced by over 80% in the
Lv-shcyclin Y group (Fig. 4B).
These results indicated that cyclin Y knockdown inhibited the
proliferation of breast cancer cells.
Cyclin Y knockdown induces cell cycle
arrest at G0/G1 phase
To investigate whether cyclin Y regulates cell cycle
progression directly, we then examined the cell cycle distribution
of MDA-MB-231 cells after cyclin Y knockdown by flow cytometry. As
shown in Fig. 5, compared with
control and Lv-shCon groups, the proportion of cells increased in
the G0/G1 phase and decreased in the G2/M phase, indicating that
cyclin Y could be involved in the cell cycle regulation.
Modifications of effector proteins in
cyclin Y-silenced cells
To explore the underlying signaling pathways
mediated by cyclin Y in breast cancer cells, PathScan®
intracellular signaling array kit was utilized to test whether
alterations of modifications occurred in proteins involved in cell
proliferation, growth, cell cycle, survival or apoptosis (Fig. 6). The detection of signaling
pathways contained the MAPK/ERK cascade, p38 and JNK MAPKs, Stat1
and Stat3, Akt, mTOR, AMPK, HSP27, p53, and caspase-3. To our
surprise, knockdown of cyclin Y in MDA-MB-231 cells resulted in a
series of phosphorylation, including Bad (Ser112), p53 (Ser15),
GSK3β (Ser9), as well as cleavage of PARP and caspase-3 (Table III). These data indicated that the
above signaling pathways could contribute to the regulation of
cyclin Y in breast cancer cell growth.
 | Table IIIRepresentative modification from
different wells in one chip. |
Table III
Representative modification from
different wells in one chip.
| Target |
Phosphorylation | Site | Modification | Lv-shcyclin Y vs.
Lv-shCon |
|---|
| 6 | Akt | Thr308 |
Phosphorylation | Upregulation |
| 7 | Akt | Ser473 |
Phosphorylation | Upregulation |
| 9 | S6 ribosomal | Ser235/236 |
Phosphorylation | Upregulation |
| 13 | p70 S6 kinase | Thr389 |
Phosphorylation | Upregulation |
| 15 | p53 | Ser15 |
Phosphorylation | Upregulation |
| 16 | p38 | Thr180/Tyr182 |
Phosphorylation | Upregulation |
| 18 | PARP | Asp214 | Cleavage | Upregulation |
| 19 | Caspase-3 | Asp175 | Cleavage | Upregulation |
Discussion
Cyclins are essential regulators of cell cycle
progression and are implicated in cancer progression. Abnormalities
in cell cycle regulatory proteins are common in breast cancers
(17). The present study indicates
that cyclin Y is highly expressed in human breast cancer specimens.
To identify cyclin Y involved in breast cancer development and
progression, lentivirus-mediated RNAi was employed to knock down
cyclin Y expression in two types of breast cancer cells, the
MDA-MB-231 and MCF-7 cells. Inhibition of cyclin Y markedly
attenuated the cell proliferation and colony formation
capacity.
Xu et al have reported that cyclin Y may
function as a S-phase-related cyclin, similar to cyclin A-CDK2
(18). CDK16 can be activated by
membrane-associated cyclin Y (19).
Unlike conventional cyclin-CDK interactions, cyclin Y-CDK16 binding
not only requires the catalytic domain, but also domains within the
N-terminal region (20). To examine
the effect of cyclin Y on the cell cycle control in breast cancer,
flow cytometry analysis was performed. Knockdown of cyclin Y in
MDA-MB-231 cells resulted in G0/G1 phase cell cycle arrest, which
could contribute to cell growth inhibition, indicating that cyclin
Y may directly participate in G1/S transition. We next sought to
determine the underlying molecular mechanism by which cyclin Y
regulated breast cancer cell growth. Depletion of cyclin Y
augmented the phosphorylation of Bad, p53, GSK3β and the cleavages
of PARP and caspase-3. Phosphorylation of the pro-apoptotic protein
Bad inhibits cell proliferation and promotes apoptosis while its
inhibition increases cell growth rate (21). Phosphorylation of the
multifunctional kinase GSK-3β at Ser9 (22,23)
inhibits its activity. Inhibition of GSK-3β enhances
reovirus-induced apoptosis in colon cancer cells (24). Increased levels of cleaved caspase-3
and cleaved PARP are reliable indicators of apoptosis (25). The above results suggested that
cyclin Y knockdown could induce apoptosis by activating Bad,
GSK-3β, PARP and caspase-3 in a p53-dependent manner, which also
contribute to cell growth inhibition.
In conclusion, cyclin Y is overexpressed in breast
cancer and modulates cell growth progression via regulating cell
cycle progression and apoptosis. A lentiviral-mediated RNAi system
may be an ideal therapeutic option for breast cancer therapy.
Acknowledgments
The present study was financially supported by the
National Natural Science Foundation of China (21475063), the
Chinese Jiangsu Provincial Special Program of Medical Science
(BL2013036), and the Grand of Medicine Leading Talents of Jiangsu
Health Department of China (LJ201131).
Abbreviations:
|
CCNY
|
cyclin Y
|
|
BC
|
breast cancer
|
|
shRNA
|
short hairpin RNA
|
|
IDCs
|
invasive ductal carcinomas
|
|
ILC
|
invasive lobular carcinoma
|
|
CDKs
|
cyclin-dependent kinases
|
|
NSCLC
|
non-small cell lung cancer
|
|
DAB
|
3,3′-diaminobenzidine
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
MTT
|
methylthiazol tetrazolium
|
|
PI
|
propidium iodide
|
References
|
1
|
Banerji S, Cibulskis K, Rangel-Escareno C,
Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY,
Sougnez C, Zou L, et al: Sequence analysis of mutations and
translocations across breast cancer subtypes. Nature. 486:405–409.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Viale G: The current state of breast
cancer classification. Ann Oncol. 23(Suppl 10): x207–x210. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagano T, Hashimoto T, Nakashima A,
Hisanaga S, Kikkawa U and Kamada S: Cyclin I is involved in the
regulation of cell cycle progression. Cell Cycle. 12:2617–2624.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yue W, Zhao X, Zhang L, Xu S, Liu Z, Ma L,
Jia W, Qian Z, Zhang C, Wang Y, et al: Cell cycle protein cyclin Y
is associated with human non-small-cell lung cancer proliferation
and tumorigenesis. Clin Lung Cancer. 12:43–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Syed Khaja AS, Dizeyi N, Kopparapu PK,
Anagnostaki L, Härkönen P and Persson JL: Cyclin A1 modulates the
expression of vascular endothelial growth factor and promotes
hormone-dependent growth and angiogenesis of breast cancer. PLoS
One. 8:e722102013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dai M, Al-Odaini AA, Fils-Aimé N,
Villatoro MA, Guo J, Arakelian A, Rabbani SA, Ali S and Lebrun J:
Cyclin D1 cooperates with p21 to regulate TGFβ-mediated breast
cancer cell migration and tumor local invasion. Breast Cancer Res.
15:R492013. View
Article : Google Scholar
|
|
8
|
Lamb R, Lehn S, Rogerson L, Clarke RB and
Landberg G: Cell cycle regulators cyclin D1 and CDK4/6 have
estrogen receptor-dependent divergent functions in breast cancer
migration and stem cell-like activity. Cell Cycle. 12:2384–2394.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aaltonen K, Amini RM, Landberg G, Eerola
H, Aittomäki K, Heikkilä P, Nevanlinna H and Blomqvist C: Cyclin D1
expression is associated with poor prognostic features in estrogen
receptor positive breast cancer. Breast Cancer Res Treat.
113:75–82. 2009. View Article : Google Scholar
|
|
10
|
Wei M, Zhu L, Li Y, Chen W, Han B, Wang Z,
He J, Yao H, Yang Z, Zhang Q, et al: Knocking down cyclin D1b
inhibits breast cancer cell growth and suppresses tumor development
in a breast cancer model. Cancer Sci. 102:1537–1544. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sgambato A, Camerini A, Collecchi P,
Graziani C, Bevilacqua G, Capodanno A, Migaldi M, Masciullo V,
Scambia G, Rossi G, et al: Cyclin E correlates with manganese
superoxide dismutase expression and predicts survival in early
breast cancer patients receiving adjuvant epirubicin-based
chemotherapy. Cancer Sci. 100:1026–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shaye A, Sahin A, Hao Q, Hunt K, Keyomarsi
K and Bedrosian I: Cyclin E deregulation is an early event in the
development of breast cancer. Breast Cancer Res Treat. 115:651–659.
2009. View Article : Google Scholar
|
|
13
|
Waltersson MA, Askmalm MS, Nordenskjöld B,
Fornander T, Skoog L and Stål O: Altered expression of cyclin E and
the retinoblastoma protein influences the effect of adjuvant
therapy in breast cancer. Int J Oncol. 34:441–448. 2009.PubMed/NCBI
|
|
14
|
Gao S, Ma JJ and Lu C: Prognostic value of
cyclin E expression in breast cancer: A meta-analysis. Tumour Biol.
34:3423–3430. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang M, Gao Y, Yang T, Zhu X and Chen J:
Cyclin Y, a novel membrane-associated cyclin, interacts with PFTK1.
FEBS Lett. 583:2171–2178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang T and Chen JY: Identification and
cellular localization of human PFTAIRE1. Gene. 267:165–172. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ahlin C, Zhou W, Holmqvist M, Holmberg L,
Nilsson C, Jirström K, Blomqvist C, Amini RM and Fjällskog ML:
Cyclin A is a proliferative marker with good prognostic value in
node-negative breast cancer. Cancer Epidemiol Biomarkers Prev.
18:2501–2506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Y, Wang Z, Wang J, Li J, Wang H and Yue
W: Lentivirus-mediated knockdown of cyclin Y (CCNY) inhibits glioma
cell proliferation. Oncol Res. 18:359–364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mikolcevic P, Sigl R, Rauch V, Hess MW,
Pfaller K, Barisic M, Pelliniemi LJ, Boesl M and Geley S:
Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin
Y and is essential for spermatogenesis. Mol Cell Biol. 32:868–879.
2012. View Article : Google Scholar :
|
|
20
|
Mikolcevic P, Rainer J and Geley S: Orphan
kinases turn eccentric: A new class of cyclin Y-activated,
membrane-targeted CDKs. Cell Cycle. 11:3758–3768. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Konishi Y, Lehtinen M, Donovan N and Bonni
A: Cdc2 phosphorylation of BAD links the cell cycle to the cell
death machinery. Mol Cell. 9:1005–1016. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cross DA, Alessi DR, Cohen P, Andjelkovich
M and Hemmings BA: Inhibition of glycogen synthase kinase-3 by
insulin mediated by protein kinase B. Nature. 378:785–789. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jacobs KM, Bhave SR, Ferraro DJ, Jaboin
JJ, Hallahan DE and Thotala D: GSK-3β: A bifunctional role in cell
death pathways. Int J Cell Biol. 2012:9307102012. View Article : Google Scholar
|
|
24
|
Min HJ, Koh SS, Cho IR, Srisuttee R, Park
EH, Jhun BH, Kim YG, Oh S, Kwak JE, Johnston RN, et al: Inhibition
of GSK-3β enhances reovirus-induced apoptosis in colon cancer
cells. Int J Oncol. 35:617–624. 2009.PubMed/NCBI
|
|
25
|
Bressenot A, Marchal S, Bezdetnaya L,
Garrier J, Guillemin F and Plénat F: Assessment of apoptosis by
immunohistochemistry to active caspase-3, active caspase-7, or
cleaved PARP in monolayer cells and spheroid and subcutaneous
xenografts of human carcinoma. J Histochem Cytochem. 57:289–300.
2009. View Article : Google Scholar :
|