Introduction
Breast cancer is one of the most common cancers
among women, and its incidence is increasing every year. Breast
cancer accounts for 25–30% of all the cases of malignancy among
women in Europe and the USA. In China, breast cancer is now the
first leading malignancy among women. The invasion, metastasis and
recurrence of breast cancer affect the survival and the quality of
life of patients. Therefore, it is important to investigate the
mechanisms involved in the development, progression, invasion,
metastasis and recurrence of breast cancer for treating this
disease and improving the survival of patients. The Annexin A3
(ANXA3) family is a family of Ca2+-dependent
phospholipid- and membrane-binding proteins (1,2). ANXA3
plays a role in membrane transport and other calmodulin-dependent
activities on the membrane surface. It is involved in regulating
inflammatory responses, cell differentiation, and interactions of
cytoskeletal proteins. The expression of ANXA3 is associated with
multiple human diseases. For instance, Annexin can mediate cell
signaling pathways, cell movement, tumor invasion and metastasis,
cell apoptosis, and drug resistance in the development and
progression of tumors (3–6). As a member of the Annexin family,
ANXA3 plays an important role in tumorigenesis, cell proliferation,
apoptosis, invasion, metastasis and drug resistance (7,8).
However, the effects and roles of ANXA3 in breast cancer are still
unclear. The present study investigated the relationships between
ANXA3 and proliferation, apoptosis, migration and invasion of
breast cancer cells to elucidate the mechanisms involved in the
development, progression, invasion and metastasis of breast
cancer.
Materials and methods
Cell lines
MDA-MB-231 and MCF-7 cell lines were passaged and
cultured in RPMI-1640 medium containing 10% fetal bovine serum
(FBS) and 100 U/l of penicillin and streptomycin. The cells were
cultured in an incubator at 37°C in 5% CO2.
Reagents and equipment
Lipofectamine 2000 was purchased from Invitrogen
(Carlsbad, CA, USA). Polyvinylidene difluoride (PVDF) membrane and
agarose were obtained from Bio-Rad (Mississauga, ON, Canada).
RPMI-1640 culture medium and bovine serum albumin were procured
from Gibco-BRL (Life Technologies, Paisley, Auckland). Primers were
obtained from Sangon (Shanghai, China). The High Purity Plasmid
Miniprep kit was purchased from Tiangen Biochemical Technology Co.,
Ltd. (Beijing, China). RNAiso total RNA extraction reagent,
HiScript II First Strand cDNA Synthesis kit, and qPCR SYBR-Green
Master Mix were procured from Vazyme Biotech (Nanjing, China). The
Annexin V-PE/7-AAD kit and Matrigel matrix basement membrane were
purchased from BD (San Diego, CA, USA), and Transwell 3422 was
obtained from Corning (Tewksbury, MA, USA). The equipment used in
the present study included an Epics-XL flow cytometer (Beckman
Coulter, Miami, FL, USA), polymerase chain reaction (PCR)
instrument (Eppendorf, Hamburg, Germany), and an Mx3000P
fluorescence quantitative PCR instrument (Agilent, Santa Clara, CA,
USA).
Assessing the expression of ANXA3 mRNA
using fluorescence quantitative RT-PCR
MDA-MB-231 and MCF-7 cells that grew well and were
in the logarithmic phase were collected and washed with cold
phosphate-buffered saline (PBS) twice. Then, 1 ml of RNA isolater
was added to extract total RNA using the one-step method. cDNA was
obtained by reverse transcription, which was used as the template
for PCR. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
used as the internal reference for standardization. SYBR-Green I
was used as the fluorescent dye. Real-time PCR was performed
according to the standard protocol. The experiments for each sample
were repeated thrice. The forward primer for ANXA3 was,
5′-TCCGAAACATCTGGTGAC-3′ and the reverse primer was,
3′-TCAAGTTCTTCGTAATACCGAT-5′; the forward primer for GAPDH was,
3′-CACTACCGTACCTGACACCA-5′, and the reverse primer was
3′-ATGTCGTTGTCCCACCACCT-5′. The 2−ΔCt method was used to
calculate the relative expression level of ANXA3 mRNA (ΔCt =
CTANXA3 - CTGAPDH).
Assessing the expression of ANXA3
protein using western blotting
MDA-MB-231 and MCF-7 cells that grew well and were
in the logarithmic phase were collected and washed with cold PBS
twice. The cell lysis solution was added to collect total protein,
and the bicinchoninic acid assay was used to evaluate the protein
concentration. Sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (10%, 50 µg/well) was used for separation, and the
protein was transferred onto the PVDF membrane, which was then
incubated overnight at 4°C with 5% skimmed milk. Then, 1:500
diluted mouse anti-human ANXA3 monoclonal antibody or 1:4,000
diluted rabbit anti-human β-actin monoclonal antibody was added.
After incubating at room temperature for 1 h, the membrane was
washed thrice. Next, 1:20,000 diluted fluorochrome-labeled
secondary antibody was added and incubated at room temperature for
2 h. The membranes were imaged using the infrared imaging system.
β-actin was used as the internal reference to analyze the relative
expression level of the ANXA3 protein. The experiment was repeated
thrice.
Construction of the RNA interference
lentiviral vector
pGMlV-SC5 was used for constructing the RNA
interference lentiviral vector. Restrictive endonucleases
BamHI (GGATCC) and EcoRI (GAATTC) were used to
linearize the vectors, and then target RNAi sequences were
connected to construct the lentiviral vector with target RNAi
sequences.
Selection of the shRNA target: the RNAi sequences
for ANXA3 gene were designed using the public network.
Multiple targeting RNAi sequences were designed in the present
study, and then the sequences with the best kinetic parameters were
selected, according to the experiences and the evaluation by the
software, for the consequent experiments. The shRNA target
sequences are listed in Table
I.
 | Table I.shRNA targets for the ANXA3
gene. |
Table I.
shRNA targets for the ANXA3
gene.
| No. | Target sequence |
|---|
| 1 |
GGACAAGCAGGCAAATGAAGG |
| 2 |
GCATTATGGCTATTCCCTATA |
| 3 |
GAGATGACATTAGTTCCGAAA |
Design of the shRNA primer: oligomeric
single-stranded DNA for the shRNA was designed and synthesized
according to the gene sequences. The oligo sequences are shown in
Table II.
 | Table II.Sequences of the oligo DNA for
shRNA. |
Table II.
Sequences of the oligo DNA for
shRNA.
| Oligo | Sequence of the
oligomeric single-stranded DNA (5′ to 3′) |
|---|
| 5753H_ANXA3-sh1-T
(BamHI) |
GATCCGGACAAGCAGGCAAATGAAGGTTCAAGAGACCTTCATTTGCCTGCTTGTCCTTTTTTG |
| 5753H_ANXA3-sh1-B
(EcoRI) |
AATTCAAAAAAGGACAAGCAGGCAAATGAAGGTCTCTTGAACCTTCATTTGCCTGCTTGTCCG |
| 5754H_ANXA3-sh2-T
(BamHI) |
GATCCGCATTATGGCTATTCCCTATATTCAAGAGATATAGGGAATAGCCATAATGCTTTTTTG |
| 5754H_ANXA3-sh2-B
(EcoRI) |
AATTCAAAAAAGCATTATGGCTATTCCCTATATCTCTTGAATATAGGGAATAGCCATAATGCG |
| 5755H_ANXA3-sh3-T
(BamHI) |
GATCCGAGATGACATTAGTTCCGAAACTCGAGTTTCGGAACTAATGTCATCTCTTTTTT |
| 5755H_ANXA3-sh3-B
(EcoRI) |
AATTAAAAAAGAGATGACATTAGTTCCGAAACTCGAGTTTCGGAACTAATGTCATCTCG |
Construction of the reconstructed RNAi
lentiviral plasmid
The oligomeric single-stranded DNA was annealed to
obtain a double-stranded shRNA oligo which was inserted into the
shRNA lentiviral vector to construct the reconstructed shRNA
lentiviral plasmid, and then competent DH5α cells were transfected.
Several single colonies were selected and cultured in the flask.
The positive clones were identified by sequencing. The plasmids
were extracted after the sequencing confirmed that the plasmids
were successfully constructed. After the concentration was
measured, the plasmids were stored at −20°C until use.
Transfection of the MDA-MB-231 cells
with small hairpin RNA-containing plasmids
The MDA-MB-231 cells were transfected with small
hairpin RNA (shRNA)-containing plasmids to silence the ANXA3
gene. The reagents used were as follows. i) Reagent 1: 6 µl of
Lipofectamine 2000 was mixed with 244 µl of RPMI-1640 culture
medium (FBS- and antibiotic-free), and then placed at room
temperature for 5 min; ii) reagent 2: 10 µl of shRNA plasmid
(containing 4 µg plasmid; ANXA3-sh1-3 and negative control
plasmids) was mixed with 240 µl of RPMI-1640 culture medium (FBS-
and antibiotic-free) and then placed at room temperature for 5
min.
The procedures for the cell transfection were as
follows. MDA-MB-231 cells in the logarithmic phase were collected
and cultured in 6-well plate at the density of
4×105/well. After the cells reached 85–90% confluence,
transfection was performed. The cells were washed with RPMI-1640
medium (FBS- and antibiotic-free) twice, and 2 ml of FBS- and
antibiotic-free RPMI-1640 culture medium was added into each well.
The reagents 1 and 2 were mixed gently, placed at room temperature
for 20 min, and then added into the wells. The medium was changed
to complete culture medium 6 h later, and a fluorescence microscope
was used to observe the cells at 48 h after the transfection.
Untreated cells (blank control group) and cells treated with only
Lipofectamine (liposome group) were used as the controls.
Evaluating transfection efficiency
using flow cytometry
The cells were collected at 48 h after the
transfection and washed with PBS once. Then, the green fluorescence
in the cells was measured using flow cytometry. The percentage of
cells expressing green fluorescence (in all the cells observed) was
calculated as the transfection efficiency.
Evaluating the expression of ANXA3
mRNA using quantitative reverse transcription-PCR
The cells were collected at 48 h after the
transfection, washed with PBS twice, and then treated (as described
in the section ‘Measuring the expression of ANXA3 mRNA using
fluorescence quantitative RT-PCR’). The 2−ΔCt method was
used to calculate the relative expression level of ANXA3 mRNA (ΔCt
= CTANXA3 - CTGAPDH), which was used to
assess the silencing effects of the plasmids on the ANXA3
gene. The plasmid with the best interference effects was selected.
The experiments were repeated thrice.
Identifying stably transfected cells
with puromycin
The experiments showed that the ANXA3 silencing
effects of the plasmid ANXA3-sh2 were the best. Thus, this plasmid
was chosen for the consequent siRNA experiments. After the
MDA-MB-231 cells were transfected with the ANXA3-sh2 plasmid and
the negative control plasmid for 48 h, the cells were 1:10 diluted
with RPMI-1640 medium and cultured in a 6-well plate at the density
of 5×104/well. Then, 600 ng/ml of puromycin was added
into each well. The non-transfected MDA-MB-231 cells were used as
the blank control. After the cells were cultured for 14 days, the
surviving cells were considered as stably transfected cells. The
cells transfected with ANXA3-sh2 were named as MDA-MB-231-Sh, while
the cells transfected with the negative control plasmid were named
as MDA-MB-231-NC. Both the MDA-MB-231-Sh and MDA-MB-231-NC cells
were cultured in complete culture medium containing 300 ng/ml of
puromycin.
Assessing the expression of ANXA3
protein using western blotting in the transfected cells
The MDA-MB-231, MDA-MB-231-NC and MDA-MB-231-Sh
cells that grew well and were in the logarithmic phase were
collected, washed with cold PBS twice, and then treated (as
described in the section ‘Measuring the expression of ANXA3 protein
using western blotting’) for western blotting.
Assessing cell cycle distribution
using flow cytometry
The MDA-MB-231, MDA-MB-231-NC and MDA-MB-231-Sh
cells that grew well and were in the logarithmic phase were
collected, washed with cold PBS twice, and fixed with 70% ethanol
at 4°C for 24 h. Then, the cells were again washed with PBS twice,
and the cell density was adjusted to 1×107/ml. Next, 100
µl of cell suspension was collected, and 1 ml of propidium iodide
was added. After incubating at 4°C for 30 min, flow cytometry was
performed and MultiCycle AV software (Beckman Coulter) was used to
analyze the cell cycle.
Assessing cell apoptosis using flow
cytometry
The MDA-MB-231, MDA-MB-231-NC and MDA-MB-231-Sh
cells that grew well and were in the logarithmic phase were
collected and washed with cold PBS twice. The cell density was
adjusted to 1×107/ml. Then, 100 µl of cell suspension
was collected, washed with cold PBS once, and suspended in 100 µl
of 1X binding buffer. Afterwards, 10 µl of Annexin V-PE was added,
and the mixture was placed on ice for 15 min in the dark. Next, 380
µl of 1X binding buffer and 10 µl of 7-AAD were added, incubated on
ice for 15 min in the dark, washed with cold PBS once, and
suspended with 1 ml of PBS. The apoptosis of the cells was measured
using flow cytometry. The EXPO32 ADC software (Beckman Coulter) was
used to analyze the immunofluorescence data and evaluate the
apoptosis rate.
Evaluating cell migration using the
wound healing assay
Transverse lines were drawn at the back of the
6-well culture plate evenly with a marking pen, with the distance
of 0.5 cm between each two lines. Five lines were drawn for each
well. Then, ~5×105 cells were seeded in each well, which
reached 100% confluence after overnight incubation. On the second
day, a 10-µl pipette tip was used to make scratches vertical to the
lines drawn by the marking pen (the same pipette tip was used to
make scratches for all the wells). The cells were washed thrice
with PBS to remove the unattached cells, and then the serum-free
culture medium was added. The cells were cultured in an incubator
at 37°C and 5% CO2. Images of the cells were captured at
0 and 24 h.
Evaluating cell invasion using the
Transwell assay
Matrigel and serum-free RPMI-1640 culture medium
were mixed (1:8) and used to cover the upper well of the Transwell
filter apparatus. Then, 60 µl of diluted Matrigel was added into
each well and incubated at 37°C for 4 h to allow the Matrigel to
turn white. The residual fluid was removed from the culture medium.
Next, 50 µl of serum-free RPMI-1640 culture medium was added into
each well and incubated at 37°C for 30 min. The MDA-MB-231,
MDA-MB-231-NC and MDA-MB-231-Sh cells that grew well and were in
the logarithmic phase were collected, starved in the serum-free
medium for 12 h, and then suspended in the serum-free RPMI-1640
medium. Then, 200 µl of cell suspension (containing
2×104 cells) was added into the upper well, and 600 µl
of complete culture medium was added into the lower well of the
Transwell filter apparatus. The cells were cultured at 37°C in 5%
CO2 for 24 h, the upper well was removed, the cells were
cleared from the upper well with a cotton swap, crystal violet
staining was performed, and the cells that migrated from the upper
well to the lower well were counted under a ×200 microscope in five
randomly selected visual fields.
Results
ANXA3 mRNA and protein levels in the
MDA-MB-231 and MCF-7 cells
The ANXA3 mRNA and protein levels in the MDA-MB-231
and MCF-7 cells were measured using fluorescence quantitative
RT-PCR and western blotting, respectively. The results showed that
the relative expression level of the ANXA3 mRNA was 0.0696±0.0248
in the MDA-MB-231 cells, which was significantly higher than the
level in the MCF-7 cells (0.0236±0.0149; P<0.01; Fig. 1). The relative expression level of
ANXA3 protein was also significantly higher in the MDA-MB-231 cells
than that in the MCF-7 cells (P<0.01; Fig. 2). The expression of ANXA3 mRNA and
protein was in a parallel trend in the cells.
Construction of the shRNA interference
lentiviral vector
The lentiviral vectors were successfully
constructed. The sequencing results of the shRNA interference
vector are shown in Figs. 3 and
4. The comparison showed that the
sequences of the inserted segments were completely identical to the
sequences of the designed oligo, suggesting that the lentiviral
vectors were successfully constructed.
Silencing of the ANXA3 gene in the
MDA-MB-231 cells by the transfection of shRNA plasmid
After the cells were transfected for 48 h, the
observation using the fluorescence microscope showed the expression
of green fluorescence in the cells transfected with H_ANXA3-sh1,
H_ANXA3-sh2, H_ANXA3-sh3, and negative control plasmid. However,
the non-transfected cells (blank control) and the MDA-MB-231 cells
in the liposome group were found without the expression of green
fluorescence (Fig. 5).
Evaluating transfection efficiency
using flow cytometry
After the MDA-MB-231 cells were transfected for 48
h, flow cytometry was used to evaluate the transfection efficiency.
The results showed that the transfection efficiency was 82.97±0.37,
85.25±1.07, 82.33±0.32 and 81.41±0.28% in the H_ANXA3-sh1,
H_ANXA3-sh2, H_ANXA3-sh3 and negative control groups, respectively.
The transfection efficiency in the H_ANXA3-sh2 group was the
highest, and the differences from all the other groups were
statistically significant (P<0.01; Fig. 6).
Assessing the expression of ANXA3 mRNA
in the transfected cells using fluorescence quantitative
RT-PCR
The results showed that the ANXA3 mRNA level in the
MDA-MB-231 cells transfected with H_ANXA3-sh1, H_ANXA3-sh2,
H_ANXA3-sh3, and negative control plasmid was 0.0196±0.0002,
0.0085±0.0002, 0.0220±0.0035, and 0.0661±0.0057, respectively,
while the ANXA3 mRNA level in the blank control group and liposome
group was 0.0692±0.0050 and 0.0652±0.0118, respectively. The
expression of ANXA3 mRNA was significantly lower in the MDA-MB-231
cells transfected with H_ANXA3-sh2 than in the other groups
(P<0.05; Fig. 7), suggesting
that the ANXA3 gene silencing effect of H_ANXA3-sh2 was the
highest. Therefore, H_ANXA3-sh2 was chosen for the consequent
experiments.
Identifying stably transfected cells
using puromycin
After treatment with 600 ng/ml puromycin for 14
days, all cells in the blank control group died, while a large
number of cells transfected with ANXA3-sh2 and negative control
plasmids were viable (Fig. 8). The
surviving cells were the stably transfected cells. The cells
transfected with ANXA3-sh2 and negative control plasmids were named
as MDA-MB-231-Sh and MDA-MB-231-NC cells, respectively.
Assessing the expression of ANXA3
protein in the transfected cells using western blotting
The results showed that the expression of ANXA3
protein was significantly lower in the MDA-MB-231-Sh cells than
that in the MDA-MB-231-NC and MDA-MB-231 cells (P<0.01; Fig. 9).
Evaluating the cell cycle of the
transfected cells using flow cytometry
The results showed that the percentage of cells in
the G0/1 phase was significantly higher in the
MDA-MB-231-Sh group than in the MDA-MB-231-NC and MDA-MB-231 groups
(P<0.05), while the proliferation index (PI; the percentage of
the cells in the S and G2/M phases among all the cells,
which reflects the proliferation activities of the cells) was
significantly lower in the MDA-MB-231-Sh cells than in the
MDA-MB-231-NC and MDA-MB-231 cells (P<0.05). However, the
percentage of the cells in the G0/1 phase and PI in the
MDA-MB-231-NC and MDA-MB-231 cells were not significantly different
(P>0.05; Fig. 10).
Evaluating the cell apoptosis rate
using flow cytometry
The results showed that the cell apoptosis rate was
significantly higher in the MDA-MB-231-Sh cells than that in the
MDA-MB-231-NC and MDA-MB-231 cells (P<0.01). However, the
apoptosis rate in the MDA-MB-231-NC and MDA-MB-231 cells was not
significantly different (P>0.05; Fig. 11).
Evaluating cell migration using the
wound healing assay
The wound healing assay showed that the migration
ability of the MDA-MB-231-Sh cells was significantly lower than
those of the MDA-MB-231-NC and MDA-MB-231 cells (Fig. 12).
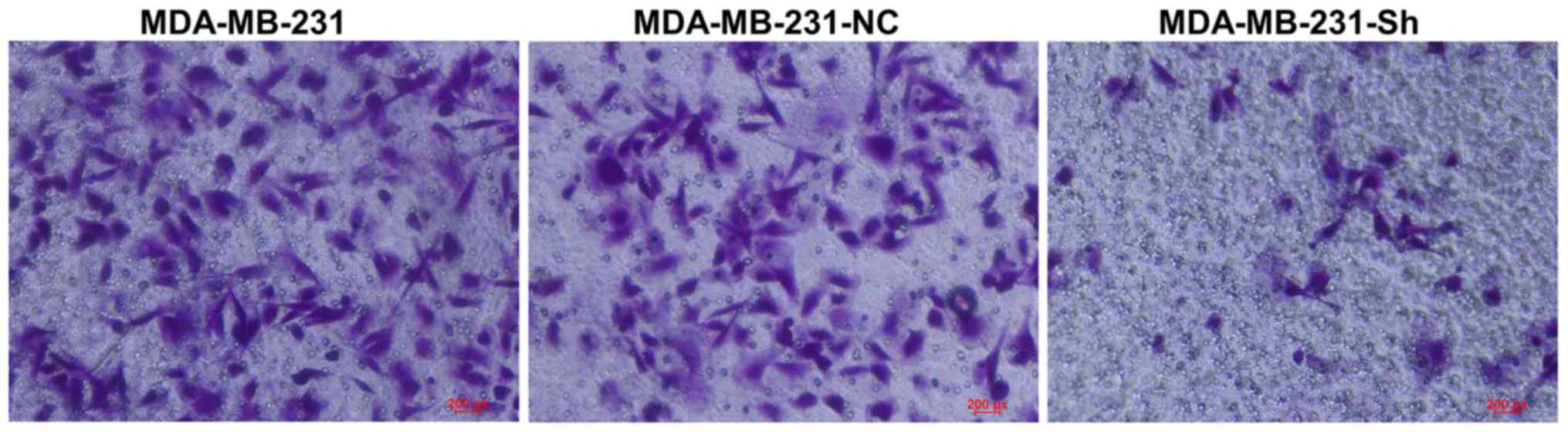
Evaluating cell invasion using the
Transwell assay
The Transwell assay showed that the number of
MDA-MB-231-Sh cells invading the membrane was significantly lower
than the number of invading MDA-MB-231-NC and MDA-MB-231 cells
(Fig. 13).
Discussion
Annexin A3 (ANXA3) is a member of the Annexin
family. Previous studies have shown that the ANXA3 gene is
located at the 4q13-q22 of the human chromosome, and consists of
323 amino acid residues (9,10). The change in expression of
ANXA3 has important effects on the development, progression,
drug resistance and metastasis of tumors. To date, only few studies
have investigated the relationships between ANXA3 and
malignant tumors, and the depth of these studies was not comparable
with the investigations on other ANXA members such as ANXA1 and
ANXA2. Liu et al (7) used
fluorescence two-dimensional differential gel electrophoresis and
liquid chromatography-mass spectrometry to screen the gastric
cancer differentiation-associated protein, and found that
ANXA3 was closely related with the differentiation of
gastric cancer cells. Xie et al (8) investigated the expression of
ANXA3 in 60 patients with rectal cancer using
immunohistochemical technology and found that the expression of
ANXA3 protein in the rectal cancer tissues was upregulated. In
addition, the overexpression of ANXA3 protein predicted the poor
prognosis of the patients. Yip et al (6) used fluorescence quantitative PCR to
compare the expression of seven biomarkers in the blood of North
American and Malaysian patients with rectal cancers, and found that
the level of ANXA3 in the blood was significantly higher in the
patients with rectal cancer than the level in the healthy controls.
Specifically, the level of ANXA3 in the blood was increased
2.06-fold in the Malaysian patients and 1.71-fold in the North
American patients, both of which were statistically significant.
These findings suggest that ANXA3 could be used as a biomarker for
the diagnosis, treatment and outcome prediction for colorectal
cancer. In a study performed by Lam et al (11), proteomic technologies were used to
compare rectal cancer tissues and normal mucosa. The results showed
that the expression of ANXA3 in the rectal cancer tissues was
significantly upregulated. In a study in China, Zhu et al
(12) reported that the RNA and
protein levels of ANXA3 were higher in drug-resistant cell lines
than in parental HT-29 cells. Zong et al (13) also found that the abnormal
expression of ANXA3 was associated with the resistance of
colorectal cancer to oxaliplatin, suggesting that ANXA3 could be
used as a biomarker for drug-resistance. However, these studies
were non-comprehensive, and hence further detailed studies are
needed. Madoz-Gúrpide et al (14) found that the expression rate of
ANXA3 in colorectal cancers was as high as 63%, which was
significantly higher than the rate in normal tissues. Baine et
al (5) found that the
expression of ANXA3 in pancreatic cancer tissues was 1.71-fold of
the level in healthy subjects. Liu et al (15) found that the expression of ANXA3 in
lung cancer tissues with lymph node metastases was significantly
higher than the expression in lung cancer tissues without lymph
node metastases. Liang et al (16) reported that the expression of ANXA3
in the liver cancer cell line Hca-F with a high metastasis
potential to lymph nodes was significantly upregulated, and the
expression level was 2.3-fold of the expression in the Hca-P cell
line with a low metastasis potential to lymph nodes, suggesting
that ANXA3 directly participates in the metastasis of liver cancer.
Other studies (17,18) also demonstrated that the expression
of ANXA3 was positively associated with the axillary lymph node
metastasis of tumors. These findings suggest that ANXA3 could play
an important role in the development, proliferation, migration and
metastasis of malignant tumors and/or tumor cells, which is in
agreement with the present findings concerning the expression of
ANXA3 in breast cancers. To date, few studies have investigated the
expression of ANXA3 in breast cancers. The present study mainly
investigated the effects of the expression of ANXA3 on the
proliferation, migration and invasion of breast cancer MDA-MB-231
cells. The present study used fluorescence quantitative RT-PCR and
western blotting to measure the expression of ANXA3 mRNA and
protein in two breast cancer cell lines (MDA-MB-231 and MCF-7). The
results showed that the expression levels of ANXA3 mRNA and protein
were significantly higher in the MDA-MB-231 cells than these levels
in the MCF-7 cells, suggesting that the expression of ANXA3 in
highly invasive breast cancer cells is increased. Three
ANXA3 gene-targeting shRNA plasmids were successfully
constructed and used to transfect the MDA-MB-231 cells for 48 h
using the Lipofectamine transfection method. The results showed
that the ANXA3 mRNA silencing effect of the ANXA3-sh2 plasmid was
the highest (87.72%). Then, puromycin was used to identify the
stably transfected cells, in which the expression of ANXA3 protein
was found to be significantly lower than the expression in the
MDA-MB-231 cells using western blotting. The results of the flow
cytometry showed that the percentage of the cells in the
G0/1 phase and the apoptosis rate were both
significantly higher, while the PI was significantly lower in the
MDA-MB-231-Sh cells than that in the MDA-MB-231-NC and MDA-MB-231
cells, suggesting that ANXA3 plays an important role in cell
proliferation. The results of the wound healing assay showed that
the migration ability of MDA-MB-231-Sh cells was significantly
lower than that of the MDA-MB-231 cells. The Transwell assay also
showed that fewer MDA-MB-231-Sh cells passed through the membrane
compared with MDA-MB-231 cells, suggesting that ANXA3 also
participated in the migration and metastasis of breast cancer
cells, indicating that inhibiting the expression of ANXA3
significantly reduced the proliferation, migration, and invasion of
breast cancer cells, and thus induced cell apoptosis and inhibited
cell proliferation, migration and invasion. The findings of the
present study were in agreement with the results reported by Zeng
et al (17). The researchers
used RNA interference technology to reduce the expression of ANXA3
in MCF-7 and MDA-MB-435 cells, and found that the proliferation of
cancer cells was effectively inhibited and the apoptosis of the
cells was evidently increased. In addition, the findings also
suggested that the effects on cancer cell apoptosis could be
associated with the effects of ANXA3 on the regulation of the
expression of BCL-2/Bax. In a study performed in China, Zhang et
al (19) upregulated the
expression of ANXA3 in gastric cancer MGC803 cells and found that
upregulating the expression of ANXA3 not only enhanced the cell
proliferation and colony formation ability, but also significantly
inhibited the apoptosis of gastric cancer cells. These findings
proved the participation of ANXA3 in regulating the proliferation
and apoptosis of cancer cells, and thus in developing malignant
tumors. However, the exact mechanisms underlying these effects
still need to be investigated.
The present study used RNA interference technology
to silence the ANXA3 gene in breast cancer MDA-MB-231 cells.
It found that ANXA3 could regulate the proliferation,
apoptosis, invasion, and migration of breast cancer cells. Thus,
this study provides the experimental basis for investigating the
mechanisms involved in the development, progression, invasion and
metastasis of breast cancer.
References
|
1
|
Bandorowicz-Pikuła J, Woś M and Pikuła S:
Participation of annexins in signal transduction, regulation of
plasma membrane structure and membrane repair mechanisms. Postepy
Biochem. 58:135–148. 2012.(In Polish). PubMed/NCBI
|
|
2
|
Fatimathas L and Moss SE: Annexins as
disease modifiers. Histol Histopathol. 25:527–532. 2010.PubMed/NCBI
|
|
3
|
Branishte T, Arsenescu-Georgescu C,
Tomescu MC, Braniste A and Mitu F: Annexins, calcium-dependent
phospholipid binding proteins in irreducible heart failure. Rev Med
Chir Soc Med Nat Iasi. 117:648–653. 2013.PubMed/NCBI
|
|
4
|
Bianchi C, Bombelli S, Raimondo F,
Torsello B, Angeloni V, Ferrero S, Di Stefano V, Chinello C, Cifola
I, Invernizzi L, et al: Primary cell cultures from human renal
cortex and renal-cell carcinoma evidence a differential expression
of two spliced isoforms of Annexin A3. Am J Pathol. 176:1660–1670.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baine MJ, Chakraborty S, Smith LM, Mallya
K, Sasson AR, Brand RE and Batra SK: Transcriptional profiling of
peripheral blood mononuclear cells in pancreatic cancer patients
identifies novel genes with potential diagnostic utility. PLoS One.
6:e170142011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yip KT, Das PK, Suria D, Lim CR, Ng GH and
Liew CC: A case-controlled validation study of a blood-based
seven-gene biomarker panel for colorectal cancer in Malaysia. J Exp
Clin Cancer Res. 29:1282010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Li Y, Tan BB, Zhao Q, Fan LQ, Zhang
ZD and Li ZX: Technique appraisement of comparative proteomics and
screening of differentiation-related protein in gastric carcinoma.
Hepatogastroenterology. 60:633–637. 2013.PubMed/NCBI
|
|
8
|
Xie YQ, Fu D, He ZH and Tan QD: Prognostic
value of Annexin A3 in human colorectal cancer and its correlation
with hypoxia-inducible factor-1α. Oncol Lett. 6:1631–1635.
2013.PubMed/NCBI
|
|
9
|
Wozny W, Schroer K, Schwall GP, Poznanović
S, Stegmann W, Dietz K, Rogatsch H, Schaefer G, Huebl H, Klocker H,
et al: Differential radioactive quantification of protein abundance
ratios between benign and malignant prostate tissues: Cancer
association of annexin A3. Proteomics. 7:313–322. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan XD, Pan LY, Yuan Y, Lang JH and Mao N:
Identification of platinum-resistance associated proteins through
proteomic analysis of human ovarian cancer cells and their
platinum-resistant sublines. J Proteome Res. 6:772–780. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
F Lam F, Jankova L, Dent OF, Molloy MP,
Kwun SY, Clarke C, Chapuis P, Robertson G, Beale P, Clarke S, et
al: Identification of distinctive protein expression patterns in
colorectal adenoma. Proteomics Clin Appl. 4:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu LQ, Shao JX, Sun JY, et al: Expression
of ANXA3 in colorectal cancer cell lines. Modern Medical Journal.
43:267–271. 2015.
|
|
13
|
Zong MZ, Feng WT, Du N, Ye LL, Tao SD, Fu
XH, He JD and Zhou JW: Screening and identifying
oxaliplatin-resistance-associated proteins in colorectal cancer
cell lines. Tumor. 33:223–228. 2013.
|
|
14
|
Madoz-Gúrpide J, López-Serra P,
Martínez-Torrecuadrada JL, Sánchez L, Lombardía L and Casal JI:
Proteomics-based validation of genomic data: Applications in
colorectal cancer diagnosis. Mol Cell Proteomics. 5:1471–1483.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu YF, Xiao ZQ, Li MX, Li MY, Zhang PF,
Li C, Li F, Chen YH, Yi H, Yao HX, et al: Quantitative proteome
analysis reveals annexin A3 as a novel biomarker in lung
adenocarcinoma. J Pathol. 217:54–64. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang RC, Neo JC, Lo SL, Tan GS, Seow TK
and Chung MC: Proteome database of hepatocellular carcinoma. J
Chromatogr B Analyt Technol Biomed Life Sci. 771:303–328. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng C, Ke Z, Song Y, Yao Y, Hu X, Zhang
M, Li H and Yin J: Annexin A3 is associated with a poor prognosis
in breast cancer and participates in the modulation of apoptosis in
vitro by affecting the Bcl-2/Bax balance. Exp Mol Pathol. 95:23–31.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu ZM, Qiu HJ and Fang SY: The expression
of Annexin A3 in breast cancer and its significance. Zhong Liu Xue
Za Zhi. 20:705–709. 2014.
|
|
19
|
Zhang DY, Wang LD and Zeng C: Effects of
high Annexin A3 expression on apoptosis and proliferation of
gastric cancer MGC-803 cells. Lin Chuang Yu Shi Yan Bing Li Xue Za
Zhi. 31:607–610. 2015.
|