Introduction
Lung cancer is one of the leading causes of
cancer-related deaths worldwide in both males and females, with
non-small cell lung cancer (NSCLC) accounting for ~87% of all lung
cancer cases (1). At diagnosis,
most patients present with advanced NSCLC stages, mainly due to the
lack of effective early diagnosis techniques. Presently,
chemotherapy is a major treatment to prolong disease-free survival
for these patients. However, little success has been made in
improving patient survival rates, as the 5-year survival rates have
remained at ~18.2% (2,3). Researchers have reported the
occurrence of intrinsic or acquired resistance, as the main
challenge associated with chemotherapeutic resistance in cancer
cells (4). Cisplatin-based
chemotherapy is frequently used in the treatment of NSCLC. The
resistance obtained following continuous therapy limits the benefit
of cisplatin in cancer treatment, but the exact mechanism of
cisplatin resistance in lung cancer remains unknown.
CXCR4, a seven-transmembrane G protein-coupled
receptor, is a physiological receptor for stromal-derived-factor-1
(5) which is highly expressed in
various types of human tumors, including lung cancer (6,7),
esophageal cancer (8), gastric
cancer (9), endometrial cancer
(10) and breast cancer (11). A number of studies have demonstrated
the vital role of CXCR4 in cancer cell survival, proliferation,
invasion and metastasis, thus high expression of CXCR4 has been
shown to be associated with a poor prognosis in cancer patients.
Nakamura et al reported that CXCR4 signaling is crucial for
drug resistance in bone marrow-disseminated tumor cells (12). While Li et al observed that
the overexpression of CXCR4 was significantly associated with
cisplatin-based chemotherapy resistance in epithelial ovarian
cancer (13). Although this
suggests a possible role for CXCR4 in development of chemotherapy
resistance, the precise role of CXCR4 in cisplatin resistance of
lung cancer remains largely unexplored.
In this study, we detected the expression of CXCR4
in NSCLC patients and cell lines, and demonstrated the possible
effect of CXCR4 knockdown on cisplatin-induced apoptosis. In
addition, we also explored the possible molecular mechanisms by
which CXCR4 possibly regulates cisplatin resistance in NSCLC.
Materials and methods
NSCLC tissue samples
A total of 64 lung cancer specimens were collected
from patients with primary NSCLC surgical resection at Renmin
Hospital of Wuhan University from May 2012 to April 2015. None of
the subjects had received any chemotherapy and radiotherapy prior
to surgery. All of these patients were treated with cisplatin-based
chemotherapy after surgery according to the NCCN guideline every 3
weeks for four cycles. Patients were followed-up by telephone until
recurrence/metastasis or until May 2016. All patients were
monitored by chest computed tomography scan, abdominal
ultrasonography and brain magnetic resonance imaging every 3 months
in the first year after surgery, or immediately when a
recurrence/metastasis was suspected. A whole-body fludeoxyglucose
positron emission tomography/CT was performed when needed. The WHO
grades and clinical features of these patients were summarized in
Table I. Patients with disease
progression or recurrence 12 months after surgery were defined as
being chemoresistance, whereas those without recurrence or
recurrence >12 months after surgery were defined as
chemosensitive. There were 32 NSCLC patients who are sensitive to
cisplatin treatment and 32 NSCLC patients with cisplatin
chemoresistance. This study was carried out in accordance with the
principles of the Helsinki Declaration and approved by the ethics
committee of the Renmin Hospital of Wuhan University.
 | Table I.Comparison of clinical
characteristics between patients with low and high CXCR4 cell
content. |
Table I.
Comparison of clinical
characteristics between patients with low and high CXCR4 cell
content.
| Clinical
factor | Total patients
(n=64) | High expression
(n=46) | expression (n=18)
Low | P-value |
|---|
| Age (years) |
|
|
|
|
|
≤65 | 15 | 12 | 3 | 0.424 |
|
>65 | 49 | 34 | 15 |
|
| Gender |
|
|
|
|
|
Male | 44 | 35 | 9 | 0.043 |
|
Female | 20 | 11 | 9 |
|
| T stage |
|
|
|
|
| T2 | 43 | 26 | 17 | 0.004b |
| T3 | 14 | 13 | 1 |
|
| T4 | 7 | 7 | 0 |
|
| N stage |
|
|
|
|
| N0 | 36 | 24 | 12 | 0.490b |
| N1 | 7 | 6 | 1 |
|
| N2 | 21 | 16 | 5 |
|
| M stage |
|
|
|
|
| M0 | 62 | 46 | 16 | 0.076a |
| M1 | 2 | 0 | 2 |
|
| Histology |
|
|
|
|
|
Adenocarcinoma | 39 | 26 | 13 | 0.247 |
|
Squamous carcinoma | 25 | 20 | 5 |
|
|
Drug-resistance |
|
|
|
|
|
Yes | 32 | 27 | 5 | 0.026 |
| No | 32 | 19 | 13 |
|
NSCLC cell lines
A549 was obtained from American Type Culture
Collection, cisplatin resistant A549 cell line (A549/DDP) was
obtained from Fudan Institutes of Biomedical Sciences Cell Center,
and the cells were maintained in Dulbecco's modified Eagle's medium
with high glucose (Gibco, USA) supplemented with 10% fetal bovine
serum (FBS; Hyclone, USA), containing 100 U/ml penicillin and 100
mg/ml streptomycin. Both cell lines were cultured in a 5%
CO2 air incubator at 37°C and passaged using 0.25%
trypsin-EDTA (Gibco) when they reached confluence. Cisplatin was
purchased from Sigma Chemical Co. (USA).
Immunohistochemistry
Paraffin-embedded tissue sections were dewaxed and
rehydrated, and antigen retrieval was performed by microwaving in
10 mM sodium citrate buffer, pH 6.0, for 20 min. Sections were then
incubated with 3% hydrogen peroxide for 30 min at room temperature
to block endogenous peroxidase, then blocked in 10% normal goat
serum for 0.5 h. Immunostaining was performed by incubating with
anti-CXCR4 (1:500, ab124824, Abcam Corp., UK) or anti-CYP1B1
(1:1,000, ab32649, Abcam Corp.) at 4°C overnight. Slides were then
washed in PBS and incubated with secondary antibody (anti-rabbit
detection system; Boster, China) for 30 min at 37°C. Staining was
visualized with 3, 3-diaminobenzidine and counterstained with
hematoxylin.
Quantitative RT-PCR
Total RNA was extracted from A549 and A549/DDP cells
using TRIzol (Invitrogen, USA) according to the manufacturer's
instructions and quantified by Nanodrop 2000 (Thermo Scientific,
USA). RNA (1 µg) was reverse-transcribed to cDNA with random
primers using the Promega reverse transcriptase kit (USA) according
to the manufacturer's protocol. To assess gene expression, cDNAs
were amplified with the SYBR® Premix Ex Taq™ II (Tli
RNaseH Plus) (Takara, Japan) using the QuantStudio 6 Flex Real-Time
PCR system (Life Technologies, USA). The relative level was
calculated by the comparative Ct method (2−∆∆Ct). The
sequences of specific primers were as follows: CXCR4, forward,
5′-CCGTGGCAAACTGGTACTTT-3′; reverse, 5′-GACGCCAACATAGACCACCT-3′.
CYP1B1, forward, 5′-AAGTTCTTGAGGCACTGCGAA-3′; reverse,
5′-GGCCGGTACGTTCTCCAAAT-3′.
Western blot analysis
Cells were washed with ice cold PBS and lysed by
RIPA supplemented with protease inhibitor PMSF on ice. The
extracted proteins were separated by 12% SDS-polyacrylamide gel
electrophoresis and transferred to PVDF membranes (Millipore, USA).
The membrane was blocked with 5% non-fat milk TBS-T (0.1% Tween-20,
100 nM Tris-HCl, 0.9% NaCl) and incubated with anti-CXCR4 (1:200,
sc-6190, Santa Cruz, USA) or anti-CYP1B1 (1:1,000, ab32649, Abcam
Corp.) with gentle shaking at 4°C overnight. After washing three
times, the membranes were incubated with HRP-conjugated secondary
antibodies for 2 h at room temperature. The protein bands were
visualized with ECL plus western blotting detection reagents
(Thermo Scientific).
Immunofluorescence assay
Cells were fixed and permeabilized with 2%
paraformaldehyde and 0.5% Triton X-100. After overnight incubation
with anti-CXCR4 (1:100, sc-6190, Santa Cruz), the specimens were
rinsed thoroughly and treated with anti-goat antibodies,
respectively, conjugated with Alexa Fluor 488 and Alexa Fluor 594
(Invitrogen), diluted 1:400 (1 h, 37°C). Nuclei were stained using
0.3 µM DAPI (Molecular BioProbes, USA). The digital images were
then captured with a cooled CCD camera and processed with the help
of Photoshop (Adobe) software.
siRNA transfect assay
Cells were plated 24 h before transfection. At
30–50% confluence, cells were transfected using Lipofectamine™ 2000
(Invitrogen) with siRNA duplexes specific for human CYP1B1
(RiboBio, China) or control negative control (NC) siRNA. The
following were the sequences of siRNAs used in the study, CXCR4
siRNA: sense, GUGGCAAACUGGUACUUUG; antisense, CAAAGUACCAGUUUGCCAC.
CYP1B1siRNA: sense, GCAUGAUGCGCAACUUCUU; antisense,
AAGAAGUUGCGCAUCAUGC. In addition, mock control was included where
cells were treated with Lipofectamine 2000 alone. The siRNA
experiment was carried out for 48 h to analyse the RNA expression
level by qRT-PCR and 72 h to analyse the protein expression level
by western blot analysis.
Apoptosis assay
Cells were dual stained with the Annexin V-FITC
apoptosis detection kit (BD Biosciences, USA) according to the
manufacturer's protocol. Stained cells were immediately analyzed by
flow cytometry (FACSAriaIII, BD Biosciences).
Cell proliferation assay
A549 and A549/DDP cells were seeded in 96-well plate
at the density of 1.0×104 cells per well overnight for
adherence. The next day cells were transfected with CXCR4-siRNA or
CYP1B1-siRNA. After 48 h of transfection, 10 µl CCK-8 solutions was
added to each well at indicated times and incubated for another 2
h. The absorbance of each well was obtained from Perkin-Elmer 2030
Victor × multilabel plate reader (Perkin-Elmer, Waltham, MA, USA)
at 450 nm.
Gene expression omnibus dataset
correlation analysis
We downloaded GSE30219 (14) and GSE41271 (15,16)
which were the largest NSCLC datasets from Gene expression omnibus
(http://www.ncbi.nlm.nih.gov/geo/) to
verify the correlation of CXCR4 and CYP1B1. GSE30219 included 272
NSCLC patients and GSE41271 included 263 NSCLC patients.
Statistical analysis
Each assay was performed in triplicate and repeated
a minimum of three times. Statistical analysis was performed using
SPSS 19.0 or GraphPad Prism 5 software. Data are reported as means
± SD. Statistical differences were analyzed by Student's t-test for
data between control and treated groups, or a one-way analysis of
variance for data from multiple groups, with the level of
significance set at P<0.05.
Results
CXCR4 expression closely correlated
with NSCLC cisplatin resistance
To investigate the association between CXCR4 and
cisplatin resistance in NSCLC, we first detected the expression
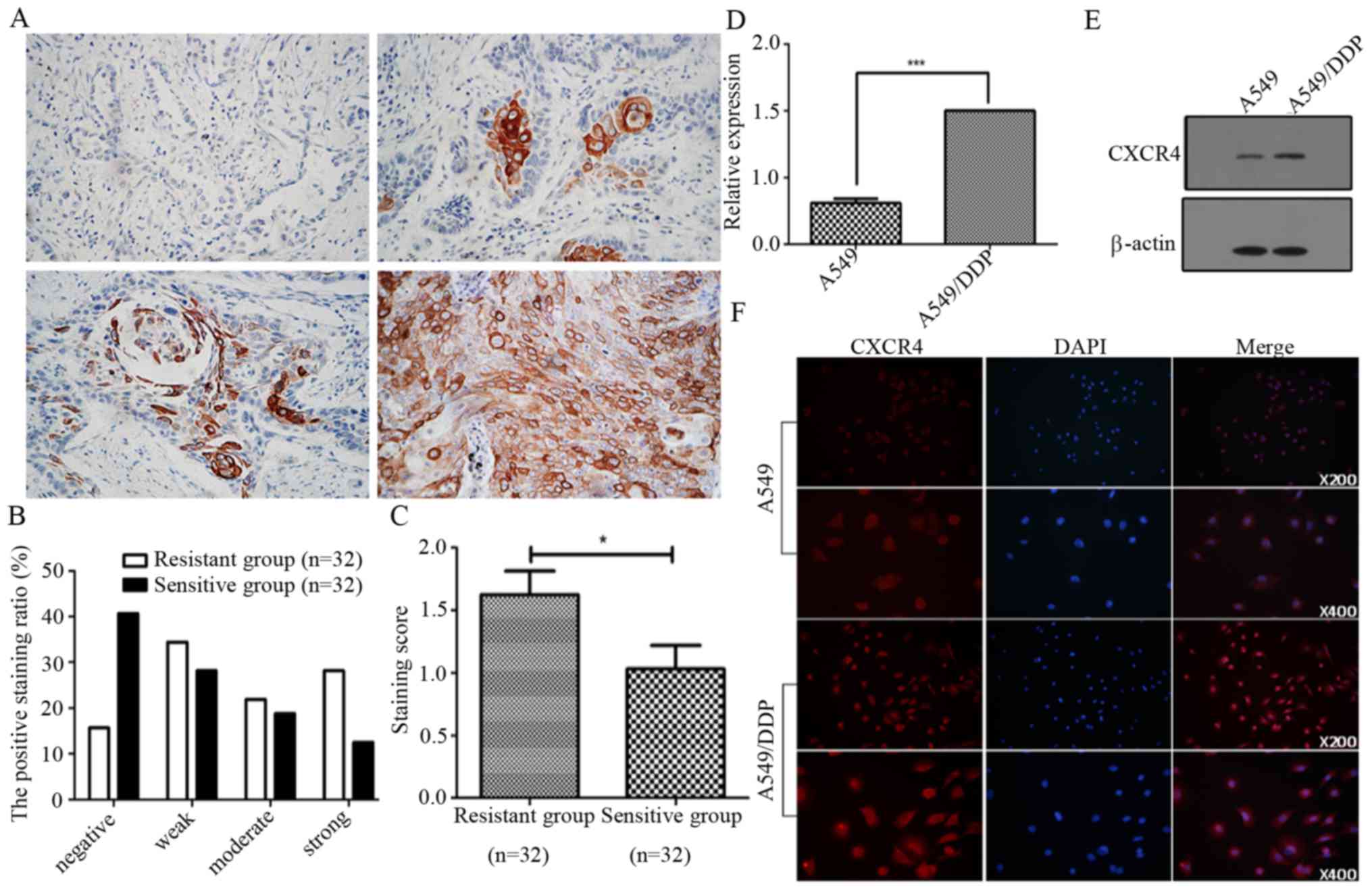
levels of CXCR4 in 64 NSCLC patients by immunohistochemistry. We
found that the levels of CXCR4 were significantly increased in
cancer samples from cisplatin-resistant patients, compared to that
in samples from cisplatin-sensitive patients. We categorized the
NSCLC-sensitive group into negative (40.6%), weak (28.1%), moderate
(18.8%) and strong (12.5%) and resistance group into negative
(15.6%), weak (34.4%), moderate (21.9%) and strong (28.1%),
respectively (Fig. 1A and B). The
staining scores being 1.63±1.07 in the cisplatin-resistant group
and 1.03±1.06 in the sensitive group (Fig. 1C). Furthermore, we investigated the
expression levels of CXCR4 in A549 and A549/DDP cells by qPCR,
western blot analysis and immunofluorescence. As shown in Fig. 1D and E, the expression levels of
CXCR4 mRNA and protein in A549/DDP were significantly higher than
that in A549 cells. These results indicate that CXCR4 may be
involved in NSCLC cancer cisplatin resistance.
Clinical characteristics associated
with CXCR4 expression in NSCLC patients
We carried out a comparative analysis of
clinicopathological characteristics and associated CXCR4
expression. As shown in Table I,
75% (n=49) of the patients were >65 years old, with ~68% (n=44,
P=0.043) being male patients, who accounted for ~76% (n=35) of all
patients with high CXCR4 expression. CXCR4 high expression was
observed to be mostly associated with T2N0M0 cancer stages,
indicating an increased tumor size at T2 stage (P=0.004), with no
nearby lymph node invasion and no metastatic spread to other
regions. CXCR4 high expression was also associated with a slightly
higher percentage prevalence in adenocarcinoma as compared to
squamous carcinoma and a higher drug resistance levels (P=0.026).
This therefore suggests that high CXCR4 expression levels may be
influenced by patient age and sex, thereby having an effect on the
tumor staging, cancer subtype and drug resistance
characteristics.
CXCR4 knockdown enhances
cisplatin-induced apoptosis and inhibits cell growth
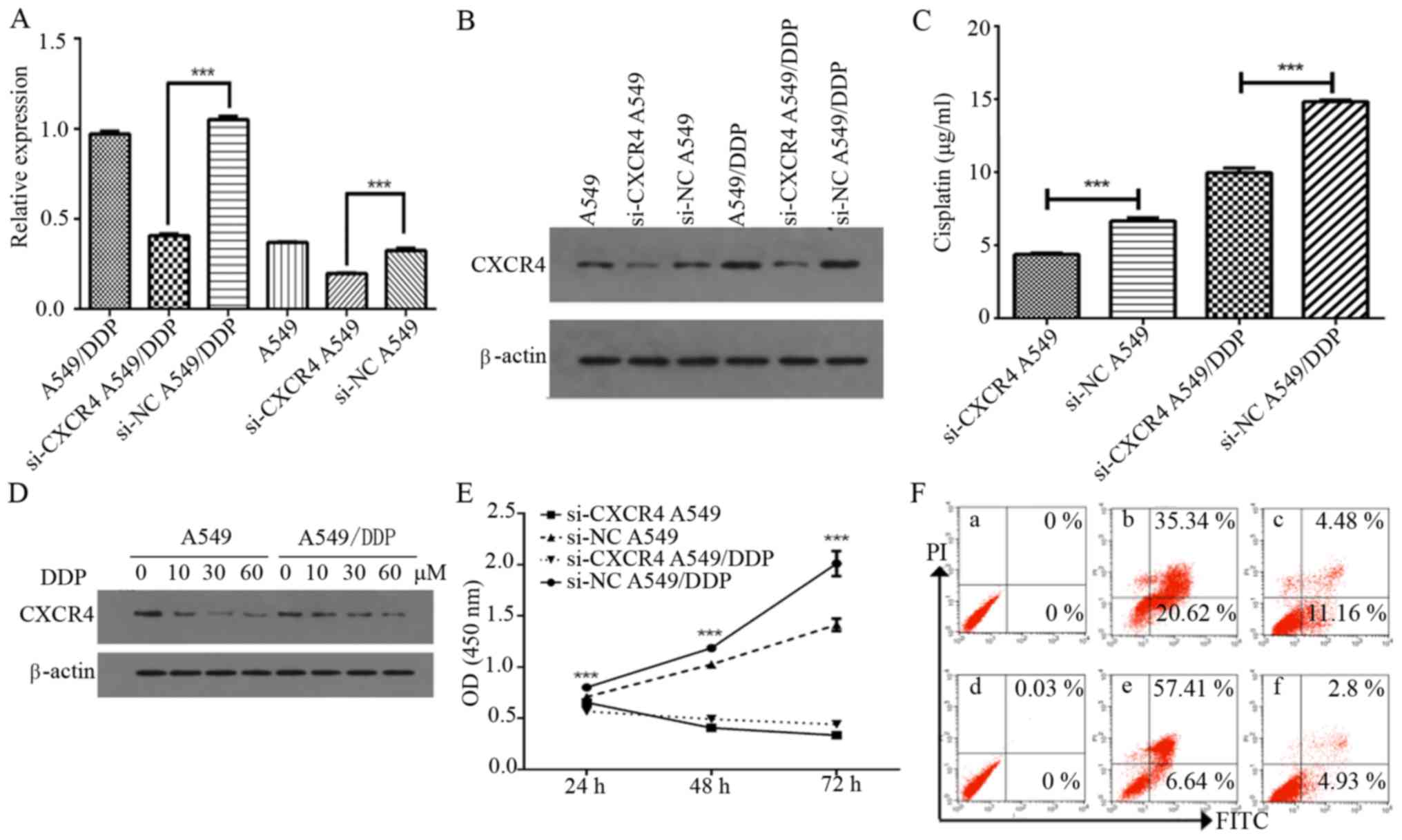
To investigate whether CXCR4 mediates chemotherapy
resistance in A549 and A549/DDP cells, we investigated the effect
of CXCR4 siRNA on CXCR4 expression. CXCR4 siRNA was observed to
inhibit the mRNA and protein expression levels of CXCR4 in A549 and
A549/DDP cells (P<0.0001) (Fig. 2A
and B). We further investigated whether CXCR4 knockdown using
CXCR4 siRNA had an effect on cisplatin sensitivity in NSCLC cells,
of which it was observed that the half-maximal inhibitory
concentration (IC50) after 48 h was 6.665±0.215 and
4.373±0.077 µg/ml for A549 and si-CXCR4 A549 cells, respectively
(P<0.0001), while it was found to be 14.837±0.099 and
9.969±0.318 µg/ml for A549/DDP and si-CXCR4 A549/DDP cells,
respectively (P<0.0001). Both the A549 and A549/DDP si-CXCR4
transfected cells were more sensitive to cisplatin treatment than
the non-transfected A549 and A549/DDP cells, indicating that CXCR4
knockdown increases sensitivity to cisplatin (Fig. 2C).
In addition, cisplatin effectively inhibited CXCR4
expression in both cells in a dose-dependent manner (Fig. 2D). CCK-8 assay was further applied
to confirm the effect of CXCR4 knockdown on proliferation in A549
and A549/DDP cells. As shown in Fig.
2E, si-CXCR4 significantly suppressed cell proliferation in
A549 and A549/DDP from 24 to 72 h (P<0.0001). Flow cytometry
results indicated that A549 and A549/DDP transfected with negative
control siRNA had an apoptotic cell percentage of ~11.16 and 4.93%,
respectively, while the percentage of apoptotic cells in the CXCR4
siRNA cells was 20.62 and 6.64%, respectively (Fig. 2F). These results also suggest that
knockdown of CXCR4 with siRNAcontributes to the recovery of
cisplatin-induced apoptosis in A549 and A549/DDP cells.
CXCR4 promotes cisplatin resistance
through upregulation of CYP1B1
Increasing evidence has shown that many cytotoxic
drugs are metabolized by cytochrome p450 (CYP) enzymes (17–25),
we therefore explored the correlation of CXCR4 with CYP-related
molecules (CYP1A1, CYP1A2, CYP1B1, CYP2C8, CYP2C9, CYP2C18,
CYP2C19, CYP2E1, CYP3A4, CYP3A5, CYP3A7 and CYP3A43) by
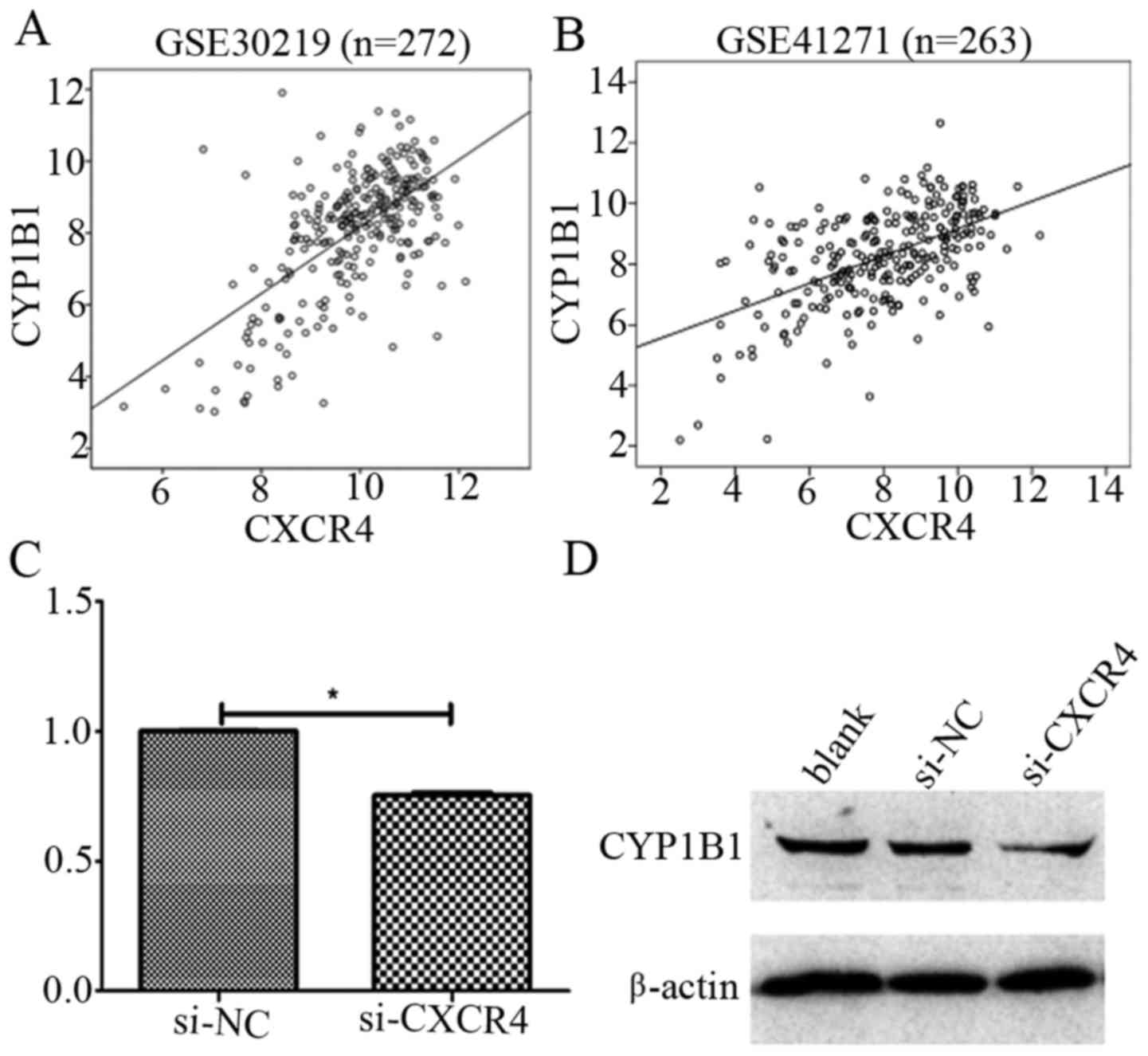
bioinformatics (Table II). Our
results indicated a positive correlation between CXCR4 and CYP1B1
with r=0.616 (P<0.0001) and r=0.538 (P<0.0001) in GSE30219
and GSE41271 datasets, respectively (Fig. 3A and B). Furthermore, we
investigated whether CXCR4 plays an important role in cisplatin
resistance in NSCLC by targeting CYP1B1, by examining the CYP1B1
mRNA and protein expression levels after CXCR4 knockdown by siRNA
in A549/DDP cells. It was observed that CYP1B1 mRNA and protein
level were significantly downregulated as a result of CXCR4
knockdown (Fig. 3C and D). Taken
together, these results demonstrate CXCR4 may promote cisplatin
resistance by regulating the expression levels of CYP1B1.
 | Table II.The correlation of CXCR4 and
cytochrome p450 molecules. |
Table II.
The correlation of CXCR4 and
cytochrome p450 molecules.
|
| GSE219 (n=72) r,
P-value | GSE271 (n=263) r,
P-value |
|---|
| CYP1A1 | 0.014, 0.819 | 0.027,
0.667 |
| CYP1A2 | 0.022, 0.721 | −0.07,
0.255 |
| CYP1B1 | 0.616, 0.000 | 0.538,
0.000 |
| CYP2C8 | 0.058, 0.337 | −0.114, 0.064 |
| CYP2C9 | −0.134, 0.027 | 0.028,
0.657 |
| CYP2C18 | −0.135, 0.026 | −0.028,
0.654 |
| CYP2C19 | −0.25, 0.000 | −0.173,
0.005 |
| CYP2E1 | −0.135, 0.026 | −0.087,
0.161 |
| CYP3A4 | 0.063, 0.3 | 0.033,
0.597 |
| CYP3A5 | −0.409, 0.000 | −0.118,
0.055 |
| CYP3A7 | −0.264, 0.000 | 0.025,
0.689 |
| CYP3A43 | −0.287, 0.000 | −0.204,
0.001 |
CYP1B1 regulates NSCLC cell
chemotherapy resistance
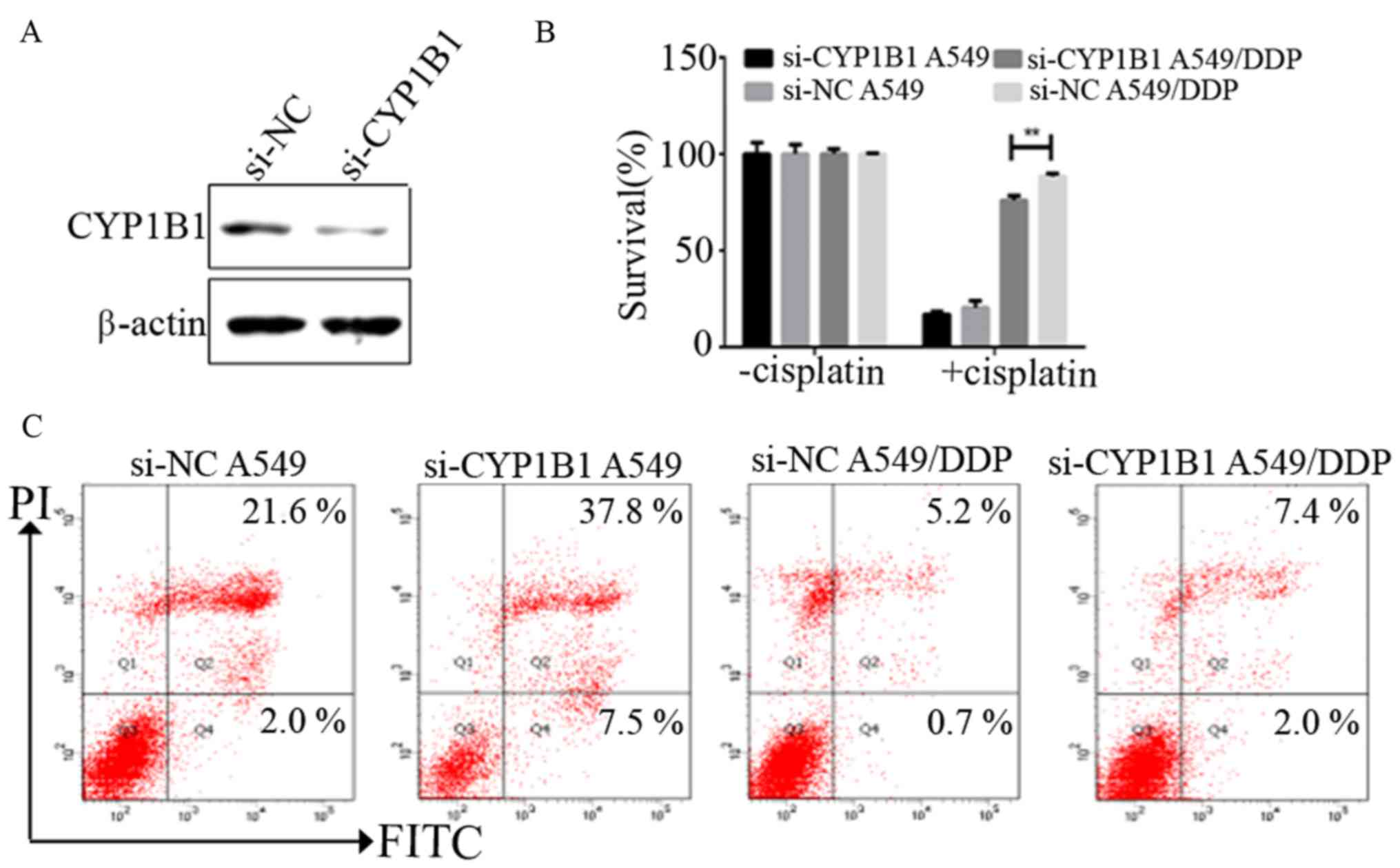
To evaluate the effects of CYP1B1 on NSCLC cell
survival and resistance to chemotherapy, we examined whether
reduction of CYP1B1 levels affects NSCLC cell survival.
Transfection with CYP1B1 siRNA resulted in a marked reduction in
endogenous levels of CYP1B1 in A549/DDP by western blot analysis
(Fig. 4A). Knockdown of CYP1B1
significantly decreased the survival rate of A549/DDP cells, but
did not decrease the survival rate of A549 cells (Fig. 4B). A549 and A549/DDP transfected
with negative control siRNA were observed to have apoptotic cell
percentages of 2.0 and 0.7%, respectively, while the percentage of
apoptotic cells was observed to increase to ~7.5 and 2.0% in CYP1B1
knockdown cells, respectively (Fig.
4C).
Taken together, it was observed that the percentage
of apoptosis was significantly increased in A549 and A549/DDP cells
after inhibiting CYP1B1 expression by siRNA, thus suggesting that
CYP1B1 may play a significant role in NSCLC cell survival and
resistance to chemotherapy.
CYP1B1 overexpression is associated
with poor survival and cisplatin resistance in NSCLC
To further investigate the association between
CYP1B1 and cisplatin resistance in NSCLC, we examined the
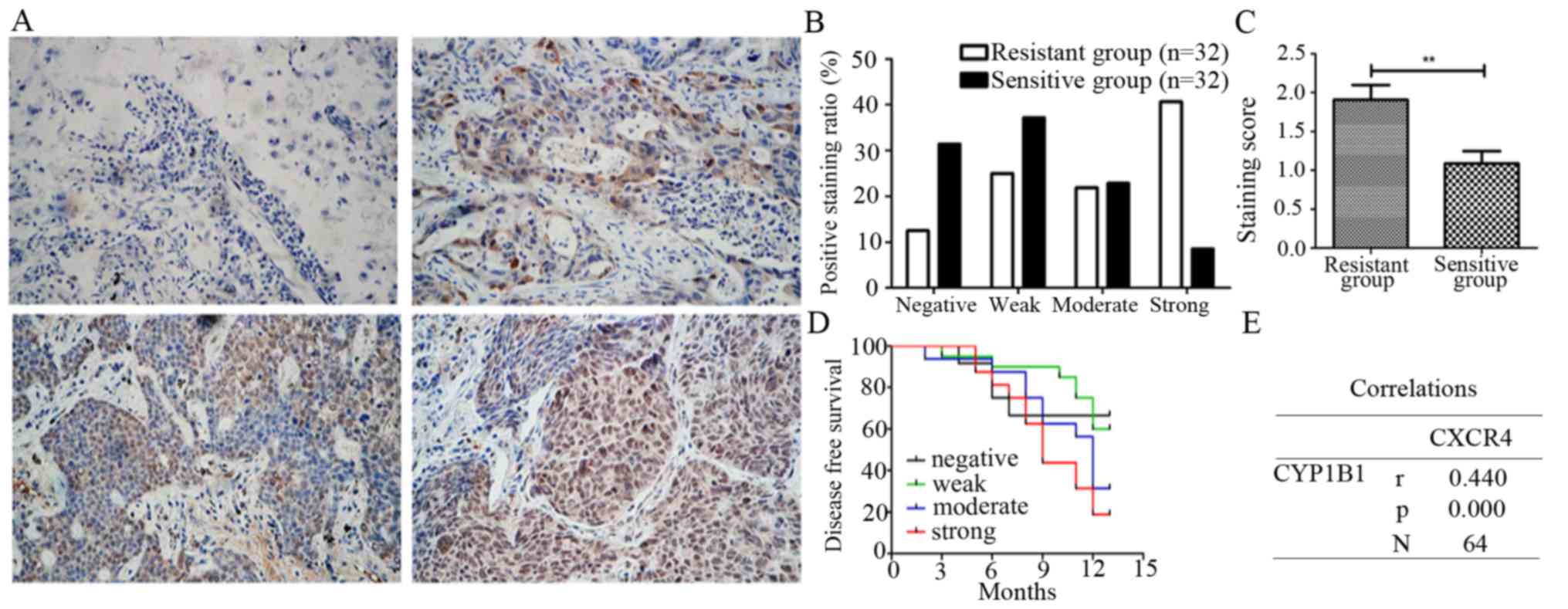
expression of CYP1B1 in 32 cisplatin-sensitive NSCLC tissues and 32
cisplatin-resistant NSCLC tissues by immunohistochemistry. The
expression levels of CYP1B1 in most cisplatin-sensitive tissues was
observed to range from weak to undetectable, while that in
cisplatin-resistant tissues ranged from weak to strong. Thus we
categorized the NSCLC cisplatin-sensitive group into negative
(25.0%), weak (37.5%), moderate (28.1%) and strong (9.4%), while
the cisplatin-resistant group was categorized into negative
(12.6%), weak (25.0%), moderate (21.9%) and strong (40.6%)
(Fig. 5A and B), the staining
scores being 1.69±1.19 in the cisplatin-resistant group and
1.08±0.97 in cisplatin-sensitive group (Fig. 5C).
Having analyzed the CYP1B1 expression levels in
cisplatin-sensitive and resistance groups, we further went on and
analyzed CYP1B1 expression with associated patient clinical
outcomes of all 64 NSCLC tissues. It was observed that expression
of CYP1B1 correlated with disease-free survival and strong
expression thus staining of CYP1B1 suggested poor survival as
compared to moderate, weak and negative CYP1B1 expression and
staining (Fig. 5D). Lastly, we
confirmed that CXCR4 has a positive correlation with CYP1B1
(r=0.44, P<0.001) in NSCLC patients (Fig. 5E). These results suggest that CYP1B1
overexpression is associated with poor survival and cisplatin
resistance in NSCLC.
Discussion
Cisplatin-based chemotherapy is one of the effective
methods used in treatment of malignancies, however, cisplatin
resistance has become a major therapeutic obstacle in clinical
practice, and as such, much attention has been paid to the
underlying mechanisms of chemotherapy resistance. In recent years,
increasing evidence has revealed that CXCR4 is involved in
chemoresistance in several types of cancers, including mantle cell
lymphoma (26), chronic myelogenous
leukemia (27) and breast cancer
(28). However, the function of
CXCR4 in NSCLC chemoresistance remains unknown. In this study, we
explored the potential role of CXCR4 in NSCLC chemotherapeutic
responses.
We investigated the possible association of CXCR4
high expression with cancer progression and poor prognosis in NSCLC
tissues. Thus, we examined the expression of CXCR4 in 64 NSCLC
patient samples. We observed that patient samples with a high CXCR4
expression significantly associated with a shorter disease-free
patient survival as seen in the cisplatin-resistant group, when
compared with the cisplatin-sensitive group, thereby suggesting
that CXCR4 high expression might be closely related with
chemoresistance. This was further amplified when we analyzed the
clinicopathological characteristics in relation to the CXCR4
expression, in which we observed that CXCR4 was mostly expressed in
male patients >65 years old and associated with the tumor size
and drug resistance levels. Other researchers have also reported
the possible association of CXCR4 expression levels with patient
sex and tumor staging (29,30).
Some studies have previously revealed the
association between CXCR4 overexpression and chemoresistance in
some cancers. Nakamura et al observed that CXCR4
overexpression promoted drug resistance in cancer cells by
upregulating transforming growth factor-β2 (12). Similarly, Sison et al
reported that CXCR4 overexpression protected pediatric acute
lymphoblastic leukemia cells from chemotherapy-induced apoptosis,
thus enhancing chemoresistance when the cells were co-cultured with
bone marrow stroma. Treatment with a CXCR4 inhibitor plerixafor
diminished the protective effect of apoptosis, thus conferring
chemosensitivity (31). High CXCR4
expression levels have also been significantly associated with
cisplatin-based chemotherapeutic resistance in epithelial ovarian
cancer (13).
In this study, by using cisplatin-resistant NSCLC
cells A549/DDP and its parental A549 cells, we demonstrated that
CXCR4 might play an important role in regulating the
chemoresistance in NSCLC, as it was observed that CXCR4 was highly
expressed in A549/DDP cells and CXCR4 knockdown resulted in reduced
proliferation and cell apoptosis induction. Analyses of the
IC50 values also indicated that CXCR4 knockdown resulted
in increased chemosensitivity of A549/DDP cells to cisplatin.
Therefore, in order to define the mechanisms of
CXCR4 associated NSCLC chemoresistance, we further downloaded two
largest datasets of NSCLC patients from gene expression omnibus and
performed a correlation analysis with CYP related-molecules. We
found only CYP1B1 had a positive correlation with CXCR4. CYP1B1
belongs to cytochrome p450 family, which is a multi-gene family of
enzymes implicated in the metabolism of a diverse range of
xenobiotics and endogenous compounds (24). Several researchers have proposed
interesting concepts, in explaining the mechanisms of CYP1B1
induced cytotoxic effect reduction of chemotherapeutic drug
(20,21,23,25),
as other have mainly focused on CYP1B1 polymorphism in cancer
process (22,32,33).
Of particular interest, Patel et al reported that CYP1B1 may
be regulated by cytokines, as interleukin-6 was observed to mediate
of CYP1B1 upregulation via DNA methylation in colorectal cancer
(34). Therefore, we further
investigated the differential expression of CYP1B1 between the
chemosensitive and chemoresistance groups, and we observed that
CYP1B1 levels were significantly increased in the chemoresistance
group. We also observed that CYP1B1 knockdown decreased the cell
survival rates, enhanced cell apoptosis and partially reversed the
chemoresistance of cisplatin in cancer cells. Additionally, we also
observed that CXCR4 mediated cisplatin resistance via regulating
CYP1B1 expression.
In conclusion, this study shows that CXCR4 mediates
cisplatin chemoresistance in NSCLC in a CYP1B1-dependent manner.
Thus, it could be used as a potential therapeutic target in NSCLC
chemoresistance patients and as a clinically useful predictor of
cisplatin response.
Acknowledgements
This study was supported by National Natural Science
Foundation of China (nos. 81270607 and 81541027) and Natural
Science Foundation of Hubei (nos. 2014CFA070 and 2015CFB653).
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
CYP1B1
|
cytochrome P450 1B1
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holohan C, Van Schaeybroeck S, Longley DB
and Johnston PG: Cancer drug resistance: An evolving paradigm. Nat
Rev Cancer. 13:714–726. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dai X, Mao Z, Huang J, Xie S and Zhang H:
The CXCL12/CXCR4 autocrine loop increases the metastatic potential
of non-small cell lung cancer in vitro. Oncol Lett. 5:277–282.
2013.PubMed/NCBI
|
|
7
|
Xie S, Zeng W, Fan G, Huang J, Kang G,
Geng Q, Cheng B, Wang W and Dong P: Effect of CXCL12/CXCR4 on
increasing the metastatic potential of non-small cell lung cancer
in vitro is inhibited through the downregulation of CXCR4 chemokine
receptor expression. Oncol Lett. 7:941–947. 2014.PubMed/NCBI
|
|
8
|
Wang T, Mi Y, Pian L, Gao P, Xu H, Zheng Y
and Xuan X: RNAi targeting CXCR4 inhibits proliferation and
invasion of esophageal carcinoma cells. Diagn Pathol. 8:1042013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nikzaban M, Hakhamaneshi MS, Fakhari S,
Sheikhesmaili F, Roshani D, Ahsan B, Kamali F and Jalili A: The
chemokine receptor CXCR4 is associated with the staging of gastric
cancer. Adv Biomed Res. 3:162014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Teng F, Tian WY, Wang YM, Zhang YF, Guo F,
Zhao J, Gao C and Xue FX: Cancer-associated fibroblasts promote the
progression of endometrial cancer via the SDF-1/CXCR4 axis. J
Hematol Oncol. 9:82016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kishima M Okuyama, de Oliveira CE,
Banin-Hirata BK, Losi-Guembarovski R, Brajão de Oliveira K,
Amarante MK and Watanabe MA: Immunohistochemical expression of
CXCR4 on breast cancer and its clinical significance. Anal Cell
Pathol (Amst). 2015:8910202015.PubMed/NCBI
|
|
12
|
Nakamura T, Shinriki S, Jono H, Guo J,
Ueda M, Hayashi M, Yamashita S, Zijlstra A, Nakayama H, Hiraki A,
et al: Intrinsic TGF-β2-triggered SDF-1-CXCR4 signaling axis is
crucial for drug resistance and a slow-cycling state in bone
marrow-disseminated tumor cells. Oncotarget. 6:1008–1019. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li J, Jiang K, Qiu X, Li M, Hao Q, Wei L,
Zhang W, Chen B and Xin X: Overexpression of CXCR4 is significantly
associated with cisplatin-based chemotherapy resistance and can be
a prognostic factor in epithelial ovarian cancer. BMB Rep.
47:33–38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sato M, Larsen JE, Lee W, Sun H, Shames
DS, Dalvi MP, Ramirez RD, Tang H, DiMaio JM, Gao B, et al: Human
lung epithelial cells progressed to malignancy through specific
oncogenic manipulations. Mol Cancer Res. 11:638–650. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Riquelme E, Suraokar M, Behrens C, Lin HY,
Girard L, Nilsson MB, Simon G, Wang J, Coombes KR, Lee JJ, et al:
VEGF/VEGFR-2 upregulates EZH2 expression in lung adenocarcinoma
cells and EZH2 depletion enhances the response to platinum-based
and VEGFR-2-targeted therapy. Clin Cancer Res. 20:3849–3861. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang I, Mitsui Y, Fukuhara S, Gill A,
Wong DK, Yamamura S, Shahryari V, Tabatabai ZL, Dahiya R, Shin DM,
et al: Loss of miR-200c up-regulates CYP1B1 and confers docetaxel
resistance in renal cell carcinoma. Oncotarget. 6:7774–7787. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hendrikx JJ, Lagas JS, Rosing H, Schellens
JH, Beijnen JH and Schinkel AH: P-glycoprotein and cytochrome P450
3A act together in restricting the oral bioavailability of
paclitaxel. Int J Cancer. 132:2439–2447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kudo T, Ozaki Y, Kusano T, Hotta E, Oya Y,
Komatsu S, Goda H and Ito K: Effect of buffer conditions on
CYP2C8-mediated paclitaxel 6alpha-hydroxylation and CYP3A4-mediated
triazolam alpha- and 4-hydroxylation by human liver microsomes.
Xenobiotica. 46:241–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laroche-Clary A, Le Morvan V, Yamori T and
Robert J: Cytochrome P450 1B1 gene polymorphisms as predictors of
anticancer drug activity: Studies with in vitro models. Mol Cancer
Ther. 9:3315–3321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pastina I, Giovannetti E, Chioni A,
Sissung TM, Crea F, Orlandini C, Price DK, Cianci C, Figg WD, Ricci
S, et al: Cytochrome 450 1B1 (CYP1B1) polymorphisms associated with
response to docetaxel in castration-resistant prostate cancer
(CRPC) patients. BMC Cancer. 10:5112010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vasile E, Tibaldi C, Leon GL, D'Incecco A
and Giovannetti E: Cytochrome P450 1B1 (CYP1B1) polymorphisms are
associated with clinical outcome of docetaxel in non-small cell
lung cancer (NSCLC) patients. J Cancer Res Clin Oncol.
141:1189–1194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wen CJ, Wu LX, Fu LJ, Shen DY, Zhang X,
Zhang YW, Yu J and Zhou HH: Preferential induction of CYP1A1 over
CYP1B1 in human breast cancer MCF-7 cells after exposure to
berberine. Asian Pac J Cancer Prev. 15:495–499. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu X, Zhang XA and Wang DW: The roles of
CYP450 epoxygenases and metabolites, epoxyeicosatrienoic acids, in
cardiovascular and malignant diseases. Adv Drug Deliv Rev.
63:597–609. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu Z, Mu Y, Qi C, Wang J, Xi G, Guo J, Mi
R and Zhao F: CYP1B1 enhances the resistance of epithelial ovarian
cancer cells to paclitaxel in vivo and in vitro. Int J Mol Med.
35:340–348. 2015.PubMed/NCBI
|
|
26
|
Chen Z, Teo AE and McCarty N: ROS-induced
CXCR4 signaling regulates mantle cell lymphoma (MCL) cell survival
and drug resistance in the bone marrow microenvironment via
autophagy. Clin Cancer Res. 22:187–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Miao H, Li W, Yao J, Sun Y, Li Z,
Zhao L and Guo Q: CXCL12/CXCR4 axis confers adriamycin resistance
to human chronic myelogenous leukemia and oroxylin A improves the
sensitivity of K562/ADM cells. Biochem Pharmacol. 90:212–225. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Luca A, D'Alessio A, Gallo M, Maiello
MR, Bode AM and Normanno N: Src and CXCR4 are involved in the
invasiveness of breast cancer cells with acquired resistance to
lapatinib. Cell Cycle. 13:148–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang M, Chen GY, Song HT, Hong X, Yang ZY
and Sui GJ: Significance of CXCR4, phosphorylated STAT3 and VEGF-A
expression in resected non-small cell lung cancer. Exp Ther Med.
2:517–522. 2011.PubMed/NCBI
|
|
30
|
Al Zobair AA, Al Obeidy BF, Yang L, Yang
C, Hui Y, Yu H, Zheng F, Yang G, Xie C, Zhou F, et al: Concomitant
overexpression of EGFR and CXCR4 is associated with worse prognosis
in a new molecular subtype of non-small cell lung cancer. Oncol
Rep. 29:1524–1532. 2013.PubMed/NCBI
|
|
31
|
Sison EA, Magoon D, Li L, Annesley CE, Rau
RE, Small D and Brown P: Plerixafor as a chemosensitizing agent in
pediatric acute lymphoblastic leukemia: Efficacy and potential
mechanisms of resistance to CXCR4 inhibition. Oncotarget.
5:8947–8958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sankhwar M, Sankhwar SN, Abhishek A, Gupta
N and Rajender S: CYP1B1 gene polymorphisms correlate with an
increased risk of urinary bladder cancer in India. Urol Oncol.
34:167. e161–168. 2016. View Article : Google Scholar
|
|
33
|
Yu PJ, Chen WG, Feng QL, Chen W, Jiang MJ
and Li ZQ: Association between CYP1B1 gene polymorphisms and risk
factors and susceptibility to laryngeal cancer. Med Sci Monit.
21:239–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Patel SA, Bhambra U, Charalambous MP,
David RM, Edwards RJ, Lightfoot T, Boobis AR and Gooderham NJ:
Interleukin-6 mediated upregulation of CYP1B1 and CYP2E1 in
colorectal cancer involves DNA methylation, miR27b and STAT3. Br J
Cancer. 111:2287–2296. 2014. View Article : Google Scholar : PubMed/NCBI
|