Introduction
Multiple myeloma (MM) is the second most common
hematologic malignancy, with an incidence higher than that of acute
leukemia. Although most MM patients are sensitive to initial
chemotherapeutic treatments, relapse often occurs due to acquired
drug resistance (1,2). The introduction of bortezomib (BTZ)
represented a breakthrough in the treatment of MM. Chemotherapy
regimens based on BTZ have become first-line recommendations for MM
patients with untreated, relapsed or refractory myeloma, including
patients who are eligible for autologous hematopoietic cell
transplantation (3,4). BTZ resistance has a strong impact on
the clinical effectiveness of MM treatments. Therefore, determining
the process by which BTZ resistance develops in MM and searching
for effective regimens to overcome resistance are critical research
areas (5).
In the present study, we established three
BTZ-resistant cell lines, U-266/BTZ, NCI-H929/BTZ and
RPMI-8226/BTZ, by using gradual induction. We examined the mRNA and
protein expression levels of UbcH10 in the resistant and their
parental cells. The data showed that UbcH10 protein was
significantly increased in the resistant cell lines while mRNA was
slightly increased, suggesting an inactivation of the
post-transcriptional regulation of UbcH10. Since microRNAs (miRNAs)
are a typical mechanism for post-transcriptional regulation
(6,7), we searched for miRNAs with a binding
site on the 3′-untranslated region (3′UTR) of UbcH10. We then
quantitatively measured the miRNAs in the resistant and parental
cells. The results indicated that hsa-miR-631 was negatively
correlated with the UbcH10 protein. A luciferase reporter assay
verified that hsa-miR-631 was able to bind to UbcH10-3′UTR and
inhibit protein expression through the seed site. The results
suggest that the inactivation of UbcH10 regulation by hsa-miR-631
may be a molecular mechanism for resistance in MM cell lines. This
study examined whether hsa-miR-631 was abnormally expressed in
BTZ-resistant MM cell lines, whether abnormal UbcH10 expression
caused by abnormal hsa-miR-631 expression was a critical factor in
BTZ resistance, whether MDR1 is involved in UbcH10-associated BTZ
resistance and whether the overexpression of hsa-miR-631 may reduce
or reverse BTZ resistance in MM cell lines.
Materials and methods
Cell culture
Three MM cell lines, U-266, NCI-H929 and RPMI-8226,
were purchased from the Chinese Academy of Sciences Cell Bank
(Shanghai, China). Cell lines were maintained in RPMI-1640 medium
containing 10% fetal bovine serum (FBS) (both from Invitrogen,
Carslbad, CA, USA). The viral packaging cell line 293T was
purchased from American Type Culture Collection (ATCC; Manassas,
VA, USA) and maintained in Dulbecco's modified Eagle's medium
(DMEM) supplemented with 10% FBS. All cells were incubated at 37°C
in a humidified 95% air and 5% CO2 incubator. U-266,
NCI-H929 and RPMI-8226 cells were cultured in a semi-suspension and
passaged by centrifugation; 293T cells were adherent and passaged
by trypsin digestion.
Establishment and verification of
BTZ-resistant MM cell lines
Three selected MM cell lines, U-266, NCI-H929 and
RPMI-8226 were cultured with BTZ in a gradient of increased
concentrations starting from 0.1 nM and doubling every three
passages, i.e., 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, 6.4, 12.8, 25.6 and
51.2 nM. Cells were cultured in a medium containing 51.2 nM BTZ for
two additional passages. These cell lines were named U-266/BTZ,
NCI-H929/BTZ and RPMI-8226/BTZ. The resistant and parental cells
were seeded in 96-well plates at 1×105 cells per well,
and medium containing BTZ was added at a final concentration of 1,
2, 4, 8, 16, 32 and 64 nM for a period of 48 h. Cell Counting Kit-8
(CCK-8) assay was employed to determine cell inhibition by BTZ and
to calculate the drug dose causing 50% growth inhibition
(IC50) values. Meanwhile, the resistant and parental
cells (1×106 each) were harvested for total RNA and
protein extraction, followed by real-time PCR and western blotting
to measure the UbcH10 mRNA, hsa-miR-631 and UbcH10 proteins.
Plasmid construction
Construction of luciferase reporter vectors
Human UbcH10 3′-UTR (259 bp) was amplified from cDNA
obtained through the reverse transcription of the total RNA of 293T
cells, using the primers (Forward/Reverse)
5′-GCTCTAGAGAAACCTACTCAAAGCAG-3′ and
5′-GCTCTAGAACCACAGCTCAAGATAAA-3′. The amplification parameters were
32 cycles of denaturation at 95°C for 10 sec, annealing at 58°C for
30 sec and extension at 72°C for 30 sec. The product was then
digested with XbaI and inserted into the pGL3-promotor
vector (Promega Corp., Madison, WI, USA). The seed region was
mutated by point mutation from 5′-CAGGTC-3′ to 5′-TCGCAG-3. The
resulting vectors were named pGL-WT-UbcH10 and pGL-MT-UbcH10.
Construction of cDNA expression vector
The CDS sequence of human UbcH10 (NM_007019.2) was
amplified using the primers (Forward/Reverse)
5′-GGAATTCGCCACCATGGCTTCCCAAAACCGCG-3′ and
5′-CGGGATCCTCAGGGCTCCTGGCTGGTG-3′, which contained an EcoRI
cutting site and Kozak sequence and a BamHI cutting site,
respectively. The cDNA was prepared by reverse transcription of RNA
isolated from 293T cells. The PCR product was digested and cloned
into a pcDH-CMV lentiviral expressing vector. Finally, the
recombinant vector was named pcDH-UbcH10.
Construction of the miR-631 expression
vector
Human genomic DNA was extracted from 293T cells and
used for amplification of the template of the precursor sequence of
miR-631. The primers (Forward/Reverse) used were
5′-GGAATTCTGGCATGCCATAGCAGCGCAG-3′ and
5′-CGGGATCCCTCCCATCTAAGCTTCCCAAAGTGT-3′. The PCR product was
digested using EcoRI and BamHI, ligated into linear
pCDH-EF1-GFP vector (System Biosciences, Mountain View, CA, USA)
and transformed into DH5α competent cells. The obtained vector was
called the pmiR-631 vector. The products of the vectors were
confirmed by DNA sequencing. Endotoxin-free DNA was prepared in all
cases.
Lentivirus packaging
One day before transfection, 293T cells were seeded
into 10-cm dishes. Then, 2 µg of each pmiR-631 or pcDH-UbcH10
vector and 10 µg pPACK Packaging Plasmid Mix (System Biosciences)
were co-transfected using Lipofectamine 2000 (Invitrogen) in
accordance with the manufacturer's instructions. The medium was
replaced with DMEM plus 1% FBS. Forty-eight hours later, the
supernatant was harvested and cleared by centrifugation at 5,000 ×
g at 4°C for 5 min, and it was passed through a 0.45-µm pore PVDF
membrane. The virus titre was determined by gradient dilution. The
packaged lentiviruses were named Lv-miR631 or Lv-UbcH10.
Verification of the binding site of hsa-miR-631
on UbcH10-3′UTR by luciferase reporter assay
TargetScan was used to predict the theoretic target
(seed region) of miR-631 in mRNA sequence of UbcH10 (NM_007019.3).
Chemically synthesized miR631-mimics, inhibitor and negative
control (NC) were obtained from Shanghai Sangon Biotech Co., Ltd.
(Shanghai, China).
293T cells were transfected with the miR631-mimics,
inhibitor or NC as well as pGL-WT-UbcH10 and pGL-MT-UbcH10 using
Lipofectamine 2000 according to the manufacturer's instructions.
Forty-eight hours after transient transfection, the cells were
harvested and luciferase assays were performed. The relative
luciferase activities (ratios of firefly and renilla
luciferase activity) of lysates were measured using a
Dual-Luciferase Reporter system (Promega Corp.).
Effects of expression of miR-631 or UbcH10 on BTZ
resistance and MDR1 expression in MM cells
The cells were divided into five groups: control,
Lv-control, Lv-miR631, Lv-UbcH10, and Lv-miR631/Lv-UbcH10. The cell
lines in logarithmic phase U-266/BTZ, NCI-H929/BTZ and
RPMI-8226/BTZ were seeded into 6-well plates at 5×105
cells per well. Lentiviral solution at an MOI of 30 was added the
next day. Seventy-two hours later, the infection efficiency was
examined by observation of fluorescent markers. The cells were
divided into three parts: one was seeded into 96-well plates and
cultured in media containing various concentrations of BTZ (1–32
nM). After 48 h, a CCK-8 assay was used to calculate the
IC50 values of BTZ before and after genetic
intervention. Next, other cells were cultured in a medium
containing 25 nM BTZ, and apoptosis was examined by double staining
assay after 48 h. The third group of cells was collected and
subjected to extraction of total RNA and protein, followed by
real-time PCR and western blotting to measure the miR-631 and
UbcH10 RNAs and the UbcH10 and MDR1 proteins, respectively.
Assessment of cell viability and IC50
values
The three cell lines and their genetically
engineered lines were seeded in 96-well plates at 5×104
cells per well, and BTZ was added to a final concentration at 1, 2,
4, 8, 16 or 32 nM for a period of 48 h, followed by a CCK-8 assay
for cell viability. Briefly, 10 µl CCK-8 solution was added, and
the cells were cultured under normal conditions for an additional 4
h before measuring absorbance at 450 nm. The cell inhibition ratio
was calculated, based on the IC50 values at 48 h.
Detection of apoptosis
Seventy-two hours after viral infection, U-266/BTZ,
NCI-H929/BTZ and RPMI-8226/BTZ cells were seeded into 6-well plates
at 1×105 cells per well in medium containing 10 µM BTZ
and were cultured for 48 h. The cells were collected and measured
for apoptosis using flow cytometry (FACSCalibur) after treatment
using the Annexin V-FITC Apoptosis Detection kit II (cat no.
556570) (both from BD Biosciences).
Effect of hsa-miR-631 overexpression on the
half-life of MDR1 protein in resistant MM cells
Seventy-two hours after viral infection, U-266/BTZ
cells were re-suspended in RPMI-1640 medium with 10% FBS, reseeded
to 6-well plates at 2×105 cells per well and cultured
overnight. Then, the medium was replaced with serum-free RPMI-1640
medium containing either 50 µM MG132 or 100 µg/ml cycloheximide
(CHX), and the cells were incubated under normal conditions. The
cells were collected at 0, 1, 2, 4 and 8 h after the drug
treatment, and total protein was extracted for MDR1 detection by
western blotting.
Measurement of mRNA levels
Total RNA was isolated with TRIzol reagent
(Invitrogen) according to the manufacturer's instructions. RNA was
reverse transcribed into cDNA using M-MLV Reverse Transcriptase and
oligo(dT)18 primer (both from Takara Bio, Inc., Otsu,
Japan). The following specific primers (Forward/Reverse) were used
in quantitative PCR of human UbcH10 and β-actin: UbcH10,
5′-AAGACCTGAGGTATAAGCTC-3′ and 5′-CCACTTTTCCTTCAGGATGTC-3′;
β-actin, 5′-CCTGTACGCCAACACAGTGC-3′ and 5′-ATACTCCTGCTTGCTGATCC-3′.
The lengths of the amplified products were 143 and 211 bp.
Real-time PCR was performed using SYBR® Premix Ex Taq™
kit and the TP800 system (both from Takara Bio, Inc.). cDNA from
200 ng total RNA was used as the template. The PCR reactions were
conducted using 40 cycles of denaturation at 95°C for 10 sec,
annealing at 60°C for 20 sec and extension at 72°C for 20 sec.
To test the miR-631 levels, total RNA (2 µg) was
used for cDNA preparation with the M-MLV reverse transcription kit
and specific primers: U6 snRNA (NM_001101.3),
5′-TACCTTGCGAAGTGCTTAAAC-3′; and miR-631,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGAGGAGA-3′. RNA
contents were detected using fluorescent dye PCR (Takara Bio, Inc.)
in accordance with the manufacturer's instructions. The following
primers (Forward/Reverse) were used for quantification of human U6
snRNA and miR-631: U6 snRNA, 5′-GTGCTCGCTTCGGCAGCACAT-3′ and
5′-TACCTTGCGAAGTGCTTAAAC-3′, which produced a segment of 112 bp;
and miR-631, 5′-GCCGGCGCCCGAGCTCTGGCTC-3′ and
5′-AGACCTGGCCCAGACCTCAGC-3′, which produced a segment of 73 bp. The
PCR systems included Takara SYBR Premix Ex Taq 10 µl, forward and
reverse primers (20 µM) 0.2 µl each, and cDNA 2 µl added with
dH2O to 20 µl. The cycling parameters were 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec and
extension at 72°C for 20 sec.
The mRNA levels of UbcH10 were normalized to the
expression of an endogenous housekeeping gene, β-actin, using the
ΔΔCt method. U6 snRNA was used as a reference to normalize the
miR-631 levels using the 2−ΔΔCt method. Each RNA sample
was run in triplicate.
Detection of proteins
Protein was extracted from the cells using M-PER
mammalian protein extraction reagent (Pierce Biotechnology, Inc.,
Rockford, IL, USA). Equal amounts of protein (25 µg per lane), as
estimated by a bicinchoninic acid (BCA) protein assay kit (Pierce
Biotechnology, Inc.), were loaded onto (11%) SDS-PAGE gels and
transferred onto nitrocellulose membranes. The blots were probed
with a monoclonal antibody against human UbcH10 (1:1,000), MDR1
(1:300) and β-actin (1:1,200), followed by the secondary
HRP-conjugated anti-mouse/rabbit antibody (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA). After washing, bands
were detected by chemiluminescence and imaged with X-ray film, and
relative optical densities were analyzed with the image processing
software (TotalLab). Relative contents of proteins were calculated
by dividing the optical density of the target band with the optical
density of the β-actin band.
Data analysis
All data are expressed as the mean ± SD and were
analyzed by one-way ANOVA. Least significant difference (LSD) was
used for multiple comparisons between any two means. P-values
<0.05 were considered statistically significant. All statistical
analysis was performed using SPSS 13.0 software.
Results
Establishment and confirmation of
BTZ-resistant myeloma cell lines
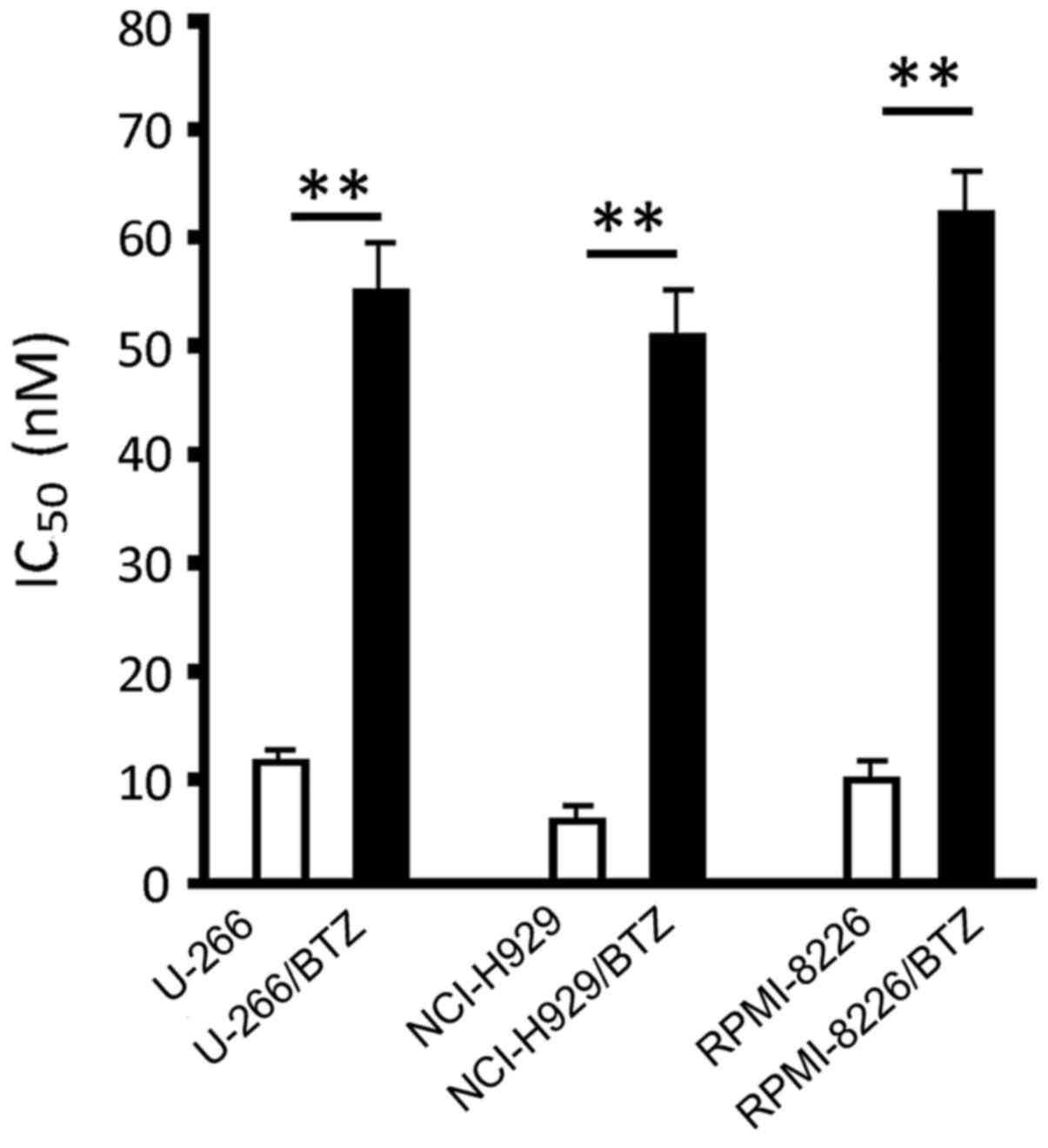
We established the three BTZ-resistant cell lines
U-266/BTZ, NCI-H929/BTZ and RPMI-8226/BTZ by gradually increasing
the concentration of BTZ in the culture medium for ~3 months. The
results of IC50 measurement showed that the
IC50 values in the U-266/BTZ, NCI-H929/BTZ and
RPMI-8226/BTZ cells increased from 11.10±1.24, 6.08±0.71 and
10.02±1.62 nM in their parental cells to 55.62±4.88, 49.12±4.32,
and 61.21±5.82 nM, respectively. These figures represented
statistically significant differences between the resistant cells
and their parental cells (P<0.01) (Fig. 1).
Examination of miR-631 and UbcH10
expression in resistant myeloma cells
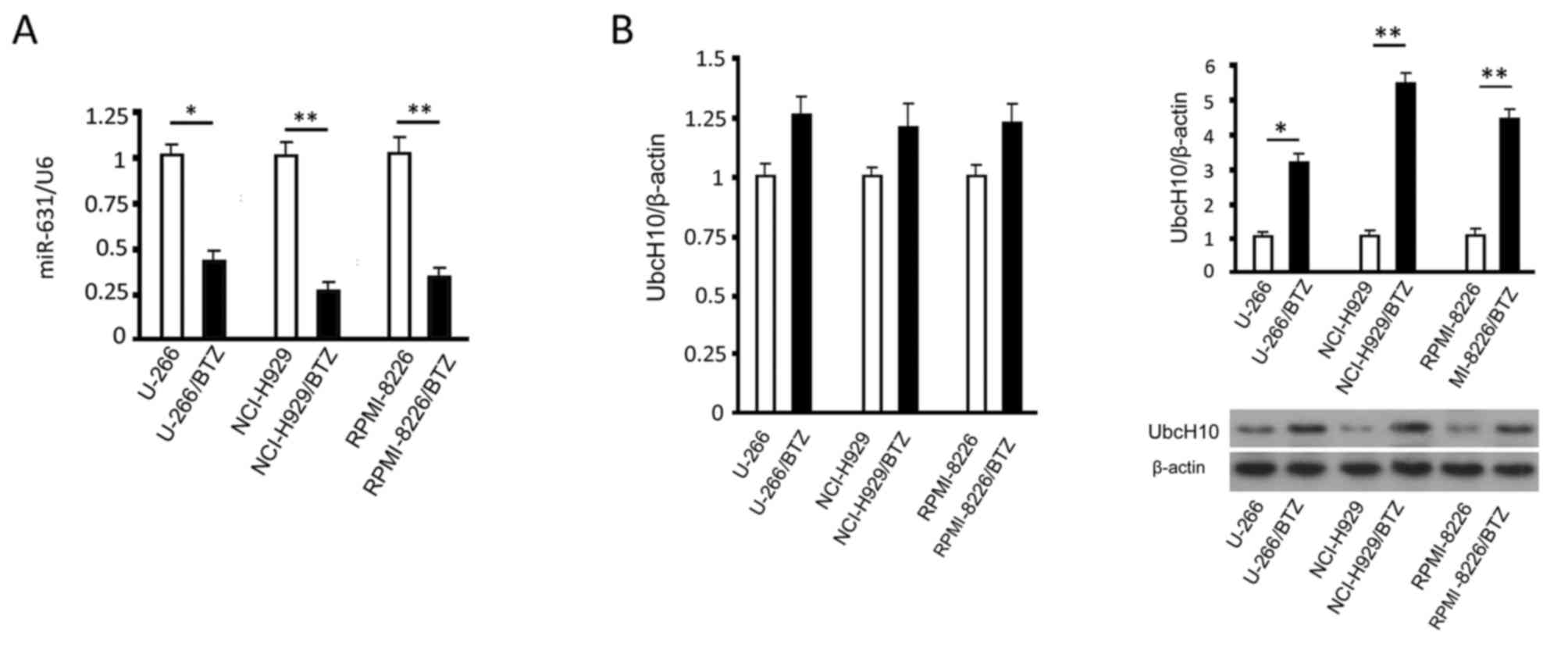
Quantitative results showed that the miR-631 levels
in resistant myeloma cells were significantly lower than those in
the parental cells (P<0.05) (Fig.
2A). Western blotting results indicated that the Ubc10 protein
levels were higher in the resistant myeloma cells than these levels
in the parental cells (P<0.05) (Fig.
2B). While UbcH10 mRNA increased in the resistant cells, there
was no significant difference between the resistant cells and their
parental cells (P>0.05) (Fig.
2B).
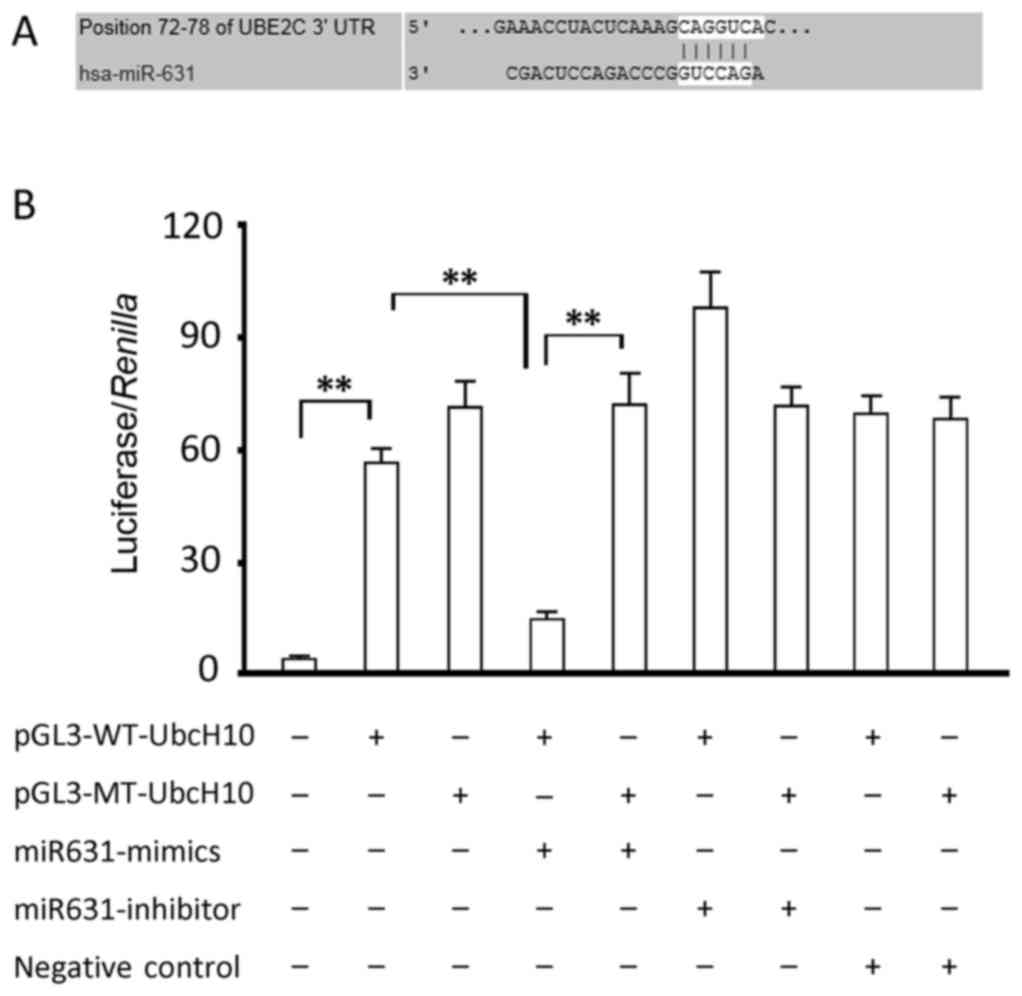
Prediction of hsa-miR-631 binding site
in UbcH10-3′UTR and verification by luciferase reporter assay
The TargetScan analysis demonstrated a theoretic
binding site (seed region) 5′-CAGGUC-3′ in the 3′UTR region of the
UbcH10 gene from bases 72–78. We cloned 3′UTR of UbcH10 into the
pGL-3 promoter luciferase reporter vector for verification
purposes. Luciferase activity detection indicated that the
miR631-mimic significantly inhibited intercellular luciferase
activity (P<0.05, compared to the group transfected with
luciferase expression vector alone), and that miR631 inhibitor
slightly increased the luciferase activity without reaching
statistical significance (Fig. 3).
However, neither produced a change in luciferase activity in cells
transfected with the luciferase expression vector carrying a
mutated binding site. Compared to the group transfected with only
luciferase expression vector, cells transfected with miR631-NC
showed a similar luciferase activity, indicating that RNA
transfection had no effect on luciferase activity. These data
suggest that the binding site of miR-631 in UbcH10 aligns with the
predicted sequence.
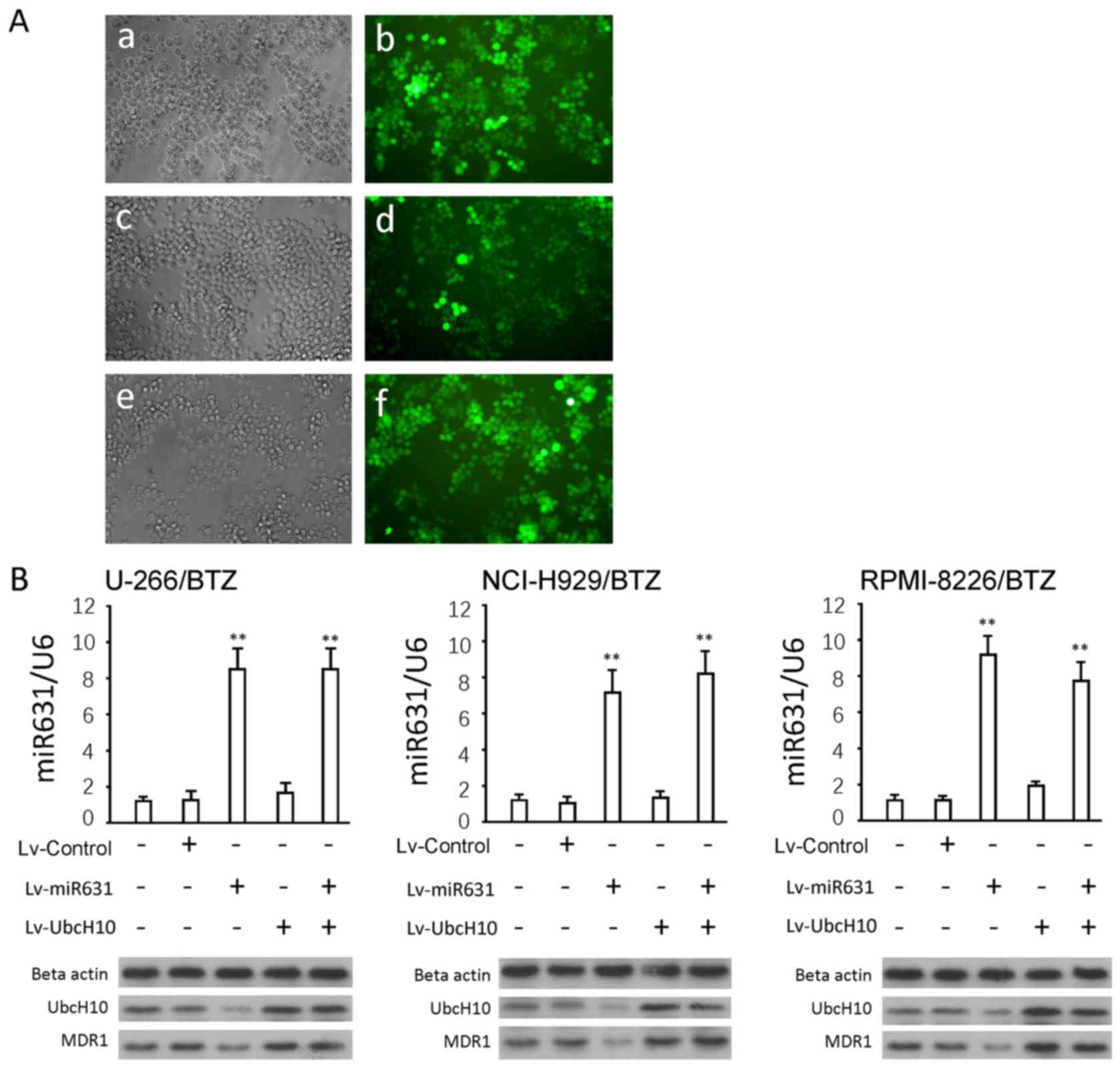
Overexpression of miR-631 and UbcH10
in resistant cells using a lentiviral approach
Genetic intervention was conducted in the three
resistant cell lines using a lentiviral approach. The gene delivery
efficiency was close to 100%, according to the GFP levels (Fig. 4A). Lv-miRNA631 infection
significantly increased mature miR-631 levels in both resistant
cell lines and parental cells (P<0.01) (Fig. 4B). Western blotting results
demonstrated that Lv-miR631 significantly decreased UbcH10 protein
(P<0.01 vs. control). Lv-UbcH10 infection significantly
increased UbcH10 protein (P<0.01 vs. control), as did the
combination of Lv-miR631 and Lv-UbcH10. No obvious difference was
found in these groups compared to the Lv-UbcH10 infection group
(P>0.05). The changes in the MDR1 proteins were similar to those
found in UbcH10.
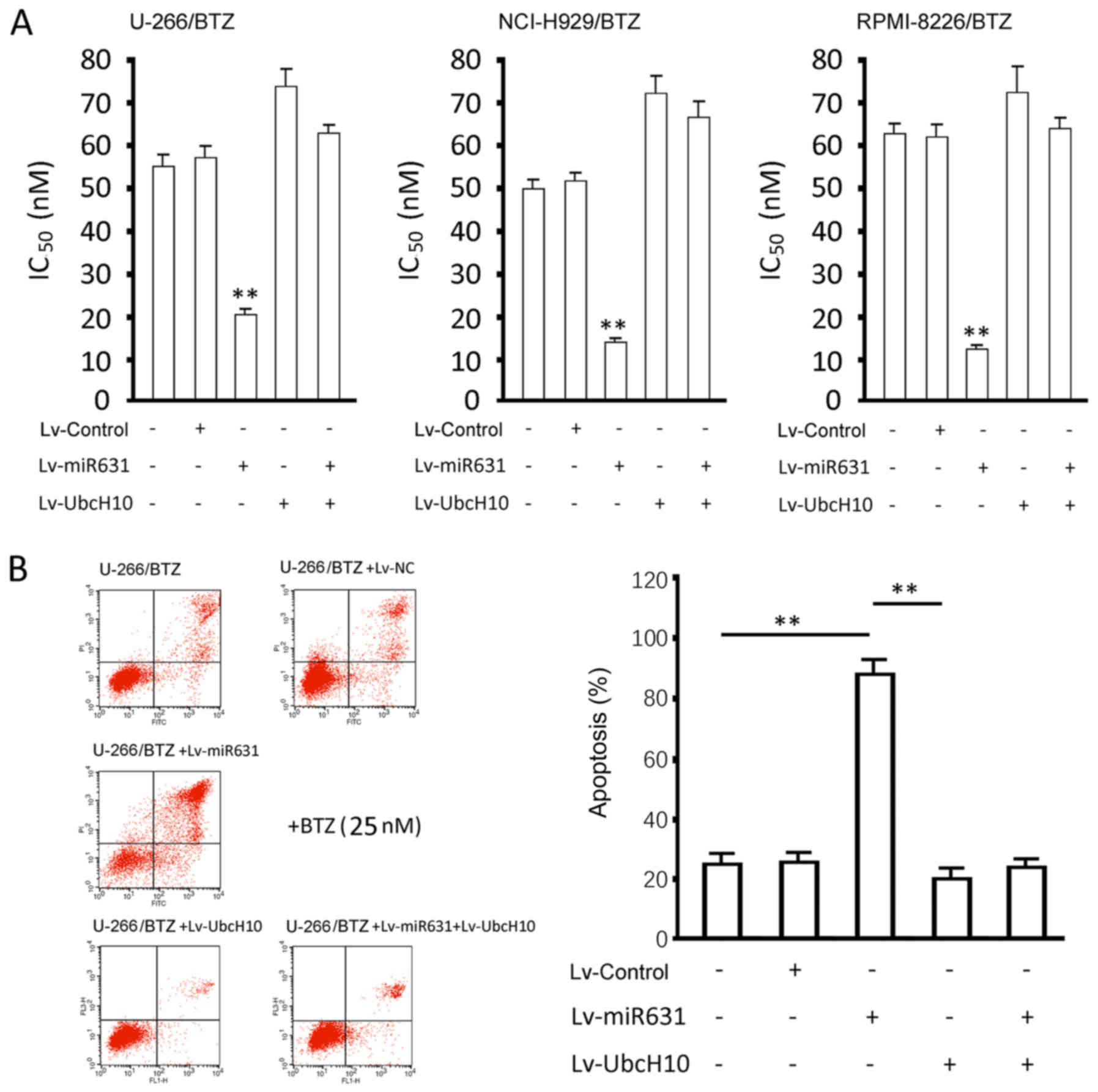
Measurements of IC50 values
and apoptosis
The overexpression of miR-631 significantly
decreased the IC50 values of the resistant cells to BTZ
(P<0.01), whereas UbcH10 overexpression slightly increased the
IC50 values, with no significant difference (P>0.05).
The overexpression of both miR-631 and UbcH10 significantly
increased the IC50 values (P<0.01, vs. control), with
no difference in the UbcH10 overexpression group (P>0.05)
(Fig. 5A).
The apoptosis rate (including early apoptosis and
late apoptosis) of the U-266/BTZ cells receiving genetic
interventions and undergoing the treatment of 25 nM BTZ for 48 h
were as follows (Fig. 5B): control,
24.63±3.02%; Lv-Control, 23.96±2.87%; Lv-miR631, 86.13±6.45%;
Lv-UbcH10, 18.73±3.92%; Lv-miR631 and Lv-UbcH10, 19.97±2.14%. The
apoptosis rate of the Lv-miR631-infected resistant cells was
significantly increased than the rates observed in the other four
groups (P<0.01), and there were no differences between the other
four groups (P>0.05). The effects of genetic intervention on
IC50 values in the NCI-H929/BTZ and RPMI-8226/BTZ cells
coincided exactly with those in the U-1996/BTZ cells (data not
shown).
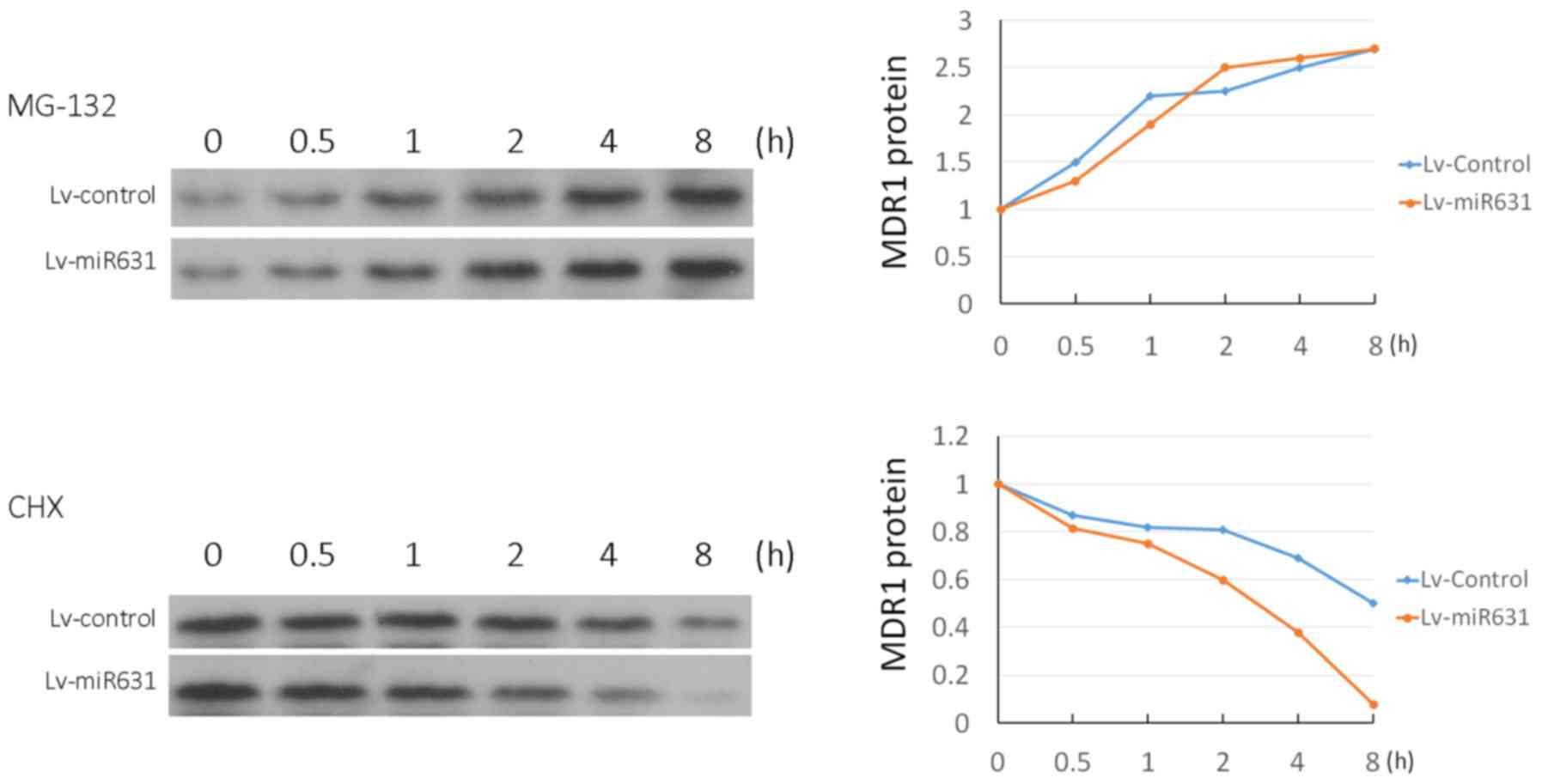
Effect of hsa-miR-631 overexpression
on the half-life of MDR1 protein in U-266/BTZ cells
When MG132 was used to inhibit protein degradation,
MDR1 increased from 0 to 8 h, while the rate of increase between
the groups showed no difference. The use of CHX to inhibit protein
synthesis resulted in a decrease in MDR1 from 0 to 8 h. The
reduction in MDR1 protein in the Lv-miR631 group was more rapid
than that in the control group (Fig.
6).
Discussion
Multiple myeloma (MM) is a common hematologic
malignancy, accounting for approximately 10% of all hematologic
malignancies (8). None of the
current treatments completely cure MM. Some commonly used
treatments for MM are chemotherapy, radiotherapy, hematopoietic
stem cell transplantation and novel molecular targeting agents
(9). Studies have shown that BTZ, a
reversible inhibitor of 26S proteasome chymotryptic activity,
increases drug sensitivity in dexamethasone-, melphalan-, and
thalidomide-resistant myeloma cells. This sensitivity increases
drug-induced apoptosis in resistant or non-resistant myeloma cells
(4). BTZ is the first FDA-approved
proteasome inhibitor for clinical treatment of MM, and it has a
high specificity and efficiency. With an increase in clinical use
of BTZ over time, some patients develop BTZ resistance and
experience relapse, ultimately reducing the efficacy of BTZ
(10). Therefore, the search for
approaches to prevent the development of resistance or eradicate
malignant plasma cells has become a hot research topic. To date,
there are few studies on BTZ resistance, the mechanism of which
remains unknown.
Although chemotherapy is an important cancer
treatment, drug resistance creates significant challenges to
eliminate cancer cells and cause poor prognosis. Many studies have
shown that miRNAs are involved in cancer initiation and development
by regulating apoptosis, proliferation, differentiation and
metastasis as well as influencing drug resistance via their target
genes (11,12). A study of differentially expressed
miRNAs and the affiliated regulation of drug resistance-associated
genes may contribute to understanding the mechanisms of drug
resistance (13). All mutations,
misexpression and abnormal processing of miRNAs impair function and
can result in abnormal target gene expression. When a target gene
is involved in the tumor cell response to drugs, gene
chemosensitivity will be changed. Akao et al found that
miR-145 and miR-34a improved the sensitivity of DLD-1 cells to 5-Fu
(14). A recent study by Bai et
al showed that miR-21 is overexpressed in various cancers and
is involved in chemoresistance. For example, miR-21 mediates
daunorubicin (DNR) resistance in K562 cells, and miR-21 depletion
increases the sensitivity of K562/DNR cells to DNR. In addition,
miR-21-mediated drug resistance is associated with the PI3k/Akt
pathway and the PTEN protein (15).
miR-200c regulates the TGF-β/ZEB1 pathway via its target ZEB1,
resulting in trastuzumab resistance in breast cancer (16). These studies demonstrate that miRNAs
are closely related to drug resistance and regulation. Studies
indicated that hsa-miR-631, which is located on chromosome 15, is
abnormally expressed in pancreatic cancer and decreases the
migration and invasion of pancreatic cancer by inhibiting one of
its target genes, ZAP70 (17).
However, there are no studies on the association of miR-631 with
cancer drug resistance.
Approximately 80–90% of cell proteins are degraded
by the ubiquitin-proteasome pathway (UPP) (18); thus, up to 90% of cell proteins may
be a target for BTZ. The study of abnormal degradation of UPP
pathway proteins may facilitate an understanding of drug resistance
in MM. UbcH10, also known as UBE2C, is a gene located at 20q13.12
and has a key role in UPP-mediated protein degradation (19). Research has demonstrated that UbcH10
is highly expressed in many types of cancers, including breast
cancer, ovarian cancer, thyroid cancer, oesophageal cancer,
lymphoma, MM and hepatocellular carcinoma (20,21).
In addition, UbcH10 overexpression is often associated with a high
cancer grade, high proliferation and poor tumor prognosis (22,23).
There are few studies on the relation between UbcH10 and
chemosensitivity. Zhao et al (24) found that UbcH10 depletion increased
the sensitivity of lung cancer cells to 5-Fu. Wang et al
(25) found that UbcH10 silencing
reversed the cyclophosphamide resistance in breast cancer cell
lines. These results suggest that UbcH10 may be a gene therapy
target due to an influence on drug resistance. As a key part of
UPP-mediated protein degradation, UbcH10 can degrade regulators
involved in cell cycle regulation, apoptosis and DNA transcription
and repair (26). In the UPP, E1
(ubiquitin activating enzyme) first activates ubiquitin and
transfers it to E2 (ubiquitin conjugating enzyme). E2, together
with E3 (ubiquitin ligating enzyme), then recognizes the substrate
protein. The ubiquitin is then either transferred directly or
transferred to E3, where the substrate protein is degraded in the
26S proteasome (27). Zhao et
al (24) found that drug
resistance caused by UbcH10 knockdown may be associated with the
MDR1 gene. Further studies on regulatory relationships between
UbcH10 and MDR1 have not been conducted. The main objectives of the
present study were to determine the association of UbcH10 and BTZ
resistance in myeloma cells and its involvement in MDR1
expression.
We first established BTZ-resistant MM cell lines and
found that while UbcH10 protein was higher in the resistant cells
than in parental cells, there was no significant difference in
UbcH10 mRNA between the groups. This result suggests that the high
UbcH10 expression found in resistant cell lines may be due to
inactivated post-transcriptional regulation. Therefore, we sought
miRNAs that may cause abnormal expression of UbcH10. Using
bioinformatic analysis tools, we obtained miRNAs that contain a
binding site in the 3′UTR region of the UbcH10 gene and assessed
the miRNAs in both resistant cells and their parental cells. The
target miRNA hsa-miR-631 was identified by analysis and a
correlation of miRNA levels. Luciferase reporter assay verified
that hsa-miR-631 may inhibit translation by binding to
UbcH10-3′UTR. An increase in UbcH10, stemming from a decrease in
miR-631 in BTZ-resistant myeloma cells, may cause BTZ resistance.
The overexpression of miR-631 in three BTZ-resistant cell lines
reduced BTZ IC50 values. Furthermore, overexpression
enabled either resensitization or allowed low BTZ concentrations to
induce apoptosis in these cells. We next explored how UbcH10
expression altered the sensitivity to BTZ. The data showed that
UbcH10 was positively related to MDR1 expression, as it inhibits
MDR1 ubiquitination. These results collectively suggest the
presence of a miR-631/UbcH10/MDR1 pathway during the development of
BTZ resistance in MM. In this study, we also verified the
specificity of the pathway, as miR-631 overexpression reversed the
resistance to BTZ by inhibiting UbcH10 expression while promoting
the ubiquitination of MDR1 and reducing MDR1 protein levels.
Moreover, overexpression of exogenous UbcH10 under the control of a
CMV promoter while free of the miR-631 binding site inhibited the
reversion of BTZ resistance through miR-631 overexpression.
Although the present study demonstrated reduced
miR-631 in BTZ-resistant MM cell lines for the first time and
elucidated that miR-631 regulates MDR1 via UbcH10, there remain two
questions to be solved in the development of BTZ resistance in
myeloma cells. First, we must ascertain whether BTZ resistance in
myeloma cells begins with the low expression of miR-631. Second,
studies must be carried out to ascertain how UbcH10 regulates MDR1
expression and whether the regulation is direct or not. Regarding
the first question, a possible explanation may be that BTZ
induction altered the activity of a certain nuclear transcription
factor that regulates the transcription of miR-631. Studies have
revealed that BTZ resistance is mainly associated with the NF-κB
pathway, heat-shock proteins or overexpression of BCL family
members. We, therefore, plan to study the mechanism upstream of
miR-631 by analyzing differentially expressed transcription factors
in cells before and after BTZ treatment with a microarray for
transcription factors. For the second question, we may come to a
deduction from our current knowledge. It is known that UbcH10
recognizes the substrate and transfers ubiquitin activated by E1 to
the target protein, which seems unable to explain the positive
correlation we found between UbcH10 and MDR1 in BTZ-resistant
cells. In the ubiquitin-proteasome pathway, the target protein may
be labelled with K48- or K11-linked polyubiquitin chains. In
addition, a previous study showed that polyubiquitin chains
comprised of two ubiquitin units result in more effective
degradation by proteasome (28).
Therefore, we speculate that low expression of UbcH10 (not
knockout) tends to produce two-ubiquitin chains, which, compared
with four-ubiquitin chains, accelerates the degradation of
p-glycoprotein. It is only a possible hypothesis. Some E2s play
their role in specific cellular process, further study is needed to
illustrate all roles of E2s.
In conclusion, we identified that the
miR-631/UbcH10/MDR1 pathway is involved in the development of BTZ
resistance in myeloma cells. In addition, in the present study, an
increased sensitivity of myeloma cells to BTZ was accomplished by
overexpression of miR-631 using genetic engineering. These results
may help resolve the mechanisms of BTZ resistance. miR-631 may be
used as a genetic marker for the selection of therapy regimes in
MM. However, for patients with low miR-631 levels, therapies other
than those based on BTZ may be more effective.
Acknowledgements
This study was supported in part by the National
Natural Sciences Fund Project of China (NSFC no. 81470360,
81372529, 81372543 and 81300391).
References
|
1
|
Barlogie B, Shaughnessy J, Tricot G,
Jacobson J, Zangari M, Anaissie E, Walker R and Crowley J:
Treatment of multiple myeloma. Blood. 103:20–32. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anderson KC: Therapeutic advances in
relapsed or refractory multiple myeloma. J Natl Compr Canc Netw 11
(Suppl 5). 676–679. 2013.
|
|
3
|
Accardi F, Toscani D, Bolzoni M, Palma B
Dalla, Aversa F and Giuliani N: Mechanism of action of bortezomib
and the new proteasome inhibitors on myeloma cells and the bone
microenvironment: impact on myeloma-induced alterations of bone
remodeling. BioMed Res Int. 2015:1724582015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Adam Z, Sčudla V, Krejčí M, Cermáková Z,
Pour L and Král Z: Treatment of AL amyloidosis in 2012; the benefit
of new drugs (bortezomib, thalidomide, and lenalidomide). Summary
of published clinical trials. Vnitr Lek. 59:37–58. 2013.(In Czech).
PubMed/NCBI
|
|
5
|
Adachi Y, Yoshio-Hoshino N and Nishimoto
N: Gene therapy for multiple myeloma. Curr Gene Ther. 8:247–255.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tétreault N and De Guire V: miRNAs: Their
discovery, biogenesis and mechanism of action. Clin Biochem.
46:842–845. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parker JS, Roe SM and Barford D: Molecular
mechanism of target RNA transcript recognition by Argonaute-guide
complexes. Cold Spring Harb Symp Quant Biol. 71:45–50. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Abe M: Multiple myeloma. Nihon Rinsho.
67:991–995. 2009.(In Japanese). PubMed/NCBI
|
|
9
|
Abdi J, Chen G and Chang H: Drug
resistance in multiple myeloma: Latest findings and new concepts on
molecular mechanisms. Oncotarget. 4:2186–2207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abidi MH, Gul Z, Abrams J, Ayash L, Deol
A, Ventimiglia M, Lum L, Mellon-Reppen S, Al-Kadhimi Z,
Ratanatharathorn V, et al: Phase I trial of bortezomib during
maintenance phase after high dose melphalan and autologous stem
cell transplantation in patients with multiple myeloma. J
Chemother. 24:167–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abba M, Mudduluru G and Allgayer H:
MicroRNAs in cancer: Small molecules, big chances. Anticancer
Agents Med Chem. 12:733–743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Azmi AS, Bao B and Sarkar FH: Exosomes in
cancer development, metastasis, and drug resistance: A
comprehensive review. Cancer Metastasis Rev. 32:623–642. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Allen KE and Weiss GJ: Resistance may not
be futile: microRNA biomarkers for chemoresistance and potential
therapeutics. Mol Cancer Ther. 9:3126–3136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Akao Y, Khoo F, Kumazaki M, Shinohara H,
Miki K and Yamada N: Extracellular disposal of tumor-suppressor
miRs-145 and −34a via microvesicles and 5-FU resistance of human
colon cancer cells. Int J Mol Sci. 15:1392–1401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bai H, Xu R, Cao Z, Wei D and Wang C:
Involvement of miR-21 in resistance to daunorubicin by regulating
PTEN expression in the leukaemia K562 cell line. FEBS Lett.
585:402–408. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ,
Huang X, Zhao J, Gu B, Zheng GX, Yang AG, et al: MiR-200c
suppresses TGF-β signaling and counteracts trastuzumab resistance
and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int J
Cancer. 135:1356–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fu D, Liu B, Zang LE and Jiang H:
MiR-631/ZAP70: A novel axis in the migration and invasion of
prostate cancer cells. Biochem Biophys Res Commun. 469:345–351.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eldridge AG and O'Brien T: Therapeutic
strategies within the ubiquitin proteasome system. Cell Death
Differ. 17:4–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okamoto Y, Ozaki T, Miyazaki K, Aoyama M,
Miyazaki M and Nakagawara A: UbcH10 is the cancer-related E2
ubiquitin-conjugating enzyme. Cancer Res. 63:4167–4173.
2003.PubMed/NCBI
|
|
20
|
Lin J, Raoof DA, Wang Z, Lin MY, Thomas
DG, Greenson JK, Giordano TJ, Orringer MB, Chang AC, Beer DG, et
al: Expression and effect of inhibition of the
ubiquitin-conjugating enzyme E2C on esophageal adenocarcinoma.
Neoplasia. 8:1062–1071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Troncone G, Guerriero E, Pallante P,
Berlingieri MT, Ferraro A, Del Vecchio L, Gorrese M, Mariotti E,
Iaccarino A, Palmieri EA, et al: UbcH10 expression in human
lymphomas. Histopathology. 54:731–740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Berlingieri MT, Pallante P, Sboner A,
Barbareschi M, Bianco M, Ferraro A, Mansueto G, Borbone E,
Guerriero E, Troncone G, et al: UbcH10 is overexpressed in
malignant breast carcinomas. Eur J Cancer. 43:2729–2735. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Berlingieri MT, Pallante P, Guida M, Nappi
C, Masciullo V, Scambia G, Ferraro A, Leone V, Sboner A,
Barbareschi M, et al: UbcH10 expression may be a useful tool in the
prognosis of ovarian carcinomas. Oncogene. 26:2136–2140. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao L, Jiang L, Wang L, He J, Yu H, Sun
G, Chen J, Xiu Q and Li B: UbcH10 expression provides a useful tool
for the prognosis and treatment of non-small cell lung cancer. J
Cancer Res Clin Oncol. 138:1951–1961. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Pan YH, Shan M, Xu M, Bao JL and
Zhao LM: Knockdown of UbcH10 enhances the chemosensitivity of dual
drug resistant breast cancer cells to epirubicin and docetaxel. Int
J Mol Sci. 16:4698–4712. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Doherty FJ, Dawson S and Mayer RJ: The
ubiquitin-proteasome pathway of intracellular proteolysis. Essays
Biochem. 38:51–63. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Summers MK, Pan B, Mukhyala K and Jackson
PK: The unique N terminus of the UbcH10 E2 enzyme controls the
threshold for APC activation and enhances checkpoint regulation of
the APC. Mol Cell. 31:544–556. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stieglitz B, Rana RR, Koliopoulos MG,
Morris-Davies AC, Schaeffer V, Christodoulou E, Howell S, Brown NR,
Dikic I and Rittinger K: Structural basis for ligase-specific
conjugation of linear ubiquitin chains by HOIP. Nature.
503:422–426. 2013. View Article : Google Scholar : PubMed/NCBI
|