Introduction
Osteosarcoma (OS) is a common primary malignancy of
bone that occurs mostly in children or adolescents with more than
26,400 new cases every year (1,2).
Chemotherapy and surgery, or their combination, is the prevalent
treatment method and has significantly improved the prognosis of OS
(3,4). For example, the five-year survival
rate of OS without distal metastasis after chemotherapy could reach
70% (5). While, poor prognosis was
frequently observed in the metastatic patients. Moreover, the
toxicity of some chemotherapeutic drugs cannot be efficiently
prevented and their efficacy could be largely different in similar
patients (6,7). Therefore, early diagnosis and
personalized therapy would be the alternative methods for the
improvement of prognosis of primary as well as the metastatic
OS.
In the past decades, many biomarkers have been
identified to be associated with the progression of OS and one of
the most well known targets is mammalian target of rapamycin
(mTOR), a serine/threonine protein kinase, which could contribute
to the development of many types of cancers (8). mTOR was regulated by various molecules
in OS, downregulation of RSK2 was associated with the inactivating
of mTOR signaling pathways and could influence the progression of
OS (9); the variation of
microRNA-99a could induce the differential expression of mTOR in OS
and could influence its development (10). Besides, the mutations of some genes
were also shown to affect the susceptibility of OS, such as the
polymorphisms of murine double minute 2 (MDM2) (rs1690916 and
rs2279744) were associated with the risk of OS (11); the mutation of P15 gene was
significantly associated with the initiation of OS (12). In addition, some inhibitors have
been developed against these biomarkers. Icariside II is a natural
mTOR inhibitor which could destroy the energy balance through
inhibiting mTORC1-4E-BP1 axis to inhibit the progression of sarcoma
(13). While, further studies are
still needed for the understanding of the mechanisms of OS and the
improvement of its prognosis.
In this study, the DNA methylation profile was also
included besides the gene and microRNA (miRNA) expression profiles,
which play important roles in the regulation of gene expression.
DNA hypermethylation in promoter is a key characteristic in cancer
and it was considered to be able to silence the tumor suppressor
genes to induce the development of cancer (14–16).
Some DNA methylation was also observed in OS and found to be
associated with its progression, the methylation level of RECK was
gradually increased with the development of OS, while the opposite
trend of its expression profile was observed (17). In the study of Sonaglio et
al, the differential ESR1 and p14ARF gene methylation profiles
were also identified as prognostic indicators in OS (18). The combined analysis of gene, miRNA
expression and DNA methylation profiles would be helpful for the
identification of novel biomarkers for OS and its early diagnosis
and treatment.
Material and methods
Microarray datasets
The microarray datasets in this study were
downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). The gene
expression (GSE36001) and DNA methylation profiles (GSE36002) were
deposited by Kresse et al and composed with the same
samples: 19 OS cell lines (case group) and 6 normal samples
(control group, 2 osteoblast and 4 bone samples) (19). While, the miRNA expression profiles
(GSE28423) were provided by Namløs et al, compared with the
gene expression and DNA methylation profiles, only 2 osteoblast
were absent and the other samples were exactly the same, i.e. 19 OS
cell lines (case group) and 4 normal bone samples (control group)
(20). The samples for the three
types of profiles were comparable and could be used for the
subsequent analysis.
Microarray analysis
R Bioconductor packages were used for the
preprocessing of the three types of microarray datasets. For gene
and miRNA expression profiles, raw datasets were normalized based
on the preprocess Core package and the differential expression
genes (DEGs) and miRNAs (DEMIs) were screened out via the limma
package through the thres-holds of fold change >2 or <0.5 and
adjusted P-value of <0.05. IMA package was adopted for the
preprocessing of raw DNA methylation profile and the identification
of differential methylation sites (DMS) and in this study, the
methylation sites satisfied the criteria of log2 (fold change)
>0.5 or <-0.5 and adjusted P-value <0.05 were screened
out.
Functional enrichment analysis
Based on the Database for Annotation, Visualization
and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov), the Gene Ontology (GO)
terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
of DEGs and genes contained DMS (DEMs) were obtained. The
thresholds used here is P-value <0.05.
miRNA-gene regulation network
Besides the promoter methylation, miRNA is another
important regulator for gene expression. In this study, the target
genes of DEMIs were identified through the TargetScan, a database
of the miRNA-gene pairs obtained by various methods, such as
biological experiments and bioinformatics prediction. Furthermore,
we selected only the pairs that contained the DEMIs and overlapping
genes of DEGs and DEMs. The Cytoscape, a network visualization and
analysis software, was used for the construction of miRNA-gene
regulation network.
Results
DEGs, DEMIs and DMS
Compared with the control group, a total of 583
genes were found to be differentially expressed in the case group,
which contained 417 downregulated and 166 upregulated genes.
Besides, we identified 1051 DMS in the case group which
corresponding to 827 unique genes and strikingly, 1018 DMS, ~96.9%
of all the DMS, were found to be hypermethylation. In total, 231
DEMIs were identified in case group and 117 were found to be
downregulated. The top 10 significantly differentially expressed
genes and methylation sites are shown in Table I and in Table II the top 10 significantly
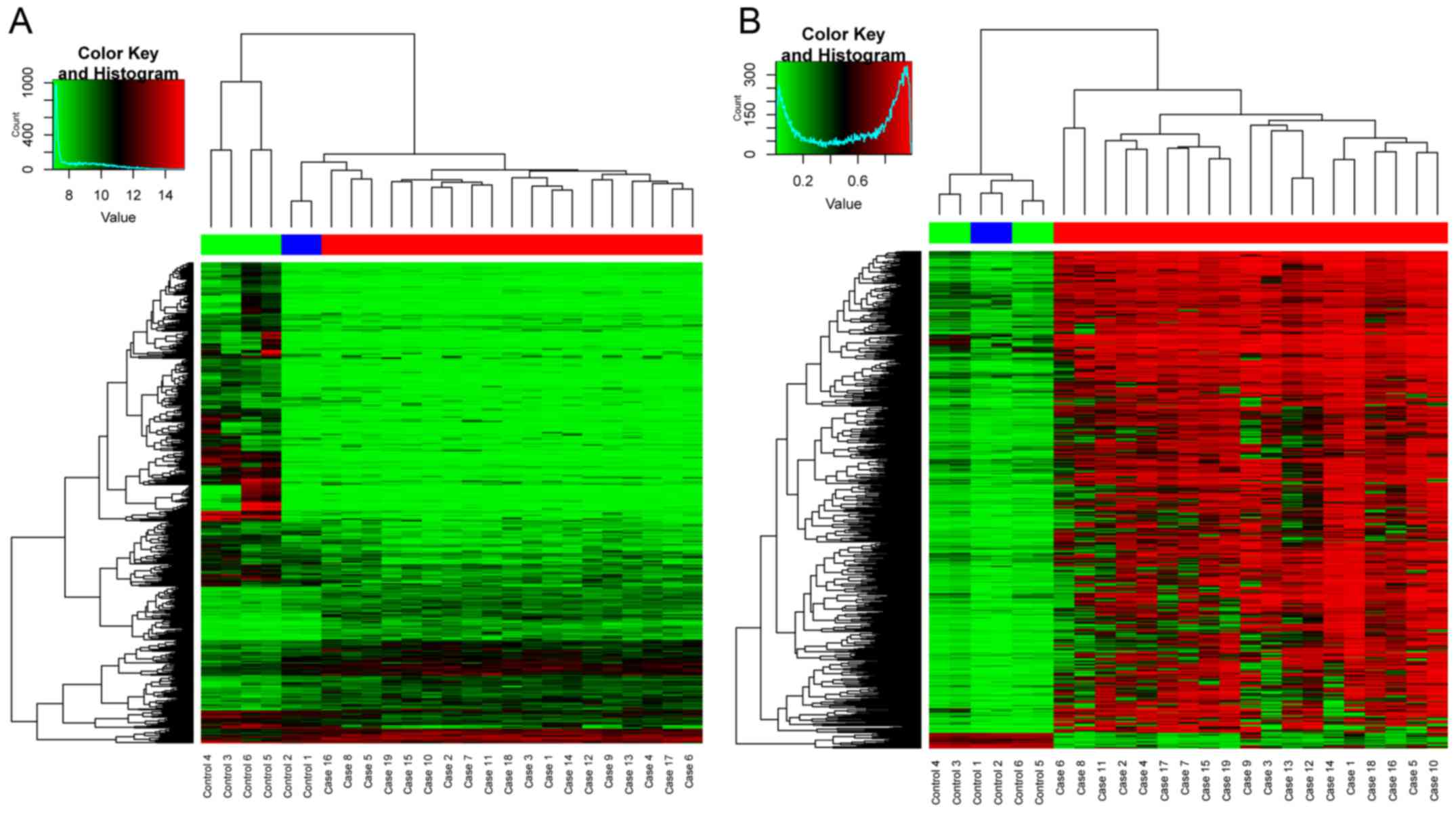
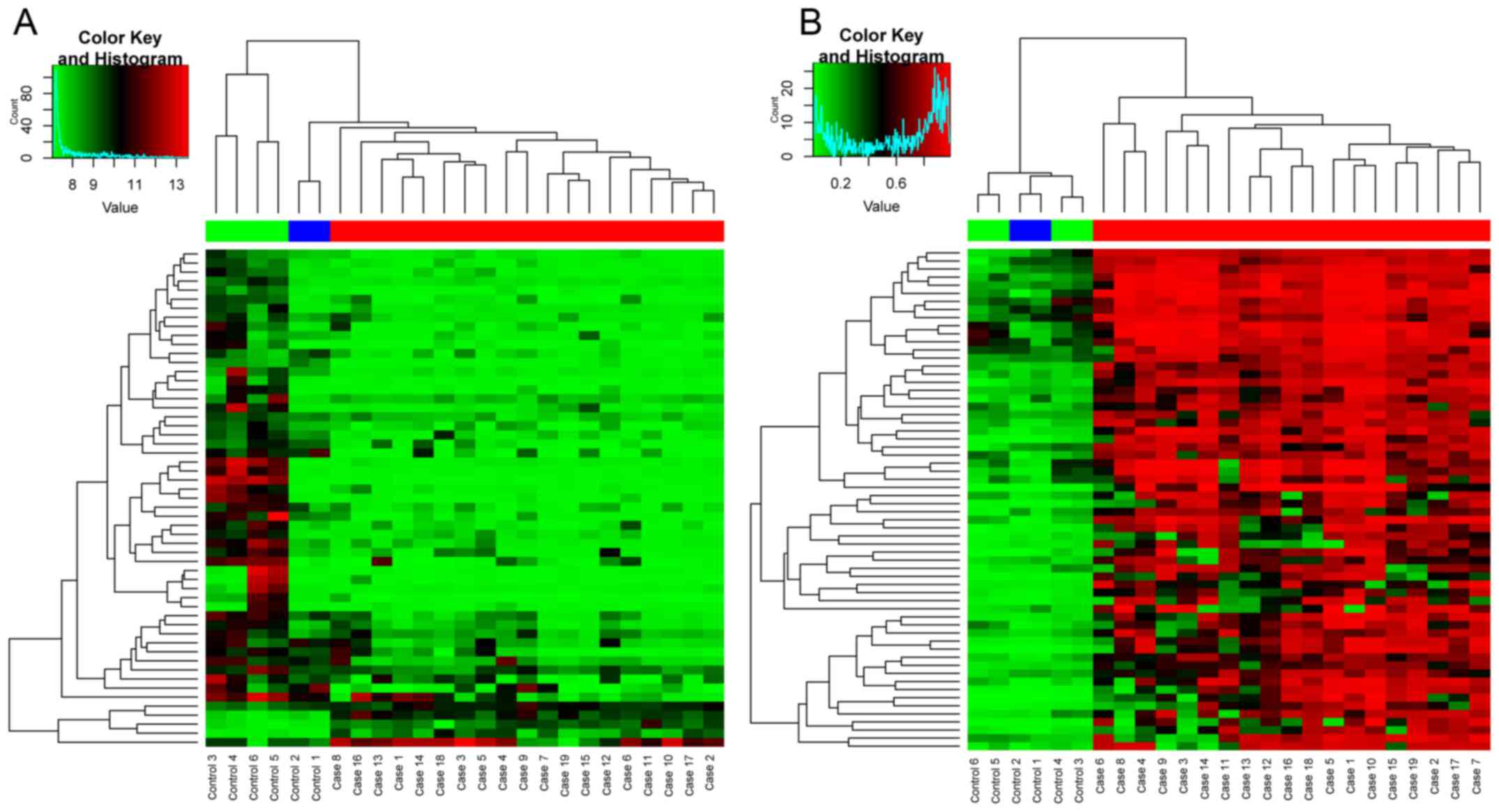
differentially expressed miRNAs. The supervised hierarchical
cluster of DEGs and DMS based on gplots package are shown in
Fig. 1. The supervised cluster of
DEMIs are shown in Fig. 2. As shown
in Figs. 1 and 2, the OS and control samples can be
separated into their types, which indicated the reliability of the
DEGs, DEMIs and DMS for the following analysis.
 | Table I.Top 10 significantly differential
expression genes and methylated sites. |
Table I.
Top 10 significantly differential
expression genes and methylated sites.
| DEG | Log2FC | Adj.P.Value | DMS | Log2FC | Adj.P.Value |
|---|
| CMKLR1 | −1.03 |
1.16×10−8 | cg03874199 | 0.90 |
1.42×10−22 |
| PTGS1 | −1.67 |
3.62×10−8 | cg18765542 | 0.87 |
2.07×10−20 |
| RPS28 | 1.15 |
5.31×10−8 | cg23130254 | 0.86 |
9.85×10−19 |
| HOXB7 | 2.09 |
9.50×10−8 | cg06274159 | 0.84 |
5.82×10−18 |
| UQCRHL | 1.91 |
9.59×10−8 | cg09522147 | 0.63 |
3.07×10−16 |
| RHPN2 | 2.15 |
4.70×10−7 | cg01615704 | 0.83 |
3.89×10−16 |
| RPS27 | 1.33 |
2.73×10−6 | cg00208967 | 0.73 |
4.01×10−16 |
| SRGN | −5.17 |
2.73×10−8 | cg08668790 | 0.87 |
1.73×10−15 |
| FOXF2 | 2.32 |
3.64×10−8 | cg09009380 | 0.67 |
1.78×10−15 |
| RPL23 | 1.70 |
4.87×10−8 | cg22396755 | 0.82 |
1.99×10−15 |
 | Table II.Top 10 significantly differential
expression miRNAs. |
Table II.
Top 10 significantly differential
expression miRNAs.
| DEMI | Log2FC | Adj.P.Value |
|---|
| hsa-miR-144 | −6.45 |
5.94×10−16 |
| hsa-miR-638 | −3.66 |
1.43×10−12 |
|
hsa-miR-1225-5p | −4.84 |
1.43×10−12 |
| hsa-miR-451 | −14.77 |
1.30×10−11 |
|
hcmv-miR-UL70-3p | −4.84 |
8.48×10−11 |
| hsa-miR-486-5p | −5.36 |
1.20×10−10 |
| hsv1-miR-LAT | −4.54 |
3.19×10−10 |
| hsa-miR-188-5p | −3.94 |
3.19×10−10 |
| hsa-miR-671-5p | −4.80 |
5.21×10−10 |
| hsa-miR-134 | −4.37 |
6.15×10−10 |
Correlation of gene expression and DNA
methylation profiles
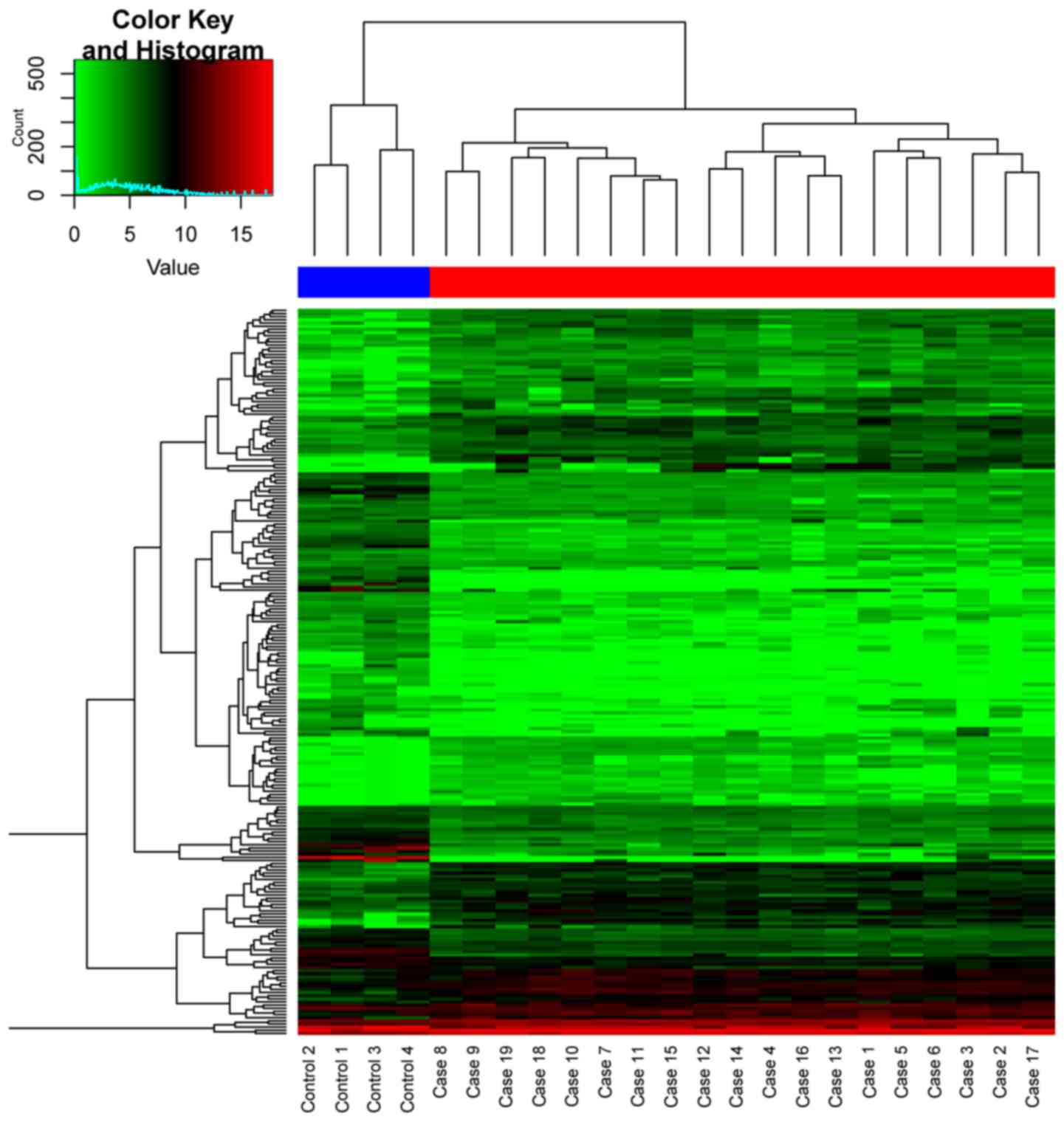
A total of 56 overlaps were identified between DEGs
and DEMs. The supervised cluster of the overlapping genes and their
corresponding DMS are shown in Fig.
3. As expected, significant negative correlation with the
minimum of −0.924 was observed between most of the methylation
profiles of DMS and their corresponding expression profile of the
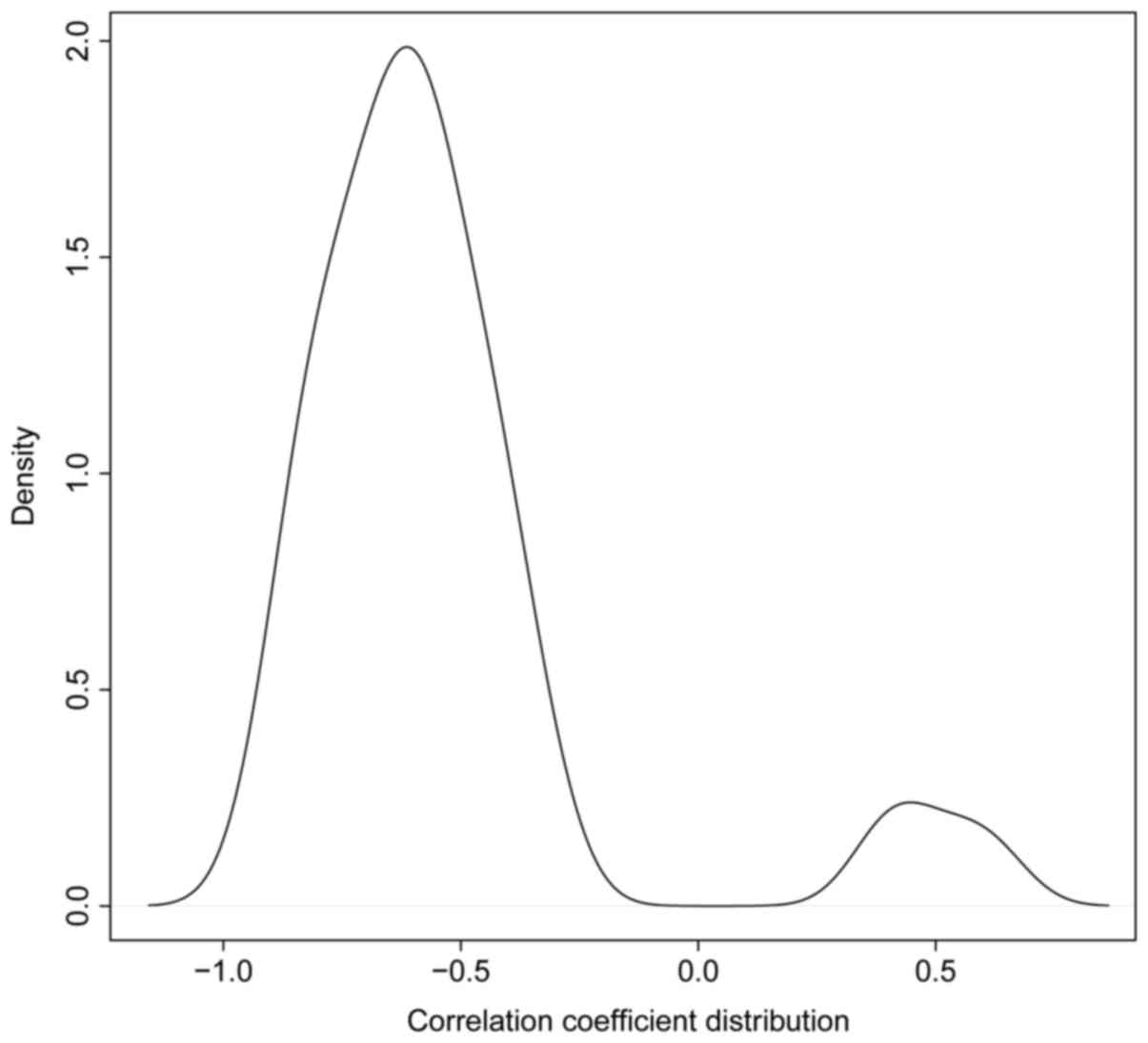
overlaps. Fig. 4 illustrates the
distribution of the correlation coefficients.
Enriched functions
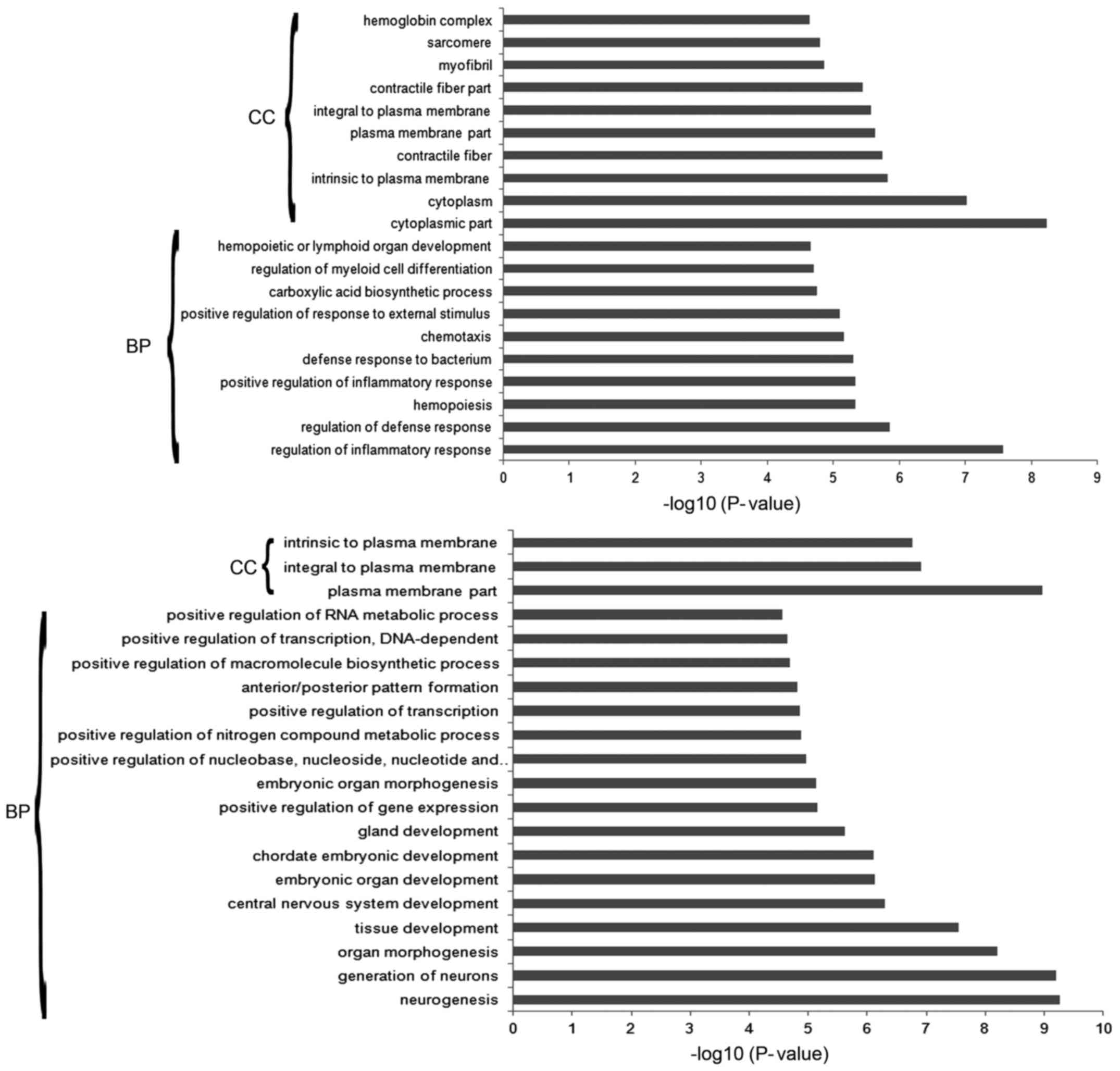
A total of 180 and 198 GO terms were found to be
enriched in the DEGs and DEMs, respectively and the top 20
significantly enriched GO terms of DEGs and DEMs are shown in
Fig. 5. The cellular component of
plasma/cytoplasm were found to be enriched in both DEGs and DEMs.
While the biological processes related to inflammatory/immune
response, cell proliferation, and so on, were enriched in DEGs, and
DEMs were mainly associated with the biological process of
regulation of gene expression, tissue and embryonic development.
Besides, 19 and 6 KEGG pathways were also enriched in DEGs and
DEMs, respectively (Tables III
and IV). Similar with the GO
terms, KEGG pathways of DEGs were mainly involved in inflammatory
and immune processes, while the enriched pathways of DEMs were
related to cancer development and cytokine signaling pathways.
 | Table III.KEGG pathways enriched in DEGs. |
Table III.
KEGG pathways enriched in DEGs.
| Pathway name | Count | P-value |
|---|
| Asthma | 11 |
3.40×10−7 |
| Intestinal immune
network for IgA production | 10 |
3.30×10−4 |
| Viral
myocarditis | 12 |
3.60×10−4 |
| Glycine, serine and
threonine metabolism | 8 |
4.20×10−4 |
| Hematopoietic cell
lineage | 13 |
5.20×10−4 |
| Systemic lupus
erythematosus | 14 |
5.60×10−4 |
| Leukocyte
transendothelial migration | 15 |
9.80×10−4 |
| Cell adhesion
molecules (CAMs) | 16 |
1.00×10−3 |
| Allograft
rejection | 8 |
1.10×10−3 |
| Graft-versus-host
disease | 8 |
1.80×10−3 |
| Type I diabetes
mellitus | 8 |
2.80×10−3 |
| Natural killer cell
mediated cytotoxicity | 15 |
3.10×10−3 |
| Complement and
coagulation cascades | 10 |
4.10×10−3 |
| Autoimmune thyroid
disease | 8 |
8.40×10−3 |
| Nitrogen
metabolism | 5 |
2.00×10−2 |
| One carbon pool by
folate | 4 |
3.50×10−2 |
| Antigen processing
and presentation | 9 |
3.70×10−2 |
| Tight junction | 12 |
4.50×10−2 |
| Pathogenic
Escherichia coli infection | 7 |
4.60×10−2 |
 | Table IV.KEGG pathways enriched in DEMs. |
Table IV.
KEGG pathways enriched in DEMs.
| Pathway name | Count | P-value |
|---|
| Neuroactive
ligand-receptor interaction | 27 |
2.86×10−4 |
| Pathways in
cancer | 29 |
2.62×10−3 |
| Small cell lung
cancer | 10 |
2.13×10−2 |
| Cytokine-cytokine
receptor interaction | 21 |
3.18×10−2 |
| Amyotrophic lateral
sclerosis (ALS) | 7 |
4.41×10−2 |
| Adipocytokine
signaling pathway | 8 |
4.50×10−2 |
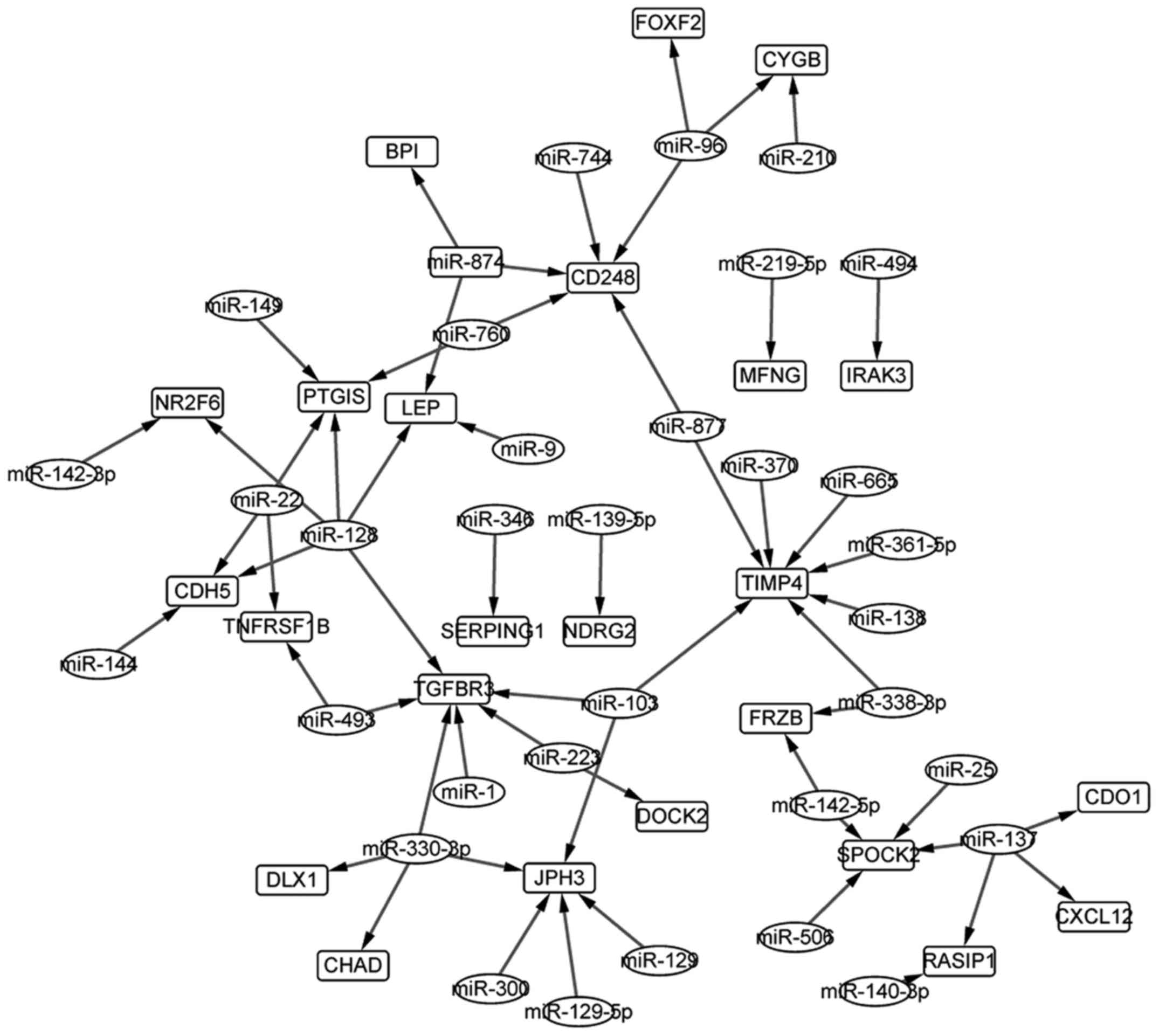
miRNA-gene regulation network
Through the TargetScan, we screened out 8596
miRNA-gene pairs that contained DEMIs, and 58 pairs of which were
found to be involved the overlapping genes of DEGs and DEMs. The
miRNA-gene regulation network composed of the 58 pairs shown in
Fig. 6. A total of 34 miRNAs and 24
genes were contained in the network and hsa-miR-128 regulated 5 of
the 24 genes. Besides, TIMP4 (TIMP metallopeptidase inhibitor 4),
found to be associated with the progression of breast cancer by
many studies, was regulated by 7 miRNAs in the miRNA-gene network,
that might indicate its important role in OS.
Discussion
OS is one of the most common malignancies in
children and adolescents with a relative high survival rate after
chemotherapy or surgery in primary tumors while not in the
metastatic ones. Identification of novel biomarkers would be
helpful in its early diagnosis and improvement of prognosis. In
this study, three types of microarray datasets: gene expression
profile, miRNA expression profile and DNA methylation profile were
downloaded for the GEO and analyzed for their relationship with the
progression of OS. This would benefit the understanding of the
mechanisms of OS and the development of novel therapeutics.
Herein, ~71.5% and 3.1% DEGs and DMS were found to
be downregulated in the OS group compared with the control ones
respectively. The expression profiles and the corresponding
methylation profiles of the overlapping genes between DEGs and DEMs
exhibited striking negative correlation as shown in Fig. 4, which might indicate their
antitumor role. In particular, LXN (latexin), a gene encoding the
only known protein inhibitor of zinc-dependent
metallocarboxypetidases, were found to be downregulated in many
types of tumors, as well as in OS in our study and it exhibited
significantly negative correlation between expression and
methylation profile (cor −0.92, P-value 4.25×10−11). In
hepatocellular carcinoma (21,22),
gastric carcinomas (23),
pancreatic cancer (24) and thyroid
cancer (25), LXN was considered to
exhibits tumor suppressor while not in OS, so it might be a novel
target for the diagnosis or treatment of OS. Besides, CMKLR1
(chemokine-like receptor 1), exhibited downregulation and
hypermethylation simultaneously in OS group, and was identified to
affect the treatment of chemotherapy in non-small cell lung cancer
(26) and the endothelial
angiogenesis (27), which might
also influence the development of OS. Some known biomarkers were
also identified in this study, such as RAC2 (28) and CD248 (29), which indicated the reliability of
our study.
The DEGs and DEMs were found to be enriched in
entirely different aspects of the biological process of ontology,
e.g. the DEGs were mainly involved in inflammatory/immune response,
and cell proliferation related processes, while DEMs were
associated with the processes of regulation of gene expression,
tissue and embryonic development. However, all of the terms are
closely associated with the development of many types of cancers.
Moreover, DNA methylation could lead to altered gene expression and
regulation and thus affect the progression of cancers (30–32).
Our study also supported the findings and expected to be helpful in
the understanding of the mechanisms of OS.
In this study, potential miRNA regulators of
overlapping genes between DEGs and DEMs were screened out through
the TargetScan online software and the miRNA expression profile
dataset of OS. Finally, the miRNA-gene regulation network was
obtained in which TIMP4 (TIMP metallopeptidase inhibitor 4) was
directly regulated by 7 miRNAs. TIMP4 was downregulated in the OS
group and exhibited significantly negative correlation between gene
expression and methylation profiles (cor −0.66, P-value
3.70×10−4). It was shown to play important roles in the
extracellular matrix and the progression of cancers, such as
cervical cancer (33), however, no
study has associated it with OS, which might indicate it as a novel
target. Besides, miRNA-128, which has been shown to influence the
risk of OS by many studies (34–36),
regulated more genes than the other miRNAs in the miRNA-gene
regulation network demonstrating the conformance of our study with
the previous ones, besides the novel findings.
In conclusion, the combined analysis of three types
of microarray datasets identified some novel biomarkers for OS and
many known ones were also confirmed in this study. Our results are
helpful for the diagnosis and treatment of OS and thus improvement
of prognosis.
References
|
1
|
Bishop MW and Janeway KA: Emerging
concepts for PI3K/mTOR inhibition as a potential treatment for
osteosarcoma. F1000Res. 5(pii): F10002016.doi:
10.12688/f1000research.8228.1. PubMed/NCBI
|
|
2
|
Picci P: Osteosarcoma (osteogenic
sarcoma). Orphanet J Rare Dis. 2:62007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Whelan J, Seddon B and Perisoglou M:
Management of osteosarcoma. Curr Treat Options Oncol. 7:444–455.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Longhi A, Errani C, De Paolis M, Mercuri M
and Bacci G: Primary bone osteosarcoma in the pediatric age: State
of the art. Cancer Treat Rev. 32:423–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wan J and Zhang X, Liu T and Zhang X:
Strategies and developments of immunotherapies in osteosarcoma.
Oncol Lett. 11:511–520. 2016.PubMed/NCBI
|
|
6
|
Vos HI, Coenen MJ, Guchelaar HJ and Te Loo
DM: The role of pharmacogenetics in the treatment of osteosarcoma.
Drug Discov Today. 21:1775–1786. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Serra M and Hattinger CM: The
pharmacogenomics of osteosarcoma. Pharmacogenomics J. May
31–2016.(Epub ahead of print). doi: 10.1038/tpj.2016.45. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu K, Dai HB and Qiu ZL: mTOR signaling in
osteosarcoma: Oncogenesis and therapeutic aspects (Review). Oncol
Rep. 36:1219–1225. 2016.PubMed/NCBI
|
|
9
|
Qiu Q, Jiang J, Lin L, Cheng S, Xin D,
Jiang W, Shen J and Hu Z: Downregulation of RSK2 influences the
biological activities of human osteosarcoma cells through
inactivating AKT/mTOR signaling pathways. Int J Oncol.
48:2508–2520. 2016.PubMed/NCBI
|
|
10
|
Zhao J, Chen F, Zhou Q, Pan W, Wang X, Xu
J, Ni L and Yang H: Aberrant expression of microRNA-99a and its
target gene mTOR associated with malignant progression and poor
prognosis in patients with osteosarcoma. Onco Targets Ther.
9:1589–1597. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bilbao-Aldaiturriaga N, Askaiturrieta Z,
Granado-Tajada I, Goričar K, Dolžan V, Garcia-Miguel P, de andoin N
Garcia, Martin-Guerrero I and Garcia-Orad A: For The Slovenian
Osteosarcoma Study Group: A systematic review and meta-analysis of
MDM2 polymorphisms in osteosarcoma susceptibility. Pediatr Res.
80:472–479. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu C and Wang W: Relationship between P15
gene mutation and formation and metastasis of malignant
osteosarcoma. Med Sci Monit. 22:656–661. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang C, Yang L, Geng YD, An FL, Xia YZ,
Guo C, Luo JG, Zhang LY, Guo QL and Kong LY: Icariside II, a
natural mTOR inhibitor, disrupts aberrant energy homeostasis via
suppressing mTORC1-4E-BP1 axis in sarcoma cells. Oncotarget.
7:27819–27837. 2016.PubMed/NCBI
|
|
14
|
Kalmár A, Péterfia B, Hollósi P, Galamb O,
Spisák S, Wichmann B, Bodor A, Tóth K, Patai ÁV, Valcz G, et al:
DNA hypermethylation and decreased mRNA expression of MAL, PRIMA1,
PTGDR and SFRP1 in colorectal adenoma and cancer. BMC Cancer.
15:7362015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qiu X, Hu B, Huang Y, Deng Y, Wang X and
Zheng F: Hypermethylation of ACP1, BMP4, and TSPYL5 in
hepatocellular carcinoma and their potential clinical significance.
Dig Dis Sci. 61:149–157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choudhury JH and Ghosh SK: Promoter
hypermethylation profiling identifies subtypes of head and neck
cancer with distinct viral, environmental, genetic and survival
characteristics. PLoS One. 10:e01298082015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Ge J, Ma T, Zheng Y, Lv S, Li Y
and Liu S: Promoter hypermethylation of the cysteine protease RECK
may cause metastasis of osteosarcoma. Tumour Biol. 36:9511–9516.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sonaglio V, de Carvalho AC, Toledo SR,
Salinas-Souza C, Carvalho AL, Petrilli AS, de Camargo B and Vettore
AL: Aberrant DNA methylation of ESR1 and p14ARF genes could be
useful as prognostic indicators in osteosarcoma. Onco Targets Ther.
6:713–723. 2013.PubMed/NCBI
|
|
19
|
Kresse SH, Rydbeck H, Skårn M, Namløs HM,
Barragan-Polania AH, Cleton-Jansen AM, Serra M, Liestøl K,
Hogendoorn PC, Hovig E, et al: Integrative analysis reveals
relationships of genetic and epigenetic alterations in
osteosarcoma. PLoS One. 7:e482622012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Namløs HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ni QF, Tian Y, Kong LL, Lu YT, Ding WZ and
Kong LB: Latexin exhibits tumor suppressor potential in
hepatocellular carcinoma. Oncol Rep. 31:1364–1372. 2014.PubMed/NCBI
|
|
22
|
Muthusamy V, Premi S, Soper C, Platt J and
Bosenberg M: The hematopoietic stem cell regulatory gene latexin
has tumor-suppressive properties in malignant melanoma. J Invest
Dermatol. 133:1827–1833. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Basang Z, Ding H, Lu Z, Ning T, Wei
H, Cai H and Ke Y: Latexin expression is downregulated in human
gastric carcinomas and exhibits tumor suppressor potential. BMC
Cancer. 11:1212011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xue ZX, Zheng JH, Zheng ZQ, Cai JL, Ye XH,
Wang C, Sun WJ, Zhou X, Lu MD, Li PH, et al: Latexin inhibits the
proliferation of CD133+ miapaca-2 pancreatic cancer
stem-like cells. World J Surg Oncol. 12:4042014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abd Elmageed ZY, Moroz K and Kandil E:
Clinical significance of CD146 and latexin during different stages
of thyroid cancer. Mol Cell Biochem. 381:95–103. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu X, Ye Y, Rosell R, Amos CI, Stewart DJ,
Hildebrandt MA, Roth JA, Minna JD, Gu J, Lin J, et al: Genome-wide
association study of survival in non-small cell lung cancer
patients receiving platinum-based chemotherapy. J Natl Cancer Inst.
103:817–825. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaur J, Adya R, Tan BK, Chen J and Randeva
HS: Identification of chemerin receptor (ChemR23) in human
endothelial cells: Chemerin-induced endothelial angiogenesis.
Biochem Biophys Res Commun. 391:1762–1768. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han JA, Kim JY and Kim JI: Analysis of
gene expression in cyclooxygenase-2-overexpressed human
osteosarcoma cell lines. Genomics Inform. 12:247–253. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun DX, Liao GJ, Liu KG and Jian H:
Endosialin-expressing bone sarcoma stem-like cells are highly
tumor-initiating and invasive. Mol Med Rep. 12:5665–5670.
2015.PubMed/NCBI
|
|
30
|
Song L and Li Y: The role of stem cell DNA
methylation in colorectal carcinogenesis. Stem Cell Rev.
12:573–583. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu P, Hu G, Luo C and Liang Z: DNA
methyltransferase inhibitors: An updated patent review (2012–2015).
Expert Opin Ther Pat. 26:1017–1030. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie P, Zang LQ, Li XK and Shu Q: An
epigenetic view of developmental diseases: New targets, new
therapies. World J Pediatr. 12:291–297. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lizarraga F, Ceballos-Cancino G, Espinosa
M, Vazquez-Santillan K, Maldonado V and Melendez-Zajgla J: Tissue
inhibitor of metalloproteinase-4 triggers apoptosis in cervical
cancer cells. PLoS One. 10:e01359292015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu X, Liang Z, Gao K, Li H, Zhao G, Wang
S and Fang J: MicroRNA-128 inhibits EMT of human osteosarcoma cells
by directly targeting integrin alpha2. Tumour Biol. 37:7951–7957.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tian Z, Guo B, Yu M, Wang C, Zhang H,
Liang Q, Jiang K and Cao L: Upregulation of micro-ribonucleic
acid-128 cooperating with downregulation of PTEN confers metastatic
potential and unfavorable prognosis in patients with primary
osteosarcoma. Onco Targets Ther. 7:1601–1608. 2014.PubMed/NCBI
|
|
36
|
Shen L, Chen XD and Zhang YH: MicroRNA-128
promotes proliferation in osteosarcoma cells by downregulating
PTEN. Tumour Biol. 35:2069–2074. 2014. View Article : Google Scholar : PubMed/NCBI
|