Introduction
Glioma is the most common brain tumor, accounting
for 80% of all primary brain and central nervous system
malignancies (1). According as the
World Health Organization (WHO) 2007 version based on the different
histological tumor types, gliomas are classified as astrocytic,
oligodendroglial, mixed oligoastrocytic, and ependymal glioma, with
malignancy grade I, II, III, and IV (2). Gliomas are not sensitive to
chemotherapy or radiotherapy, or the combination treatment of these
methods. The prognosis of malignant glioma remains very poor, and
the median overall survival of patients with glioblastoma is
limited to approximately 16 months within clinical trials (2,3). Thus,
it is necessary to find biomarkers and effective therapeutic
targets for early diagnosis and improvement of the prognosis for
glioma.
The IQ motif containing GTPase-activating protein 1
(IQGAP1) is a scaffold protein that regulates distinct cellular
processes including actin dynamics, cell adhesion, cell motility,
extracellular signals through interaction with cell adhesion
molecules, several signaling molecules and cytoskeleton components
(4–7). Many studies have revealed that IQGAP1
is upregulated in various human malignancies, such as colon cancer
(8), lung cancer (9), hepatocellular carcinoma (10) and breast cancer (11). IQGAP1 plays a critical role in
cancer cell invasion. The overexpression of IQGAP1 in colon cancer
cells correlates with poor prognosis (12), and cell motility was increased by
overexpressing IQGAP1 in breast cancer (13). Moreover, the upregulation of IQGAP1
has been found to be associated with poor prognosis of glioma and
could be a potential prognosis marker (14).
As a scaffold protein, IQGAP1 interacts with
multiple proteins to exert various roles in carcinogenesis. For
example, IQGAP1 interacts directly with RhoA/C to act as a
regulator and pro-oncogenic effector of RhoA/C in breast cancer
(13). Moreover, another study
reported IQGAP1 can bind with β-catenin to promote liver cancer
progression in vitro and in vivo (10). CDC42, one Rho family member,
contributes to oncogenic transformation, invasion, and
tumorigenesis (15). In breast
cancer, the active CDC42 and RhoA trigger the interaction of IQGAP1
with exocyst subunits (16), which
is required for matrix proteolysis and invasion of breast carcinoma
cells. We focused on the association of IQGAP1 level with CDC42
expression in human glioma, and how two proteins collaborate to
exert biological effects in glioma development.
In this study, we investigated the associated
expression level and clinical significance of IQGAP1 and CDC42 in
human glioma progression. We confirmed that IQGAP1 and CDC42 are
frequently overexpressed in glioma tissues compared with their
noncancerous counterparts, and high expression of IQGAP1 and CDC42
is associated with glioma grade and overall survival of glioma
patients. Furthermore, our study discovered that the overexpression
or knockdown of IQGAP1 in glioma cells has significant effects on
cell proliferation and migration in vitro. Our results
suggest that the elevated level of IQGAP1, CDC42 and their
associations contribute to human glioma carcinogenesis and
progression.
Materials and methods
Cell culture
The human glioma cell lines including U87 and H4
were obtained from American Type Culture Collection (Manassas, VA),
and U251 cells were obtained from the Type Culture Collection of
the Chinese Academy of Sciences (Shanghai, China). All cells were
cultured in DMEM medium supplemented with 10% fetal bovine serum
(FBS) (16000–044, Gibco), 100 U/ml penicillin, and 100 µg/ml
streptomycin. Cells were incubated in a humidified atmosphere at
37°C with 5% CO2.
Tissue sample collection
This study was approved by the Institutional Ethics
Committee of the Affiliated Hospital of Inner Mongolia Medical
University (Inner Mongolia, China). Thirty cases of human glioma
tissues (HGTs) and their paired para-cancerous brain tissues (PBTs)
were surgically resected in the Affiliated Hospital of Inner
Mongolia Medical University with the patients' informed consent.
The HGTs and their paired PBTs were immediately stored frozen in
liquid nitrogen for further use.
Bioinformatics analysis for
protein-protein interaction network
IQGAP1-interacting proteins were predicted based on
the online human protein-protein interaction (PPI) datasets
(http://www.hprd.org/). These protein interactions
were summarized from literature studies, which have been widely
validated by low-throughput and high-throughput experiments and
applied in disease research (17).
Cytoscape software was applied for visualization and analysis of
PPI networks, which provides various plugins for different
analyses. PPI networks are illustrated as graphs in Cytoscape with
the nodes representing the proteins and the edges representing
their interactions (18).
Expression plasmids, siRNAs and cell
transfection
The original IQGAP1 cDNA (gi57242794) clone was
ordered from Generay Biotechnology (Shanghai, China), and was
sub-cloned into a pCMV6 plasmid to obtain a recombinant plasmid
pIQGAP1, which was confirmed to be correct by DNA sequencing.
Glioma U87 and U251 cells were respectively seeded on a 6-well
plate for culture overnight, cells were transiently transfected
with 2.5 µg pIQGAP1 plasmids for one well using Lipofectamine 2000
(cat. no. 11668-019, Life Technologies) according to the
manufacturer's instructions.
The IQGAP1-specific siRNA (siIQGAP1) based on
literature (19) was synthesized by
RiboBio Co., Ltd. (Guangzhou, China). The siIQGAP1 sequence, 5′-UUA
UCG CCC AGA AAC AUC UUG UUG G-3′; and negative control
oligonucleotides (NC) were 5′-UUC UCC GAA CGU GUC ACG U-3′. The U87
and U251 cells were seeded on a 6-well plate to incubate with 100
nM siIQGAP1 for one well by the transfection reagent (INTERFER,
Polyplus Transfection) following the manufacturer's protocols.
Western blotting
Total cellular protein was extracted with RIPA
buffer (50 mM Tris base, 1.0 mM EDTA, 150 mM NaCl, 0.1% SDS, 1%
Triton X-100, 1% sodium deoxycholate, 1 mM PMSF). Protein samples
were separated on 10% SDS-PAGE and transferred onto the PVDF
membrane (Amersham Biosciences, Amersham, UK) to detect protein
expression level. The PVDF membrane was respectively incubated with
IQGAP1 antibody (ab133490, Abcam, 1:1000) or CDC antibody
(ab187643, Abcam, 1:10000) at 4°C overnight, followed by three
15-min washes in PBS within 0.1% Tween-20. The membranes were then
incubated with HRP-conjugated secondary antibodies at 37°C for 60
min. Detection was performed with Western blot reagent ECL
(Amersham Biosciences). Membranes were re-probed with mouse
anti-β-actin (sc-1616, Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) for normalization of signal as a control.
Cell viability
Cell viability was measured using MTT assay. After
IQGAP1 overexpression or knockdown for 48 h, 3×103
cells/well were seeded in one 96-well plate in DMEM supplemented
with 10% FBS to incubate for another 24–48 h. In addition, 20 µl of
5 mg/ml MTT solution (Sigma) was added to each well to incubate for
2–4 h at 37°C, the formazan crystals were dissolved with 150 µl
dimethyl sulfoxide (Sigma). Absorbance was determined at 490 nm on
Multiskan MK3 (Thermo Scientific, Rockford, IL, USA) immediately.
Each assay was separately performed for three replicates and all
experiments were repeated at least three times.
Transwell migration assay
After being transfected with pIQGAP1 plasmids or
IQGAP1-specific siRNA for 48 h, 1×104 cells in 100 µl
serum-free DMEM were seeded in the upper chamber of a Transwell
(Millipore, 8-mm pore size), and the bottom of the chambers was
filled with 800 µl of medium containing 10% FBS to culture another
24 h. The migrated cells moved toward medium containing 10% FBS.
The remaining cells were fixed and stained with 1% crystal violet.
Images were captured using an inverted microscope (Olympus), and
the migrated cells were counted manually. The percentage of
migration cells on the condition of IQGAP1 overexpression or
knockdown was calculated by comparison with the control treatment
with the empty plasmids or non-targeting siRNA.
Immunohistochemistry
Tissues were fixed with paraformaldehyde, embedded
with paraffin, then cut into sections of 5 µm thickness for
immunohistochemistry (IHC) analysis mainly according to literature
studies (20). The first antibody
against IQGAP1 (ab133490, Abcam), CDC (ab187643, Abcam) was
respectively used at a dilution of 1:100, 1:400. The second
antibody was a biotinylated IgG for 40 min incubate at 37°C. Tissue
slices were visualized by the 3, 3′-diaminobenzidine solution, and
cellular nuclei were slightly counterstained with hematoxylin.
Substitution of the primary antibody with phosphate-buffered saline
(PBS) was taken as a control for IHC. According to general
evaluation standards (21), the
staining intensity was scored as 0 (negative), 1 (weak), 2
(moderate) or 3 (strong). The extent of staining was monitored
based on the percentage of positive tumor cells: 0 (negative), 1
(1–25%), 2 (26–50%), 3 (51–75%) and 4 (76–100%). The final score of
0 was defined as a negative expression (−); scores of 1–3 were
accepted as a low/weak expression (+), and scores over 3 were
defined as a high/strong expression (++). The intensity and
percentage of positive cells were evaluated at least in five
separate fields at a 400-fold magnification. The IHC scores of one
tissue sample was respectively determined by two pathologists, and
the final score for a tissue sample was calculated from the average
value of the two sets of total scores. P<0.05 was considered
statistically significant.
Association analysis for protein
expression and patient survival
The patient overall survival (OS) was evaluated
using the Kaplan-Meier method (21). The 30 glioma patients were
classified into two groups based on the protein expression level,
including low IQGAP1/CDC42-expressing (n=8) and high
IQGAP1/CDC42-expressing groups (n=21). The group differences were
assessed using the log-rank test. P<0.05 was considered
statistically significant.
Statistical analysis
All statistical analyses were performed using the
SPSS software system (version 19.0; SPSS, Inc., Chicago, IL, USA).
Statistical data were expressed as the mean ± standard deviation
(SD). P<0.05 was considered to be statistically significant.
Results
IQGAP1 interacts with CDC42 by
bioinformatics analysis
The interacting proteins with IQGAP1 were analyzed
based on HPRD database online (http://www.hprd.org/). In the protein-protein
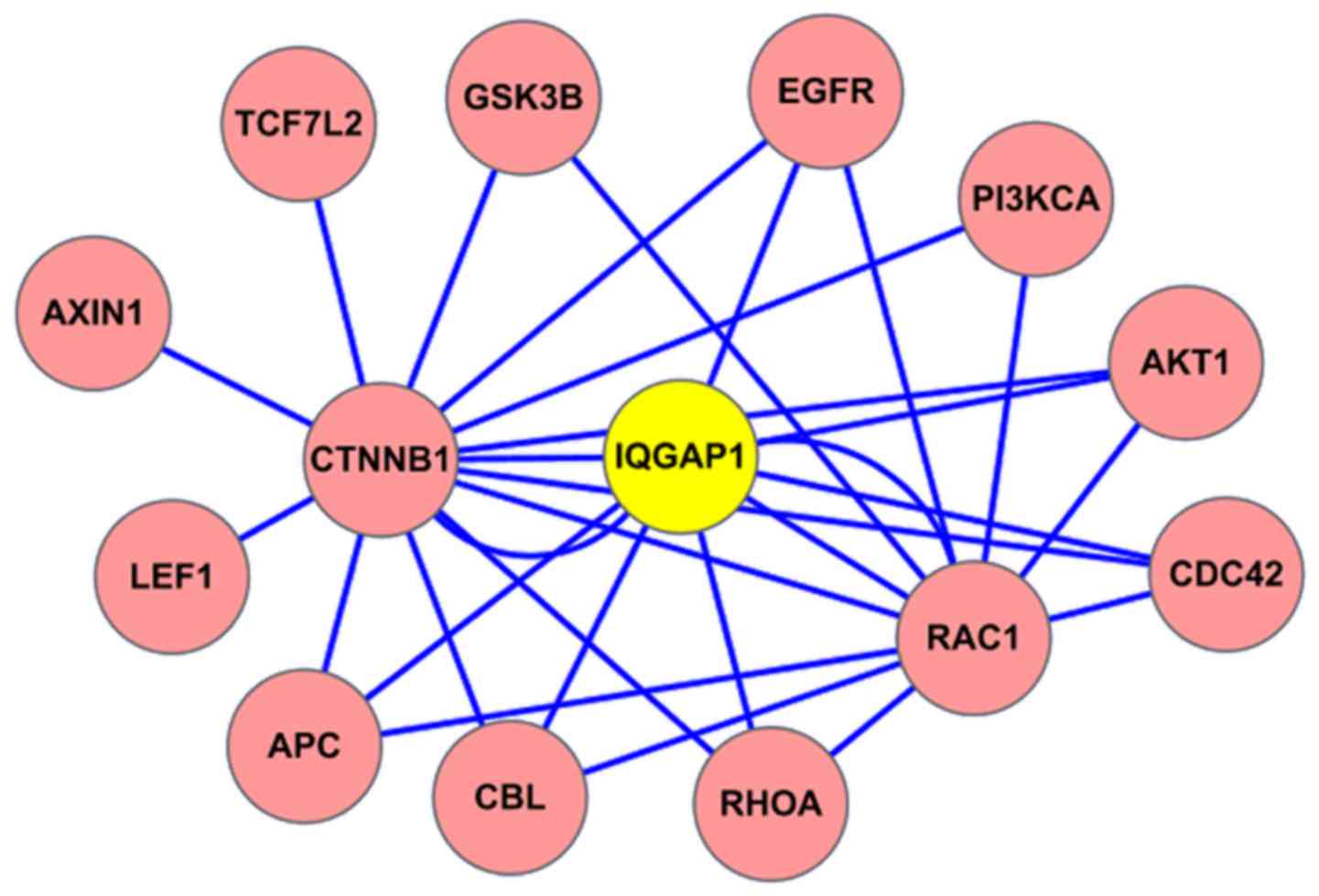
interaction (PPI) map, IQGAP1 located in the central position, and
the protein CDC42 was shown to interact with IQGAP1 (Fig. 1). Of course, the known binding
partners RhoA and Rac1 were also included within the IQGAP1
interacting protein network.
CDC42 level is linked with IQGAP1
expression in glioma cells
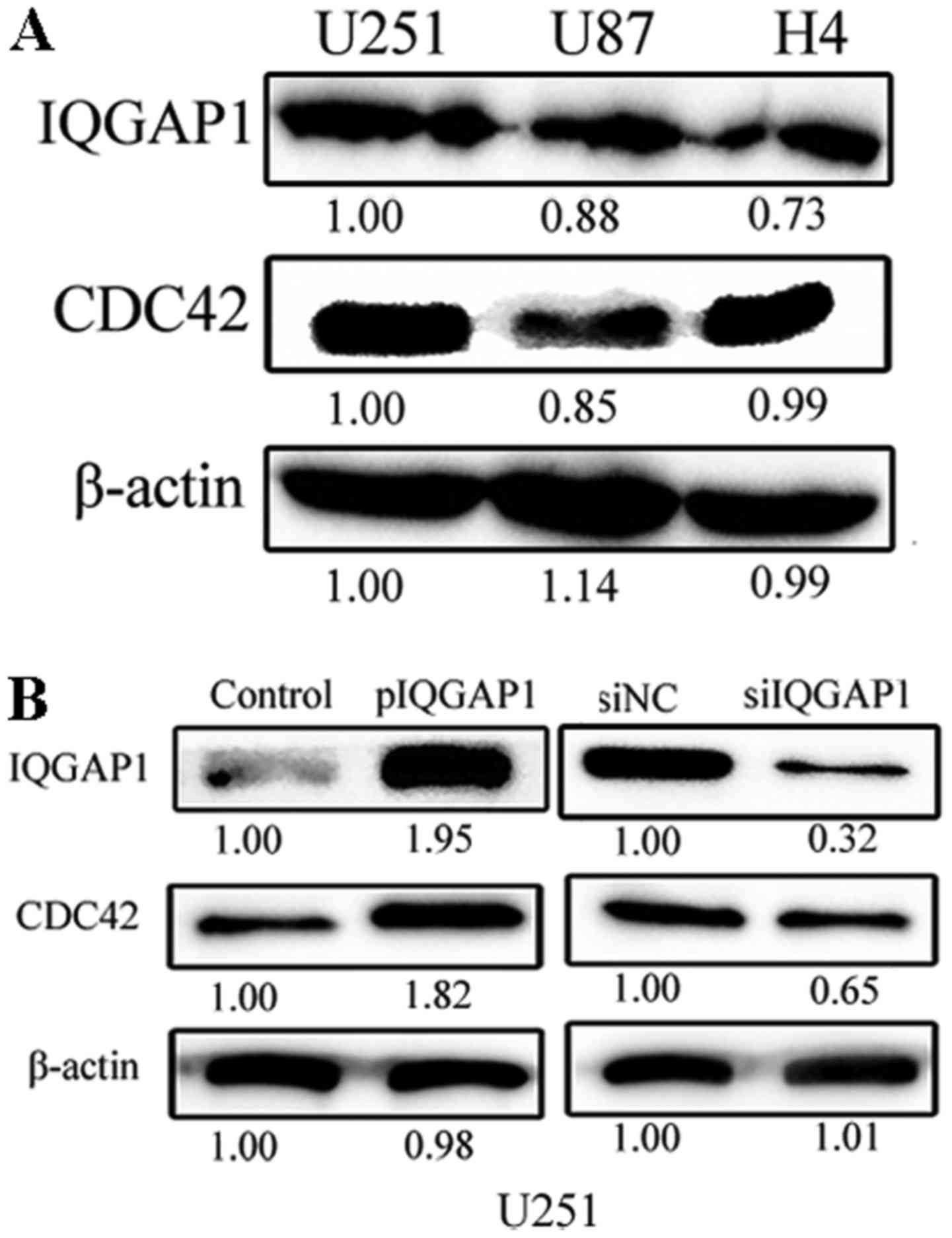
The endogenous expression levels of IQGAP1 and CDC42
are high in glioma U251, U87 and H4 cells (Fig. 2A). When IQGAP1 expression was
elevated 1.95 times in U251 cells by transient transfection of
pIQGAP1 plasmids for 48 h, the relative level of CDC42 was also
respectively increased to 1.82-fold (Fig. 2B). When IQGAP1 expression was
knocked down by siRNA treatment for 48 h, CDC42 level was
correspondingly decreased to 0.65 times in U251 cells. Similar
co-expressing relationship between IQGAP1 and CDC42 was obtained in
U87 cells (data not shown). Therefore, the expression of CDC42 is
tightly linked with IQGAP1 level in glioma cells, which also
indicates the two proteins interact with each other.
IQGAP1 overexpression promotes glioma
cell growth and migration
In order to further investigate cellular biological
influence of high IQGAP1 level, the gain- and loss-of-function
studies were performed in glioma cells. We explored this protein
biological effects on glioma U87 and U251 cells by overexpression
or knockdown of IQGAP1 in these glioma cells.
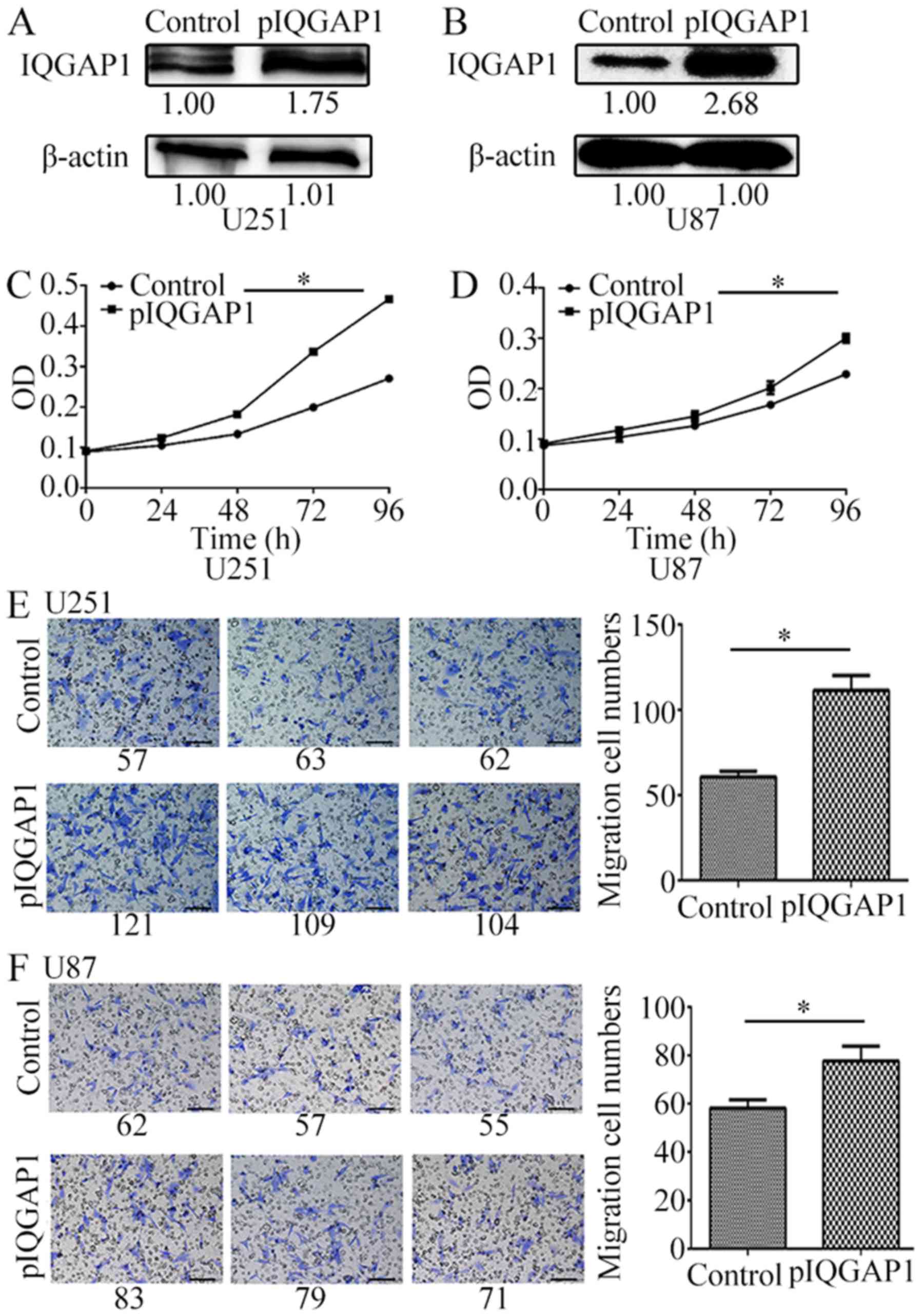
The overexpression of IQGAP1 protein (Fig. 3A and B) significantly increased
glioma cell proliferation (Fig. 3C and
D) and cell migration (Fig. 3E and
F) of U251 and U87 cells. For example, in pIQGAP1-transfected
U251 cells, cell proliferation was increased by 37, 68.7 and 72.3%
after transfection for 48–96 h compared with the vehicles (Fig. 3C). For IQGAP1-overexpressing U87
cells, cell growth curve was also obviously increased (Fig. 3D). Moreover, the quantity of cell
migration was respectively increased to 1.84, 1.34 times in U251
and U87 cells which were transiently transfected with pIQGAP1
plasmids for 72 h (Fig. 3E and
F).
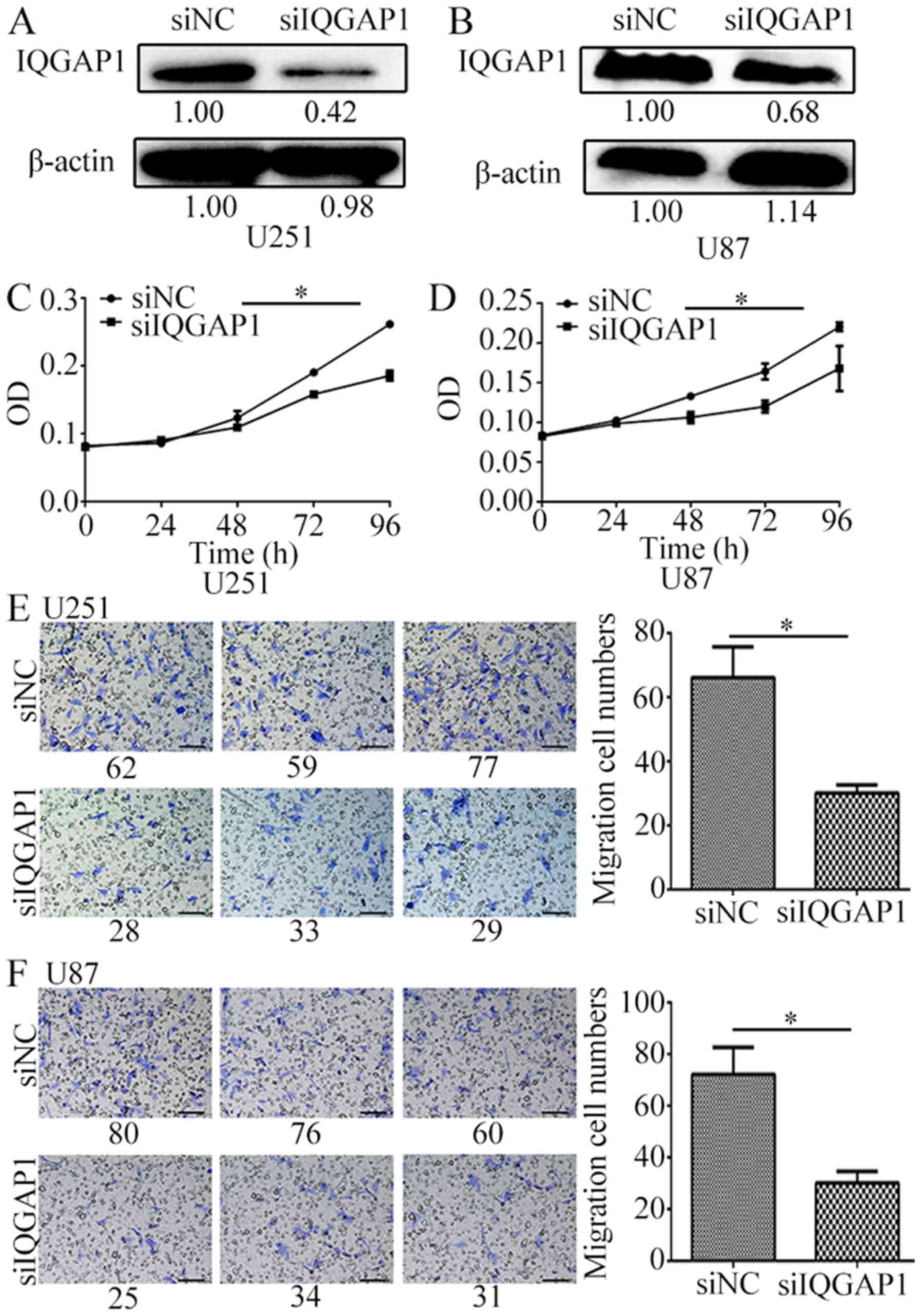
On the contrary, knockdown of IQGAP1 significantly
inhibited glioma cell proliferation rate and cell migration. When
IQGAP1 was knocked down by siIQGAP1 for 72 h in U251 and U87 cells
(Fig. 4A and B), cell growth was
decreased by 30% (Fig. 4C and D),
and cell migration number was decreased to over 2 times by
comparison with the non-targeting siRNA groups (Fig. 4E and F). These results suggest that
IQGAP1 downregulation greatly inhibits glioma cell proliferation
and migration.
IQGAP1 and CDC42 are increased in
human glioma tissues
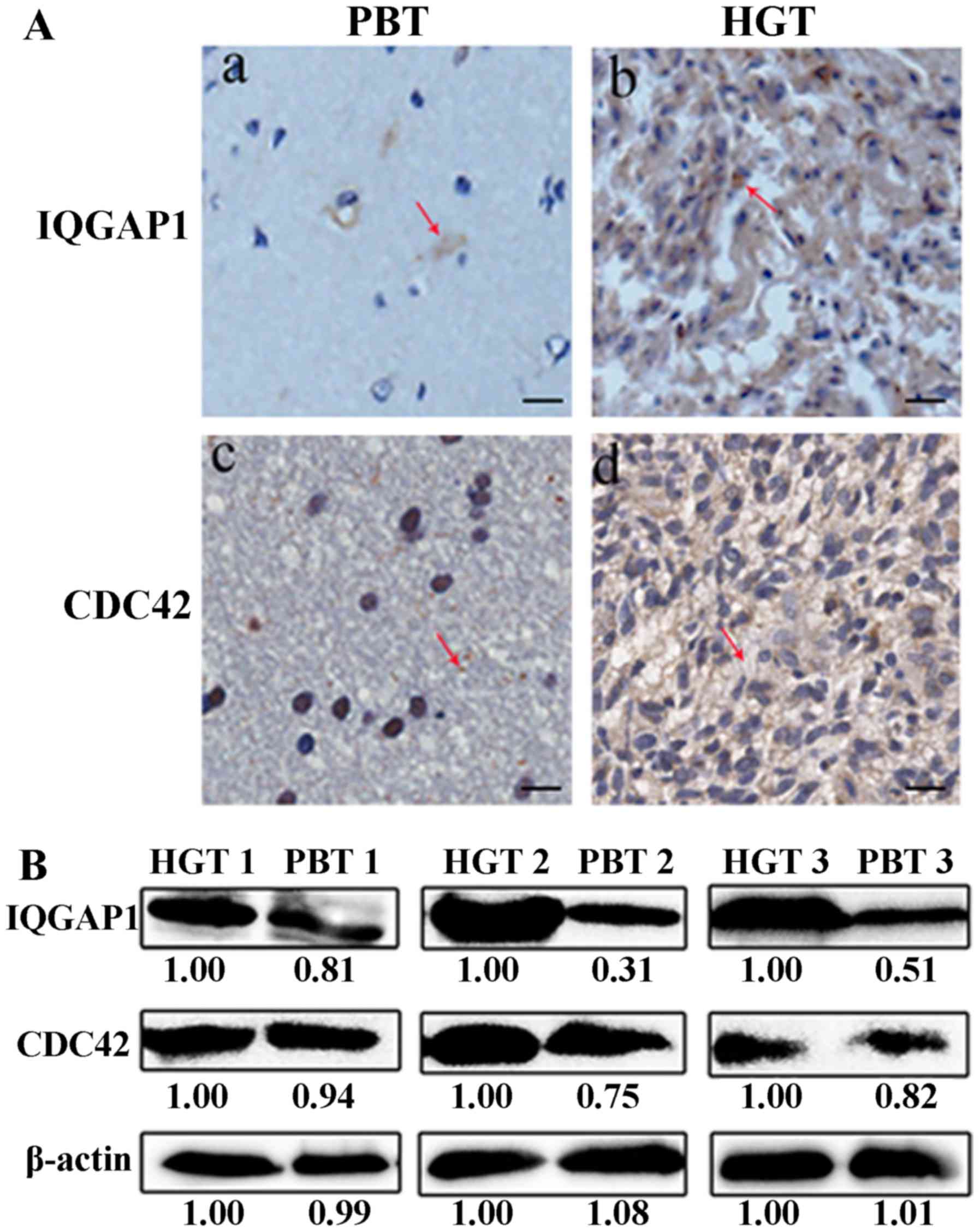
The expression level of IQGAP1 and CDC42 was greatly
elevated in human glioma tissues compared with their counterparts
by IHC analysis (Fig. 5A). The
average immunostaining score of IQGAP1, CDC42 was 4.62±0.48 and
4.40±0.47 in 30 HGTs, respectively, which was much higher than the
average staining score 1.30±0.16, 2.37±0.19 in PBTs (Table I) (P<0.01). The IHC scores and
clinical information for gliomas are provided in detail in Table II, and the IHC scores for PBTs are
listed in Table III. Among the
HGTs, more than 73.3% of glioma tissues (22 cases) showed a strong
expression of IQGAP1 with scores 5.75±0.48, and only 8 cases
(26.7%) had a weak expression of IQGAP1 with mean staining scores
1.14±0.26 (Table I). While in 30
PBTs, IQGAP1 was usually detected with a lower expression level
with an average staining score 1.30±0.16. Only 2 cases showed
strong expression with a score of 4, most of PBTs (93%) had low
IQGAP1 expression scoring 1.11±0.09. Similarly, 21 HGTs (70%) had
strong CDC42 level with scores 5.62±0.44, which was higher than the
frequency of 26.7% (8/30) with scores 3.88±0.18 in PBTs. It was
consistent with the IHC data that IQGAP1 and CDC42 had a strong
expression in 3 randomly selected HGTs compared with their
counterparts PBTs by western blot analysis (Fig. 5B).
 | Table I.The expression of IQGAP1 and CDC42
between HGTs and PBTs. |
Table I.
The expression of IQGAP1 and CDC42
between HGTs and PBTs.
|
| HGTs (n=30) | PBTs (n=30) |
|---|
|
|
|
|
|---|
| Protein | Percentage | Average score | Expression level | Percentage | Average score | Expression level |
|---|
| IQGAP1 | 100% (30/30) | 4.62±0.48 | ++ | 100% (30/30) | 1.30±0.16 | + |
|
| 26.7% (8/30) | 1.50±0.30 | + | 93.3% (28/30) | 1.11±0.09 | + |
|
| 73.3% (22/30) | 5.75±0.48 | ++ | 6.7% (2/30) | 4 | ++ |
| CDC42 | 100% (30/30) | 4.40±0.47 | ++ | 100% (30/30) | 2.37±0.19 | + |
|
| 30.0% (9/30) | 1.56±0.28 | + | 73.3% (22/30) | 1.82±0.12 | + |
|
| 70.0% (21/30) | 5.62±0.44 | ++ | 26.7% (8/30) | 3.88±0.18 | ++ |
 | Table II.Protein IHC scoring and pathological
information for human glioma tissues. |
Table II.
Protein IHC scoring and pathological
information for human glioma tissues.
|
|
|
|
|
|
| Scoring of
IQGAP1 | Scoring of
CDC42 |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Case no. | Age | Gender | TNM stage | Survival time
(months) | Survival state | A | B | Average | A | B | Average |
|---|
| 1 | 57 | Male | I | 35 | Survival | 0 | 0 | 0 | 2 | 2 | 2 |
| 2 | 34 | Male | II | 32 | Survival | 1 | 1 | 1 | 1 | 1 | 1 |
| 3 | 30 | Female | II | 36 | Survival | 2 | 3 |
2.5 | 2 | 3 |
2.5 |
| 4 | 39 | Male | II | 24 | Death | 1 | 2 |
1.5 | 2 | 2 | 2 |
| 5 | 58 | Female | II | 21 | Death | 3 | 2 |
2.5 | 1 | 1 | 1 |
| 6 | 61 | Male | III | 25 | Death | 4 | 2 | 3 | 3 | 2 |
2.5 |
| 7 | 30 | Female | III | 30 | Survival | 3 | 4 |
3.5 | 4 | 2 | 3 |
| 8 | 51 | Male | III | 33 | Survival | 3 | 3 | 3 | 3 | 3 | 3 |
| 9 | 36 | Female | III | 12 | Survival | 4 | 4 | 4 | 3 | 3 | 3 |
| 10 | 57 | Female | III | 7 | Survival | 6 | 4 | 5 | 4 | 4 | 4 |
| 11 | 73 | Female | III | 9 | Death | 3 | 3 | 3 | 4 | 4 | 4 |
| 12 | 50 | Female | III | 10 | Survival | 4 | 6 | 5 | 6 | 3 |
4.5 |
| 13 | 31 | Male | III | 11 | Survival | 3 | 4 |
3.5 | 2 | 4 | 3 |
| 14 | 56 | Male | IV | 12 | Death | 6 | 4 | 5 | 4 | 4 | 4 |
| 15 | 58 | Male | IV | 1 | Death | 8 | 9 |
8.5 | 8 | 8 | 8 |
| 16 | 70 | Male | IV |
0.5 | Death | 8 | 8 | 8 | 9 | 6 |
7.5 |
| 17 | 65 | Female | IV | 7 | Survival | 9 | 8 |
8.5 | 9 | 9 | 9 |
| 18 | 65 | Female | I | 31 | Survival | 1 | 2 |
1.5 | 2 | 2 | 2 |
| 19 | 61 | Male | II | 11 | Death | 2 | 2 | 2 | 0 | 0 | 0 |
| 20 | 38 | Female | III | 3 | Survival | 4 | 4 | 4 | 4 | 4 | 4 |
| 21 | 48 | Female | III |
2.2 | Death | 4 | 6 | 5 | 6 | 4 | 5 |
| 22 | 63 | Male | III | 12 | Death | 4 | 6 | 5 | 4 | 6 | 5 |
| 23 | 72 | Female | IV | 6 | Death | 6 | 9 |
7.5 | 6 | 8 | 7 |
| 24 | 48 | Male | IV | 8 | Survival | 6 | 6 | 6 | 6 | 6 | 6 |
| 25 | 56 | Female | IV | 9 | Survival | 6 | 6 | 6 | 6 | 8 | 7 |
| 26 | 64 | Male | IV | 11 | Death | 6 | 8 | 7 | 6 | 9 |
7.5 |
| 27 | 35 | Male | IV | 5 | Death | 9 | 9 | 9 | 8 | 8 | 8 |
| 28 | 63 | Female | IV | 6 | Death | 9 | 8 |
8.5 | 6 | 9 |
7.5 |
| 29 | 34 | Male | IV | 5 | Death | 9 | 8 |
8.5 | 8 | 8 | 8 |
| 30 | 59 | Male | I | 13 | Death | 1 | 1 | 1 | 1 | 1 | 1 |
 | Table III.Protein IHC scoring for
para-cancerous brain tissues. |
Table III.
Protein IHC scoring for
para-cancerous brain tissues.
|
| Scoring of
IQGAP1 | Scoring of
CDC42 |
|---|
|
|
|
|
|---|
| Case no. | A | B | Average | A | B | Average |
|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 2 | 1 | 1 | 1 | 2 | 2 | 2 |
| 3 | 1 | 1 | 1 | 2 | 1 |
1.5 |
| 4 | 0 | 0 | 0 | 1 | 1 | 1 |
| 5 | 4 | 4 | 4 | 6 | 3 |
4.5 |
| 6 | 1 | 3 | 2 | 4 | 4 | 4 |
| 7 | 1 | 1 | 1 | 2 | 2 | 2 |
| 8 | 1 | 1 | 1 | 3 | 2 |
2.5 |
| 9 | 1 | 1 | 1 | 1 | 1 | 1 |
| 10 | 1 | 1 | 1 | 3 | 2 |
2.5 |
| 11 | 2 | 2 | 2 | 3 | 4 |
3.5 |
| 12 | 1 | 1 | 1 | 1 | 2 |
1.5 |
| 13 | 1 | 1 | 1 | 2 | 2 | 2 |
| 14 | 1 | 1 | 1 | 3 | 1 | 2 |
| 15 | 1 | 3 | 2 | 4 | 3 |
3.5 |
| 16 | 0 | 0 | 0 | 2 | 2 | 2 |
| 17 | 1 | 1 | 1 | 1 | 1 | 1 |
| 18 | 1 | 1 | 1 | 2 | 2 | 2 |
| 19 | 1 | 1 | 1 | 1 | 1 | 1 |
| 20 | 4 | 4 | 4 | 6 | 3 |
4.5 |
| 21 | 1 | 1 | 1 | 2 | 1 |
1.5 |
| 22 | 1 | 1 | 1 | 2 | 3 |
2.5 |
| 23 | 1 | 1 | 1 | 3 | 2 |
2.5 |
| 24 | 1 | 3 | 2 | 4 | 4 | 4 |
| 25 | 2 | 2 | 2 | 4 | 4 | 4 |
| 26 | 1 | 1 | 1 | 2 | 2 | 2 |
| 27 | 1 | 1 | 1 | 3 | 2 |
2.5 |
| 28 | 1 | 1 | 1 | 2 | 4 | 3 |
| 29 | 1 | 1 | 1 | 2 | 2 | 2 |
| 30 | 1 | 1 | 1 | 2 | 2 | 2 |
Elevated IQGAP1 and CDC42 expression
are associated with glioma malignancy grade
Based on the activating roles for glioma cell growth
and migration in vitro, we further discovered the clinical
significance of the expression level of IQGAP1 and CDC42 for glioma
development. The high expression level of IQGAP and CDC42 is
positively associated with glioma malignancy (Table IV). The clinicopathological
characteristics of glioma samples included patient gender, age and
tumor TNM stage. It was clearly shown that a strong expression of
IQGAP1, CDC42 existed in 22 advanced grade gliomas with TNM stages
III–IV, with average IHC scoring 5.75±0.45 and 5.48±0.45. Whereas,
a lower level of IQGAP1 and CDC42 was present in human gliomas with
TNM stage I–II. This difference between protein expression level
with tumor grade was obvious (P<0.01). However, the expression
level of IQGAP1 and CDC42 has no linkage with glioma patient gender
or age.
 | Table IV.Correlations of the expression of
IQGAP1 and CDC42 in gliomas with clinical information. |
Table IV.
Correlations of the expression of
IQGAP1 and CDC42 in gliomas with clinical information.
|
|
| Average score | Expression
level | P-value |
|---|
|
|
|
|
|
|
|---|
| Clinicopathologic
variables | Number (n) | IQGAP1 | CDC42 | IQGAP1 | CDC42 | IQGAP1 | CDC42 |
|---|
| Gender |
|
|
|
|
|
|
|
Male | 16 | 4.50±0.76 | 4.28±0.71 | ++ | ++ |
0.8015 |
0.7927 |
|
Female | 14 | 4.75±0.59 | 4.54±0.62 | ++ | ++ |
|
|
| Age |
|
|
|
|
|
|
|
|
<56 | 13 | 4.35±0.66 | 4.08±0.60 | ++ | ++ |
0.6330 |
0.5576 |
|
≥56 | 17 | 4.82±0.70 | 4.65±0.70 | ++ | ++ |
|
|
| TNM stage |
|
|
|
|
|
|
|
|
I–II | 8 | 1.50±0.30 | 1.44±0.29 | + | + | <0.001 | <0.001 |
|
III–IV | 22 | 5.75±0.45 | 5.48±0.45 | ++ | ++ |
|
|
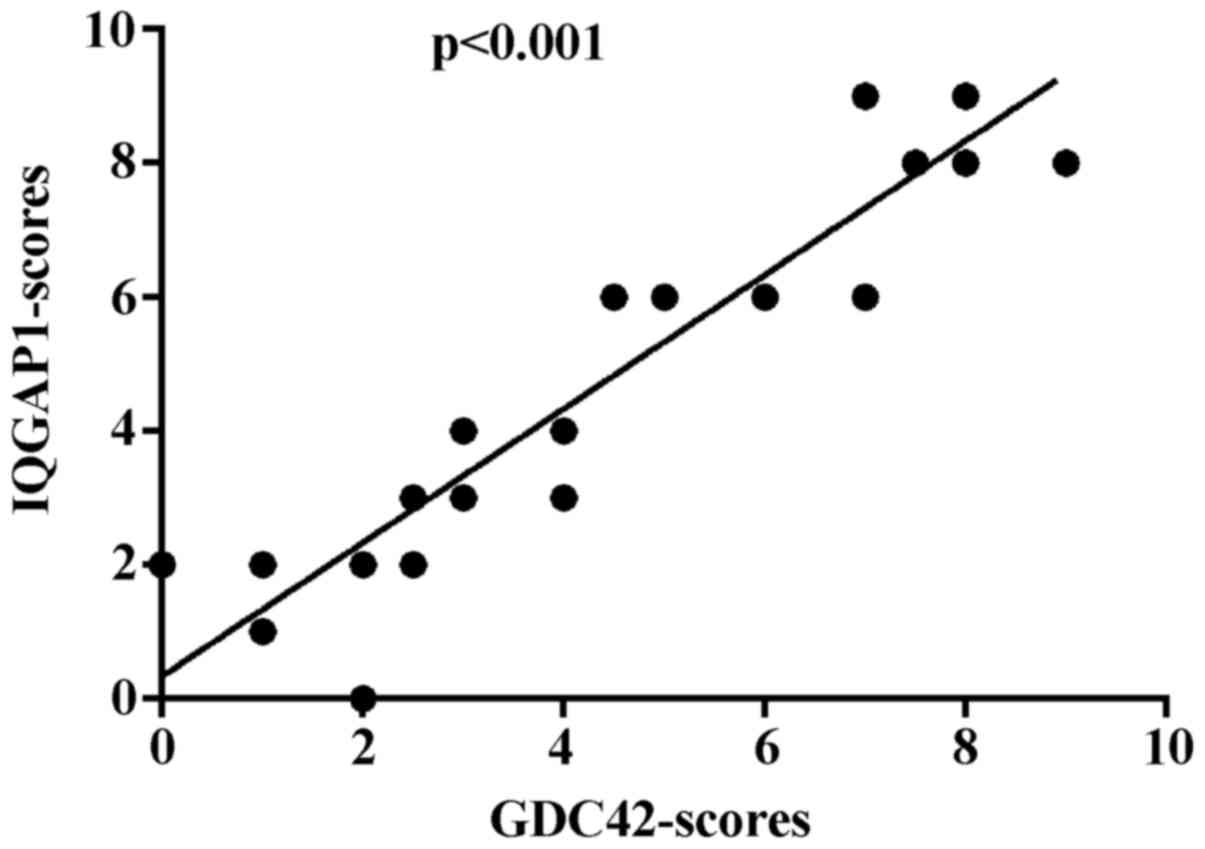
In conclusion, IQGAP1 and CDC42 show widely
increased expression in glioma, and much higher expression levels
of the two proteins are detected in high-grade glioma tissues. A
statistical analysis of the expression patterns revealed that there
is a positive correlation between IQGAP1 and CDC42 expression
(p<0.001) (Fig. 6).
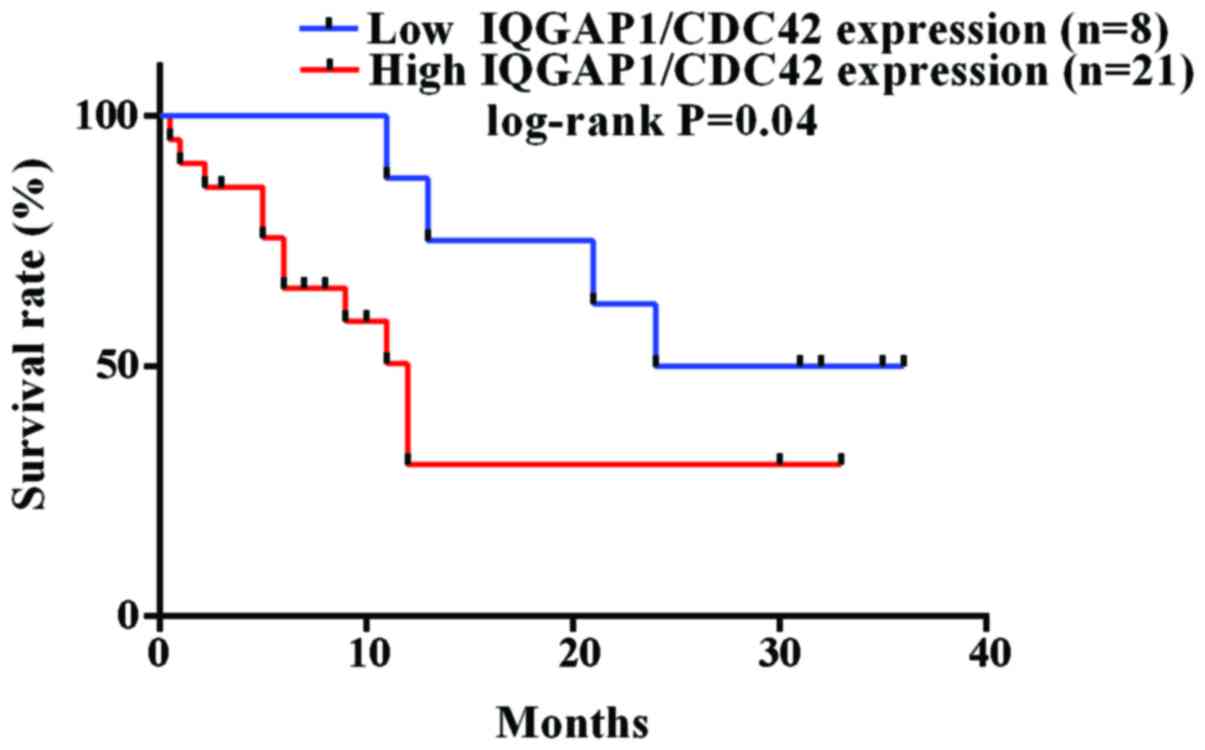
IQGAP1/CDC42 expression inversely
correlates with overall survival for glioma patients
Furthermore, the combined expression level of IQGAP1
and CDC42 was discovered to correlate with the overall survival of
glioma patients. In order to determine the prognostic significance,
30 cases of glioma patients who have exact overall survival (OS)
rates were grouped into two types, including a low (score <3)
and high (score >3) protein expression level of IQGAP1/CDC42.
Among 30 glioma patients, 21 and 8 cases were, respectively,
included into the high and low expression level of IQGAP1/CDC42.
The Kaplan-Meier estimates showed significant differences in OS
rates between patients with a low level of IQGAP1/CDC42 and those
with a high level of IQGAP1/CDC42 (P<0.05 by the log-rank test;
Fig. 7). The median overall
survival was 12.0 months for 21 patients with high expression level
of IQGAP1/CDC42, while the time was 30 months for 8 patients with
low expression of IQGAP1/CDC42.
Discussion
As a hotspot of biological therapy, biomarker is an
indicator of normal biological processes, pathogenic processes or a
pharmacological response to a therapeutic intervention. In recent
years, tumor biomarkers have been continuously reported due to the
important roles in diagnosis, therapy and prognosis for cancer
(22). As far as we know, potential
glioma biomarkers have been widely screened through various
multidisciplinary methods (22,23),
including gene chip, genome-wide approach, proteomics
identification and molecular pathophysiology analysis (21). Although several biomarkers have been
discovered to be important in the management of gliomas, including
1p19q co-deletion, MGMT promoter methylation, BRAF and IDH1
mutations, these potential biomarkers have certain limitations in
clinical application (24). For
example, it is known that the hypermethylation frequency of
O(6)-methylguanine-DNA
methyltransferase (MGMT) promoter varies widely in the different
subtypes of glioma, and the methylation of MGMT appears to be a
useful prognostic marker in the elderly patients with newly
diagnosed glioblastoma (25). MGMT
methylation is well established as a prognostic/predictive marker
for glioblastoma. However, it is not currently utilized widely in
guiding patient management (24).
It is necessary to establish more convenient and effective
biomarkers for glioma diagnosis, treatment and prognosis based on
the molecular basis of biomarker-mediated carcinogenesis.
By now, the overexpression of IQGAP1 has been
reported to be associated with certain cancerous metastasis
(6,26–28).
In our study, the correlation and biological effects between
IQGAP1, CDC42 and glioma development have been clarified. IQGAP1
and CDC42 are widely upregulated in human glioma tissues, and their
expression levels have a positive correlation with tumor
malignancy. However, high expressions of IQGAP1 and CDC42 reversely
correlate with glioma patient survival. Of course a more scale-up
human glioma samples should be further verified for the
associations of IQGAP1 level with glioma development and patient
prognosis, which is very helpful for classifying and grading
gliomas, as well as evaluating the potential predictive value based
on the protein expression.
In addition, the contribution of the overexpression
of IQGAP1 to glioma progression by promoting cell proliferation and
cell migration needs clarification. According to our bioinformatics
analysis of IQGAP1-interacting proteins based on human
protein-protein interactions database (HPRD) (http://www.hprd.org/), IQGAP1 with its interacting
proteins involve in oncogenesis-associated signaling pathways
(Fig. 1). The interacting proteins
with IQGAP1 include CDC42, RAC1, RHOA, CTNNB1 (β-catenin), APC,
GSK3B, AXIN1, and EGFR partners. Among these interaction proteins,
IQGAP1 mediates signaling by Rho family GTPases, including RAC1,
RHOA and CDC42 to regulate cell-cell adhesion and cell migration
(4). Our experimental data in
vitro and in vivo demonstrate that upregulation of
IQGAP1 and CDC42 improves cell proliferation and migration ability
of human glioma cells, whereas the knockdown of IQGAP1 reduces cell
growth and cell migration. Moreover, the protein-protein
interactions of IQGAP1 and CDC42 enhance the oncogenic effects for
glioma.
Targeting protein-protein interaction is a promising
strategy to block cancer signal transduction (29,30).
Several new glioma therapeutic targets are currently being tested
in clinical trials (31,32), providing new approaches of targeted
therapies for glioma. IQGAP1 is a scaffold protein by interacting
with CDC42, which exerts a signal integrator to play crucial roles
in affecting signal intensity and the specific cellular response to
an extracellular cue, and their abnormal levels and changes
contribute to glioma carcinogenesis and progression. Thus, it is
interesting and valuable to further look for chemical small
molecules or protein inhibitors targeting the interaction of IQGAP1
and CDC42, which is a novel strategy to develop new drugs for
glioma.
Acknowledgements
This work was financially supported by the grants
from the Natural Science Foundation of Inner Mongolia (grant no.
2014MS0856).
References
|
1
|
Dolecek TA, Propp JM, Stroup NE and
Kruchko C: CBTRUS statistical report: Primary brain and central
nervous system tumors diagnosed in the United States in 2005–2009.
Neuro Oncol. 14:(Suppl 5). v1–v49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brat DJ, Scheithauer BW, Fuller GN and
Tihan T: Newly codified glial neoplasms of the 2007 WHO
Classification of Tumours of the Central Nervous System:
Angiocentric glioma, pilomyxoid astrocytoma and pituicytoma. Brain
Pathol. 17:319–324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weller M, van den Bent M, Hopkins K, Tonn
JC, Stupp R, Falini A, Cohen-Jonathan-Moyal E, Frappaz D,
Henriksson R, Balana C, et al: European Association for
Neuro-Oncology (EANO) Task Force on Malignant Glioma: EANO
guideline for the diagnosis and treatment of anaplastic gliomas and
glioblastoma. Lancet Oncol. 15:e395–e403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Noritake J, Watanabe T, Sato K, Wang S and
Kaibuchi K: IQGAP1: A key regulator of adhesion and migration. J
Cell Sci. 118:2085–2092. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mataraza JM, Briggs MW, Li Z, Entwistle A,
Ridley AJ and Sacks DB: IQGAP1 promotes cell motility and invasion.
J Biol Chem. 278:41237–41245. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Johnson M, Sharma M and Henderson BR:
IQGAP1 regulation and roles in cancer. Cell Signal. 21:1471–1478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Watanabe T, Wang S, Noritake J, Sato K,
Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T and Kaibuchi
K: Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin
filaments during cell polarization and migration. Dev Cell.
7:871–883. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nabeshima K, Shimao Y, Inoue T and Koono
M: Immunohistochemical analysis of IQGAP1 expression in human
colorectal carcinomas: Its overexpression in carcinomas and
association with invasion fronts. Cancer Lett. 176:101–109. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miyoshi T, Shirakusa T, Ishikawa Y,
Iwasaki A, Shiraishi T, Makimoto Y, Iwasaki H and Nabeshima K:
Possible mechanism of metastasis in lung adenocarcinomas with a
micropapillary pattern. Pathol Int. 55:419–424. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin X, Liu Y, Liu J, Lu W, Liang Z, Zhang
D, Liu G, Zhu H, Xu N and Liang S: The overexpression of IQGAP1 and
β-catenin is associated with tumor progression in hepatocellular
carcinoma in vitro and in vivo. PLoS One. 10:e01337702015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jadeski L, Mataraza JM, Jeong HW, Li Z and
Sacks DB: IQGAP1 stimulates proliferation and enhances
tumorigenesis of human breast epithelial cells. J Biol Chem.
283:1008–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hayashi H, Nabeshima K, Aoki M, Hamasaki
M, Enatsu S, Yamauchi Y, Yamashita Y and Iwasaki H: Overexpression
of IQGAP1 in advanced colorectal cancer correlates with poor
prognosis-critical role in tumor invasion. Int J Cancer.
126:2563–2574. 2010.PubMed/NCBI
|
|
13
|
Casteel DE, Turner S, Schwappacher R,
Rangaswami H, Su-Yuo J, Zhuang S, Boss GR and Pilz RB: Rho
isoform-specific interaction with IQGAP1 promotes breast cancer
cell proliferation and migration. J Biol Chem. 287:38367–38378.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McDonald KL, O'Sullivan MG, Parkinson JF,
Shaw JM, Payne CA, Brewer JM, Young L, Reader DJ, Wheeler HT, Cook
RJ, et al: IQGAP1 and IGFBP2: Valuable biomarkers for determining
prognosis in glioma patients. J Neuropathol Exp Neurol. 66:405–417.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stengel K and Zheng Y: Cdc42 in oncogenic
transformation, invasion, and tumorigenesis. Cell Signal.
23:1415–1423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sakurai-Yageta M, Recchi C, Le Dez G,
Sibarita J-B, Daviet L, Camonis J, D'Souza-Schorey C and Chavrier
P: The interaction of IQGAP1 with the exocyst complex is required
for tumor cell invasion downstream of Cdc42 and RhoA. J Cell Biol.
181:985–998. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goel R, Muthusamy B, Pandey A and Prasad
TS: Human protein reference database and human proteinpedia as
discovery resources for molecular biotechnology. Mol Biotechnol.
48:87–95. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brandt DT, Marion S, Griffiths G, Watanabe
T, Kaibuchi K and Grosse R: Dia1 and IQGAP1 interact in cell
migration and phagocytic cup formation. J Cell Biol. 178:193–200.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang S, Xu Y, Shen G, Zhao X, Zhou J, Li
X, Gong F, Ling B, Fang L, Huang C, et al: Gene expression and
methylation status of 14-3-3sigma in human renal carcinoma tissues.
IUBMB Life. 60:534–540. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu W, Wang X, Liu J, He Y, Liang Z, Xia Z,
Cai Y, Zhou L, Zhu H and Liang S: Downregulation of ARHGDIA
contributes to human glioma progression through activation of Rho
GTPase signaling pathway. Tumour Biol. Oct 10–2016.(Epub ahead of
print). doi: 10.1007/s13277-016-5374-6.
|
|
22
|
Liang S and Shen G: Biomarkers of
gliomaMolecular Targets of CNS Tumors. ISBN:
978-953-307-736-9Garami M: InTech Press; Rijeka: pp. 325–342.
2011
|
|
23
|
Ma R, de Pennington N, Hofer M, Blesing C
and Stacey R: Diagnostic and prognostic markers in gliomas - an
update. Br J Neurosurg. 27:311–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
von Deimling A, Korshunov A and Hartmann
C: The next generation of glioma biomarkers: MGMT methylation, BRAF
fusions and IDH1 mutations. Brain Pathol. 21:74–87. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gerstner ER, Yip S, Wang DL, Louis DN,
Iafrate AJ and Batchelor TT: Mgmt methylation is a prognostic
biomarker in elderly patients with newly diagnosed glioblastoma.
Neurology. 73:1509–1510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong P, Nabeshima K, Nishimura N, Kawakami
T, Hachisuga T, Kawarabayashi T and Iwasaki H: Overexpression and
diffuse expression pattern of IQGAP1 at invasion fronts are
independent prognostic parameters in ovarian carcinomas. Cancer
Lett. 243:120–127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakamura H, Fujita K, Nakagawa H, Kishi F,
Takeuchi A, Aute I and Kato H: Expression pattern of the scaffold
protein IQGAP1 in lung cancer. Oncol Rep. 13:427–431.
2005.PubMed/NCBI
|
|
28
|
Balenci L, Clarke ID, Dirks PB, Assard N,
Ducray F, Jouvet A, Belin MF, Honnorat J and Baudier J: IQGAP1
protein specifies amplifying cancer cells in glioblastoma
multiforme. Cancer Res. 66:9074–9082. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
White CD, Brown MD and Sacks DB: IQGAPs in
cancer: A family of scaffold proteins underlying tumorigenesis.
FEBS Lett. 583:1817–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H, Eishingdrelo A, Kongsamut S and
Eishingdrelo H: G-protein-coupled receptors mediate 14-3-3 signal
transduction. Signal Transduct Target Ther. 1:160182016.doi:
10.1038/sigtrans.2016.18. View Article : Google Scholar
|
|
31
|
Chi AS, Sorensen AG, Jain RK and Batchelor
TT: Angiogenesis as a therapeutic target in malignant gliomas.
Oncologist. 14:621–636. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sanson M: Editorial review: Targets for
glioma treatment: from bench to bedside. Curr Opin Oncol.
20:650–651. 2008. View Article : Google Scholar : PubMed/NCBI
|