Introduction
Gliomas have been reported to be the most common
type of primary intracranial tumors and account for ~80% of all
malignant brain tumors. According to the World Health Organization
(WHO), gliomas are classified into 4 grades of malignancy (I–IV)
based on their histological features, and glioblastoma multiform
(GBM; grade IV) shows the worst prognosis, with a median survival
time of only 12–15 months and a 5-year survival rate <3%
(1). In recent years, The Cancer
Genome Atlas (TCGA) described a robust gene expression-based
molecular classification of GBMs that divided them into 4 subtypes:
proneural, neural, classical and mesenchymal, among which the
mesenchymal subtype was distinguished from the others as being
particularly malignant (2–5). Thus, identification of effective
biomarkers for the mesenchymal type of glioma is eagerly
awaited.
Ras-related GTP-binding protein 43 (RAB43) is a
member of the Ras superfamily with a molecular weight of 23 kDa.
Previous studies have shown that RAB43 is mainly located in
endoplasmic reticulum and Golgi, and plays a role as a key
regulator of vesicle movement, signal transduction and tethering
membrane events in membrane trafficking (6). Recent studies have revealed that some
of the RAB proteins are involved in the regulation of several
signal transduction pathways relating to cell invasion, cell
apoptosis and innate immune response (7,8).
Meanwhile, several members of RABs including RAB1B, RAB3D, RAB27A
and RAB38 have been demonstrated to be dysregulated in a variety of
malignant diseases and to play critical roles in tumor progression
(9–12). Nonetheless, the differential
expression, intracellular function and potential mechanism of RAB43
in tumors have not been reported.
In the present study, we compared the expression
level of RAB43 in high-grade gliomas (HGGs) and low-grade gliomas
(LGGs) and among different molecular subtypes based on the Chinese
Glioma Genome (CGGA) and 4 additional independent microarray
datasets. We also explored the relationship of RAB43 expression
with clinicopathological parameters including overall survival
(OS). Additionally, the protein expression level was validated in
52 glioma samples (WHO grade I–IV) by immunohistochemistry (IHC).
Furthermore, we explored the potential biological impact of RAB43
on the invasive and metastatic properties of glioma cells, and the
effects of RAB43 on cell migration and invasive capability were
assessed in vitro.
Materials and methods
Clinical specimens and
bioinformatics
The present study was approved by the Ethics
Committee of Qilu Hospital. Archived paraffin-embedded glioma
tissues were collected from 52 patients (WHO I–IV) who underwent
surgery at the Department of Neurosurgery, Qilu Hospital of
Shandong University (Shandong, China). Normal brain tissue samples
(n=5) were taken from trauma patients for whom partial resection of
normal brain was required as decompression treatment for their
severe head injuries. Written informed consent was obtained from
all patients. Whole genome mRNA expression microarray data and
clinical information from 310 samples [batch 1, 5 normal brain
tissue (NBT) and 220 diffuse gliomas; batch 2, 85 diffuse gliomas]
from CGGA (http://www.cgcg.org.cn/) were used
for the analysis, containing 126 grade II, 51 grade III and 128
grade IV samples histologically diagnosed according to the WHO
classification. Four external independent glioma databases (TCGA,
Rembrandt, GSE16011 and GSE4290) were included as well.
IHC
Formalin-fixed, paraffin-embedded tissues of 52
specimens including different grades of astrocytic glioma were
included. Endogenous HRP activity was blocked with 3%
H2O2. Antigen retrieval was achieved by
boiling in sodium citrate buffer (pH 6.0). After blocking with 10%
normal goat serum, immunostaining was performed using a mouse
anti-RAB43 monoclonal antibody (cat no. ab58030; Abcam, Cambridge,
UK) at 1:50 dilution. Finally, the visualized signal was developed
with 3,3′-diaminobenzidine (DAB) and the slides were counterstained
in hematoxylin. The sections incubated with normal mouse serum
instead of the primary antibody were used as negative controls. The
results of the immunohistochemical staining were evaluated by two
independent pathologists. The percentage of positive staining cells
was scored as: 0–3 (0 points for no cells stained, 1 points for
<25%, 2 points for 25–75% and 3 points for >75% of cells
stained), and the intensity of immunoreactivity was also graded on
a scale of 0–3 scored as: (0, no staining; 1, weak staining; 2,
moderate staining; and 3, strong staining). The two scores were
then multiplied to yield a total IHC score regarding the expression
of RAB43 protein in a sample. Negative cases (−) had a total score
of 0, weakly positive (+) cases had a total score of 1–2,
moderately positive (++) cases had a score of 3–4, and strongly
positive (+++) cases had a total score of 6–9.
Gene ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analysis
Correlation analysis of RAB43 in whole genome gene
expression profile was performed in the CGGA dataset (n=305). To
detect the biological processes and signaling pathways that
correlate with RAB43 expression in glioma, RAB43 positively and
negatively correlated genes (p<0.01) were analyzed by DAVID web
tool (http://david.abcc.ncifcrf.gov/home.jsp).
Cell culture
U-87 MG and U-251 human GBM cell lines were
purchased from the Culture Collection of the Chinese Academy of
Sciences (Shanghai, China), and cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS) (both from Life Technologies, (Grand Island, NY, USA) and
maintained at 37°C in an atmosphere of humidified air containing 5%
CO2.
RNA interference
Small interference RNA (siRNA) targeting RAB43
(5′-CCATTGAGACGTCTGCCAA-3′) was synthesized by GenePharma Co., Ltd.
(Shanghai, China). For transient silencing, 3×105
cells/well were seeded into 6-well plates and transfected with the
relevant siRNA (100 nmol/well) using Lipofectamine RNAiMAX reagent
(cat no. 13778150; Invitrogen, Carlsbad, CA, USA) following the
manufacturer's protocol.
Western blotting
Total proteins were extracted in lysis buffer
containing 50 mM Tris-HCl, 150 mM NaCl, 1% sodium deoxycholate,
0.1% SDS, 20 mM EDTA, 1 mM NaF and 1% Triton X-100 (pH 7.4) with
protease inhibitors. The protein concentration was determined using
the Bradford assay (Bio-Rad, Hercules, CA, USA). Lysis was run in a
10% sodium dodecyl sulfate-polyacrylamide electrophoresis
(SDS-PAGE) gel, transferred to polyvinylidene difluoride (PVDF)
membranes (Millipore, Billerica, MA, USA), incubated with
antibodies against RAB43 (cat no. ab58030; Abcam), Snail (cat no.
3879), N-cadherin (cat no. 13116), vimentin (cat no. 12020),
β-actin (cat no. 12262), MMP-2 (cat no. 13132) and MMP-9 (cat no.
13667) (all from Cell Signaling Technology, Beverly, MA, USA)
diluted at 1:1,000.
Cell invasion and migration
assays
For Transwell Matrigel invasion assays,
5×104 cells in 100 µl of serum-free medium were plated
onto the upper chamber of 24-well Transwell inserts (8-µm pores; BD
Biosciences, San Diego, CA, USA) coated with Matrigel. The lower
chamber was filled with 600 µl medium containing 20% FBS. After
24–36 h, the non-invaded cells were gently scraped off by cotton
swab. As for the migration assay, 2×104 cells in 100 µl
of serum-free medium were plated onto the upper chamber of the same
inserts and migrated for 12 h. The migrated cells were fixed with
10% formalin, and stained with crystal violet. Five random fields
of each well were photographed and cell numbers were determined by
Kodak Molecular Imaging (MI) software (Kodak, Rochester, NY,
USA).
Statistical analysis
Survival curves were estimated by the Kaplan-Meier
method and compared using the log-rank test. High or low expression
was defined as higher or lower than the median value. Expression
pattern of RAB43 in different subtypes and the associations of
RAB43 with isocitrate dehydrogenase 1 (IDH1) mutation, methylation
of O6-methylguanine-DNA methyltransferase (MGMT)
promoter, co-deletion of 1p/19q, telomerase reverse transcriptase
(TERT) loss, and alpha thalassemia/mental retardation syndrome
X-linked (ATRX) mutation were performed in the CGGA and TCGA
datasets. The one-way ANOVA test or t-test were used for all other
data comparisons using GraphPad Prism 6 software. Pearson
correlation was applied to evaluate the linear relationship between
gene expression. A two-tailed χ2 test was used to
determine the association between RAB43 expression and
clinicopathological characteristics. All data are presented as the
mean ± standard error. All tests were two-sided, and p-values
<0.05 were considered to indicate a statistically significant
result.
Results
Expression of RAB43 is associated with
the grade of progression of the gliomas
By whole-genome expression profiling data, we
compared the mRNA expression level of RAB43 in HGGs and LGGs as
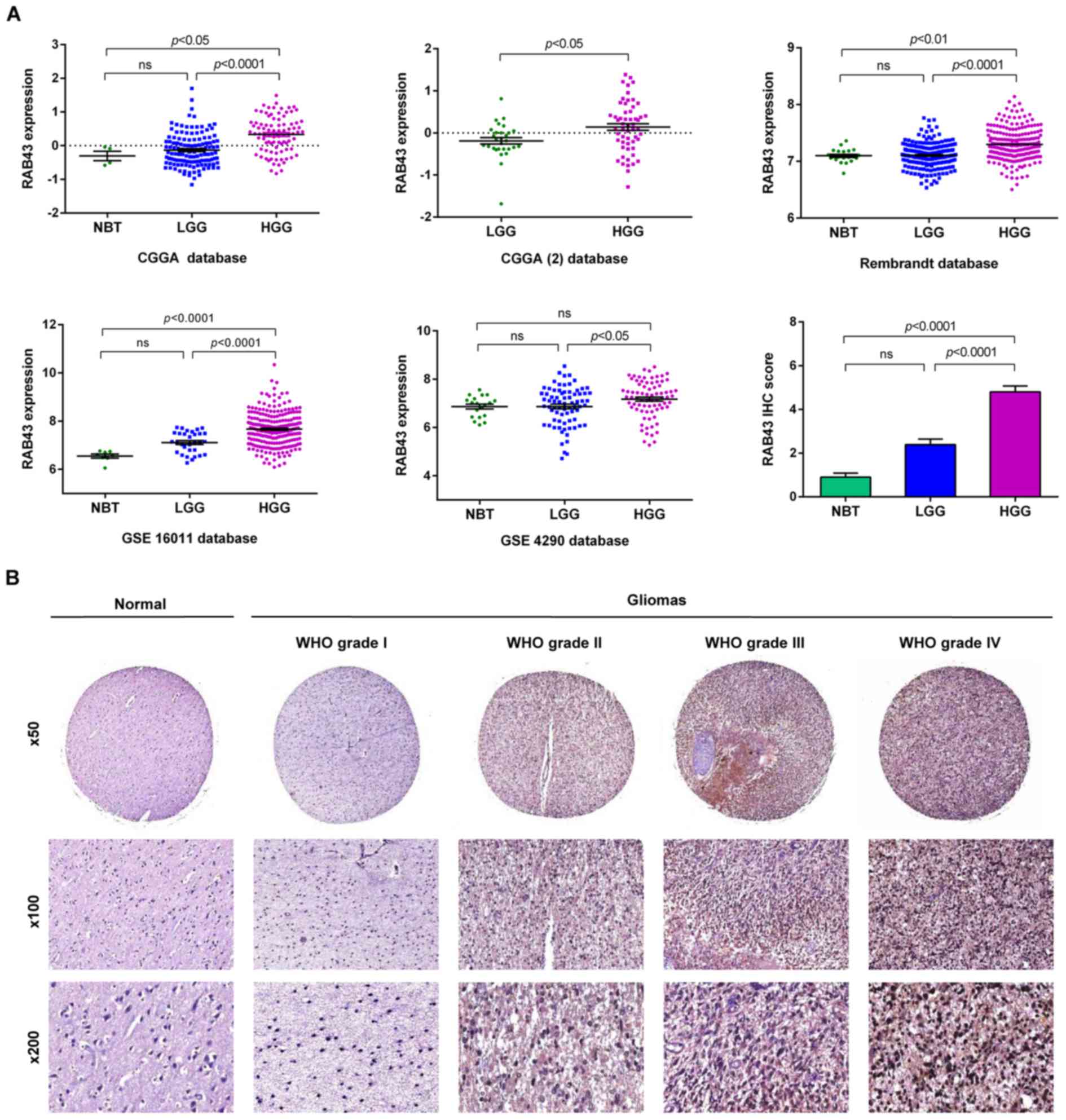
well as normal brain tissues (NBT). As shown in Fig. 1A, the results indicated that RAB43
mRNA expression level was significantly upregulated in the HGGs
compared with that noted in the LGGs both in CGGA (p<0.0001) and
3 additional datasets. There were no significant differences
between LGGs and NBT. Meanwhile, we analyzed the association of
RAB43 with the clinicopathological characteristics of the glioma
patients. As shown in Table I, the
rate of high RAB43 expression tended to increase from grade II to
IV according to the WHO classification (p<0.0001). Furthermore,
high expression of RAB43 was statistically associated with patients
with age ≥45 years (p=0.0056). It was previously demonstrated that
glioma patients with Karnofsky Performance Status (KPS) score
>80 had a better prognosis than those with a KPS of <80
(13). However, no significant
differences were identified between RAB43 expression in relation to
KPS score (p=0.0675).
 | Table I.Clinical features of the glioma
patients with differential expression of RAB43 in CGGA. |
Table I.
Clinical features of the glioma
patients with differential expression of RAB43 in CGGA.
| Variable | RAB43 high expression
(n=152) | RAB43 low expression
(n=153) | P-value |
|---|
| Age (years) |
|
| 0.0056 |
| ≥45 | 78 | 54 |
|
|
<45 | 74 | 99 |
|
| Gender |
|
| 0.4151 |
| Male | 94 | 87 |
|
|
Female | 58 | 66 |
|
| KPS |
|
| 0.0675 |
| ≥80 | 81 | 93 |
|
|
<80 | 29 | 17 |
|
| WHO grade |
|
|
<0.0001 |
| II | 30 | 96 |
|
| III | 32 | 19 |
|
| IV | 90 | 39 |
|
To further validate the expression pattern of RAB43,
we detected the protein level of RAB43 in an independent group of
52 glioma patients and 5 NBT by IHC. RAB43 protein showed a higher
expression status in the HGGs compared with the LGGs (p<0.0001),
which was consistent with our findings in the mRNA microarrays
(Fig. 1B). Thus, RAB43 was
upregulated and positively correlated with the grade of progression
both in silico and in glioma specimens.
High RAB43 expression is related to
poor clinical outcomes in gliomas
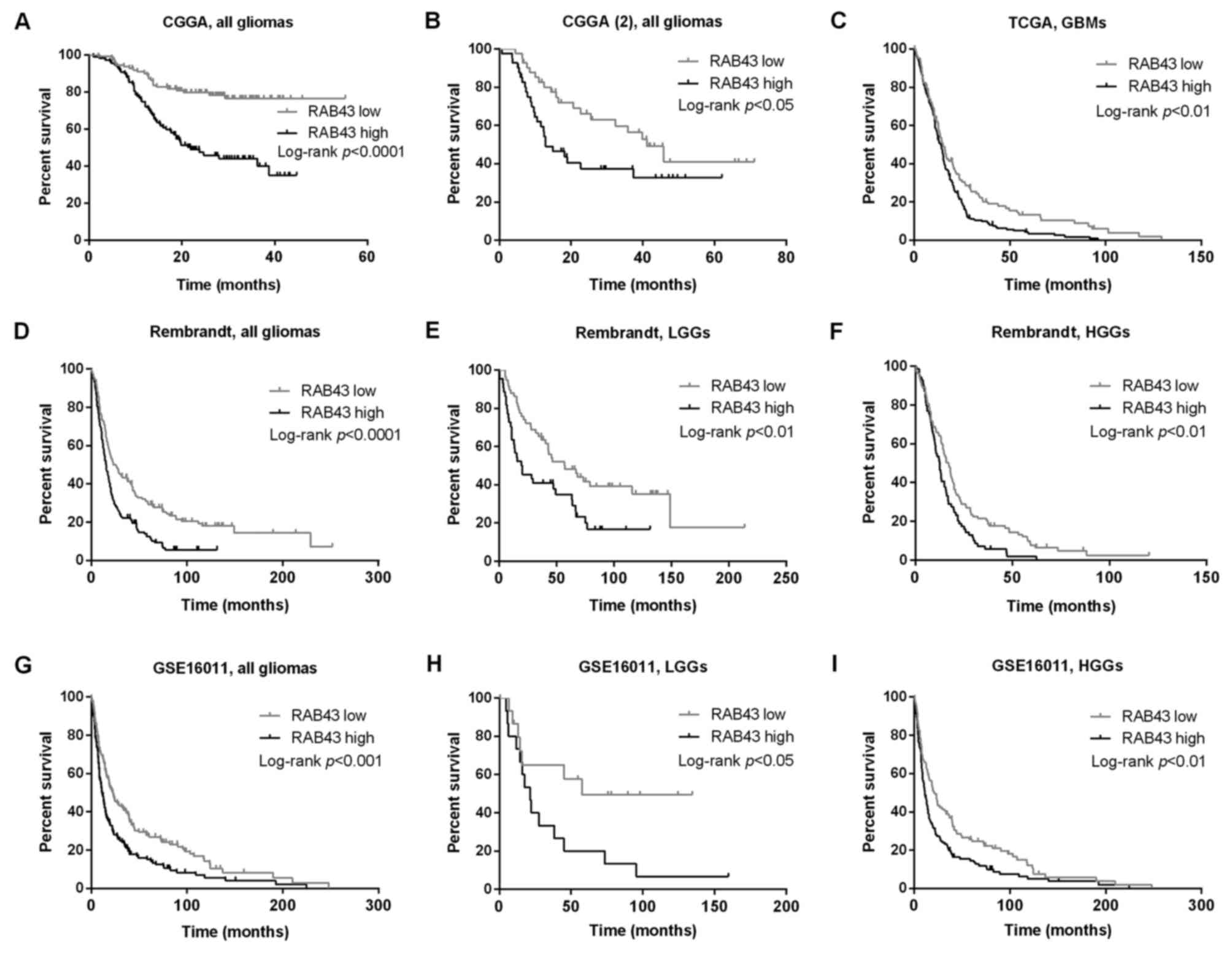
The association of RAB43 expression with prognosis
of the glioma patients was investigated through Kaplan-Meier
survival curves. We found that patients with high RAB43 expression
had a significant shorter OS time than those with low RAB43
expression (Fig. 2A and B). Similar
results were found in the TCGA database among the 348 GBM patients
(Fig. 2C). Moreover, the prognostic
values of RAB43 in GSE16011 (n=276) and Rembrandt databases (n=329)
were analyzed, and the results indicated that high RAB43 was
significantly associated with worse OS both in low- and high-grade
glioma patients (Fig. 2D-I).
RAB43 expression shows subtype
preference
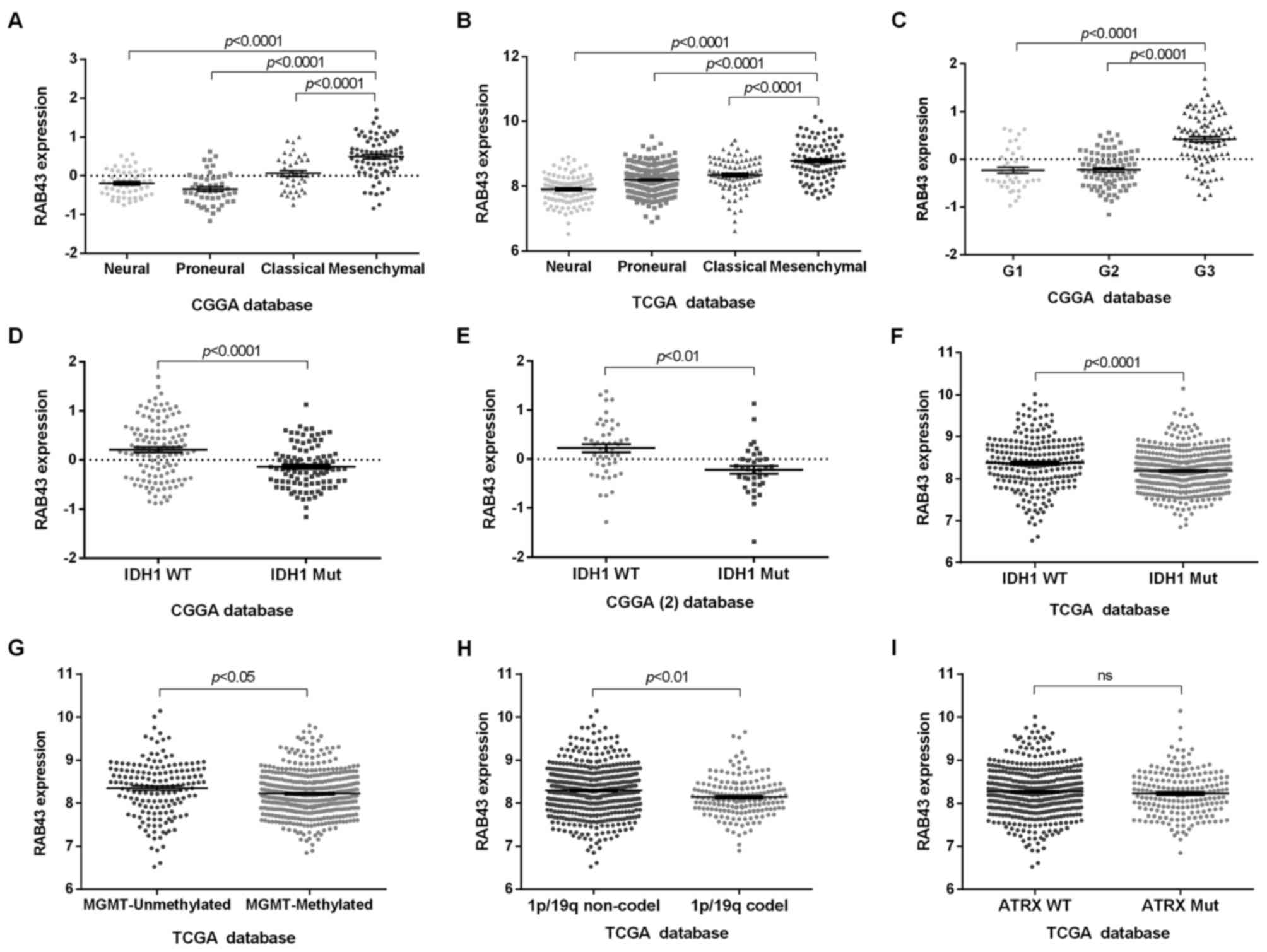
Since RAB43 was upregulated in HGGs, we further
screened its expression pattern among different molecular subtypes.
As shown in Fig. 3A-C, a high RAB43
level was associated with mesenchymal and G3 subtypes. In contrast,
the patients with low RAB43 were more likely to be of the proneural
subtype. More recently, some genetic alterations in glioma patients
have been reported to predict favorable survival, including IDH1
mutation, methylation of MGMT, co-deletion of 1p/19q, TERT loss and
ATRX mutation (14,15). We therefore analyzed whether RAB43
expression was correlated with these characteristics. As a result,
patients with wild-type IDH1 gene showed higher expression of RAB43
than those with mutant IDH1 (Fig.
3D-F). Additionally, RAB43 was observed to be upregulated in
glioma patients with unmethylated MGMT, 1p/19q non-co-deletion, but
not with the wild-type ATRX gene (Fig.
3G-I).
RAB43 is significantly associated with
tumor cell adhesion and invasion
Next, to further understand the biological
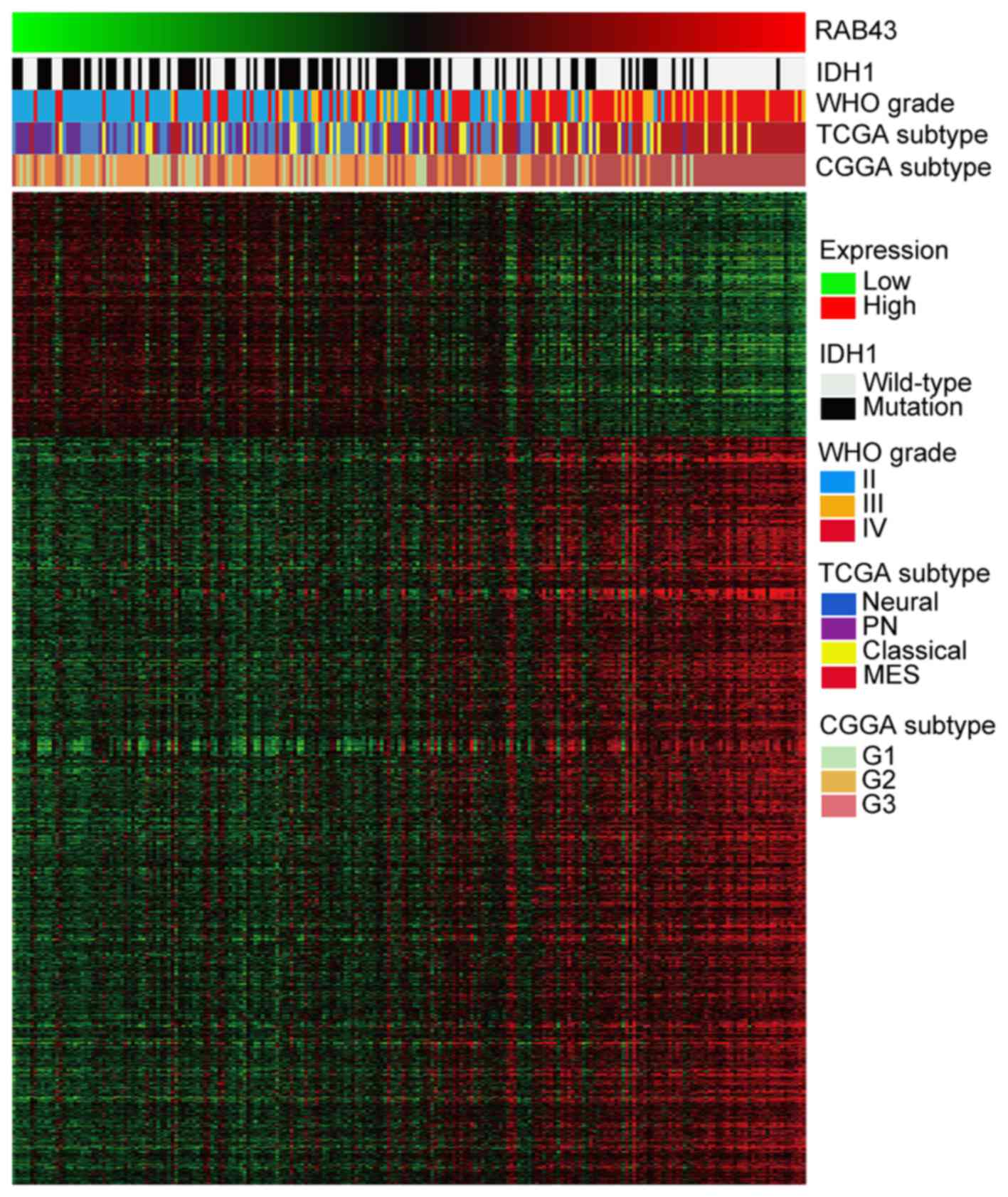
implications of RAB43 in gliomas, correlation analysis of RAB43
expression in whole genome gene profiling was performed in CGGA. As
illustrated in the heatmap in Fig.
4, 1,165 genes were positively correlated and 382 genes were
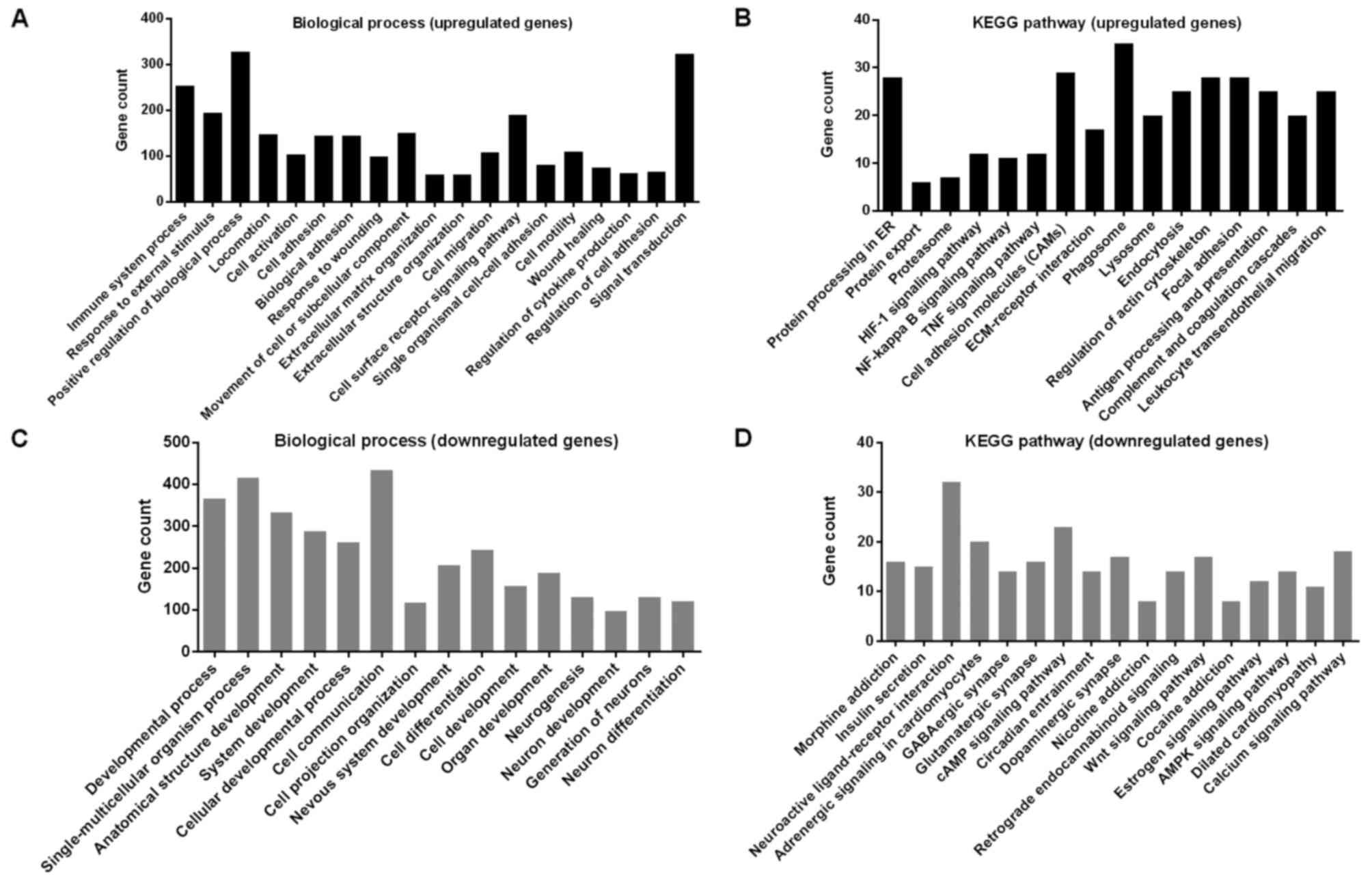
negatively correlated (p<0.01). Afterwards, GO analysis revealed
that RAB43 was strongly associated with biological processes
including response to stimulus, cell adhesion and migration,
extracellular matrix organization, as well as response to wounding
(Fig. 5A; Table II). In the KEGG analysis, the
upregulated genes were enriched in pathways related to focal
adhesion, cell adhesion molecules (CAMs) and ECM-receptor
interaction in cancer progression (Fig.
5B). In contrast, the downregulated genes were enriched in
developmental process, cell differentiation as well as neurogenesis
(Fig. 5C and D).
 | Table II.Gene sets enriched in the glioma
samples with RAB43 high expression |
Table II.
Gene sets enriched in the glioma
samples with RAB43 high expression
| GO name | GO ID | Gene count | P-value |
|---|
| Immune system
process | GO:0002376 | 253 | 1.29E-76 |
| Response to
external stimulus | GO:0009605 | 194 | 2.01E-48 |
| Positive regulation
of biological process | GO:0048518 | 328 | 4.83E-48 |
| Locomotion | GO:0040011 | 147 | 1.29E-38 |
| Cell
activation | GO:0001775 | 103 | 4.1E-37 |
| Cell adhesion | GO:0007155 | 144 | 6.11E-37 |
| Biological
adhesion | GO:0022610 | 144 | 8.46E-37 |
| Response to
wounding | GO:0009611 | 99 | 2.74E-33 |
| Movement of cell or
subcellular component | GO:0006928 | 150 | 9.24E-33 |
| Extracellular
matrix organization | GO:0030198 | 60 | 4.19E-31 |
| Extracellular
structure organization | GO:0043062 | 60 | 4.78E-31 |
| Cell migration | GO:0016477 | 108 | 8.08E-29 |
| Cell surface
receptor signaling pathway | GO:0007166 | 190 | 1.25E-28 |
| Single organismal
cell-cell adhesion | GO:0016337 | 80 | 3.18E-28 |
| Cell motility | GO:0048870 | 110 | 2.47E-27 |
| Wound healing | GO:0042060 | 74 | 2.5E-27 |
| Regulation of
cytokine production | GO:0001817 | 63 | 4.84E-26 |
| Regulation of cell
adhesion | GO:0030155 | 66 | 2.2E-24 |
| Signal
transduction | GO:0007165 | 324 | 4.47E-23 |
Moreover, referring to the aberrant expression of
RAB43 in HGGs and the mesenchymal subtype, co-expression of RAB43
with invasion or metastasis-related factors including matrix
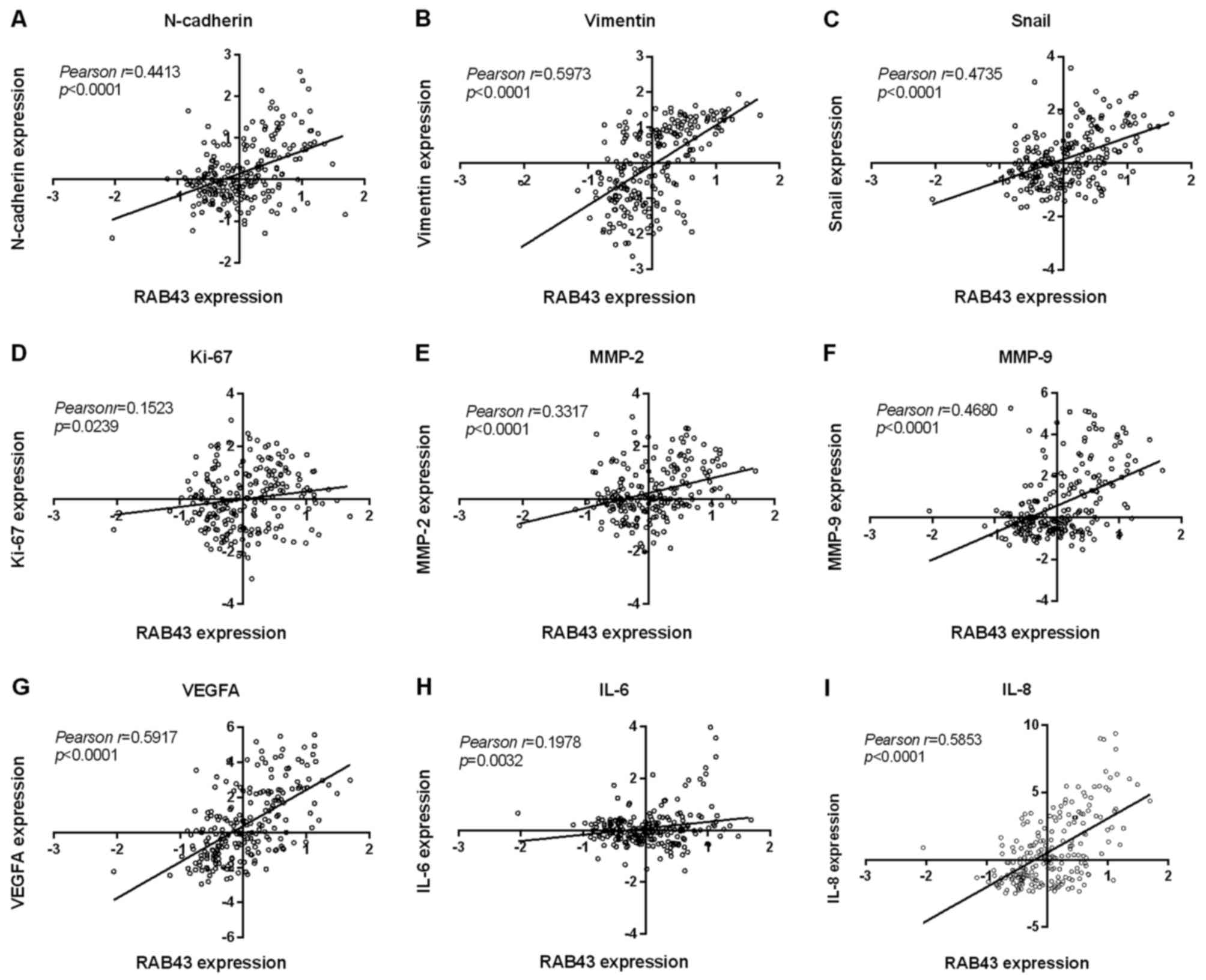
metalloproteinases (MMPs) was assessed. As shown in Fig. 6A-I, there was a significant positive
correlation between RAB43, MMP-2 and MMP-9, as well as several
mesenchymal subtype markers (vimentin, N-cadherin and Snail)
(p<0.0001, respectively). The RAB43 level was also found to be
correlated with other key genes for malignant phenotypes in glioma,
such as interleukin-6 (IL-6), IL-8, vascular endothelial growth
factor A (VEGFA) and Ki-67. Overall, these functional analyses and
correlation results implied the vital role of RAB43 as a potential
oncogene in tumor migration and invasion.
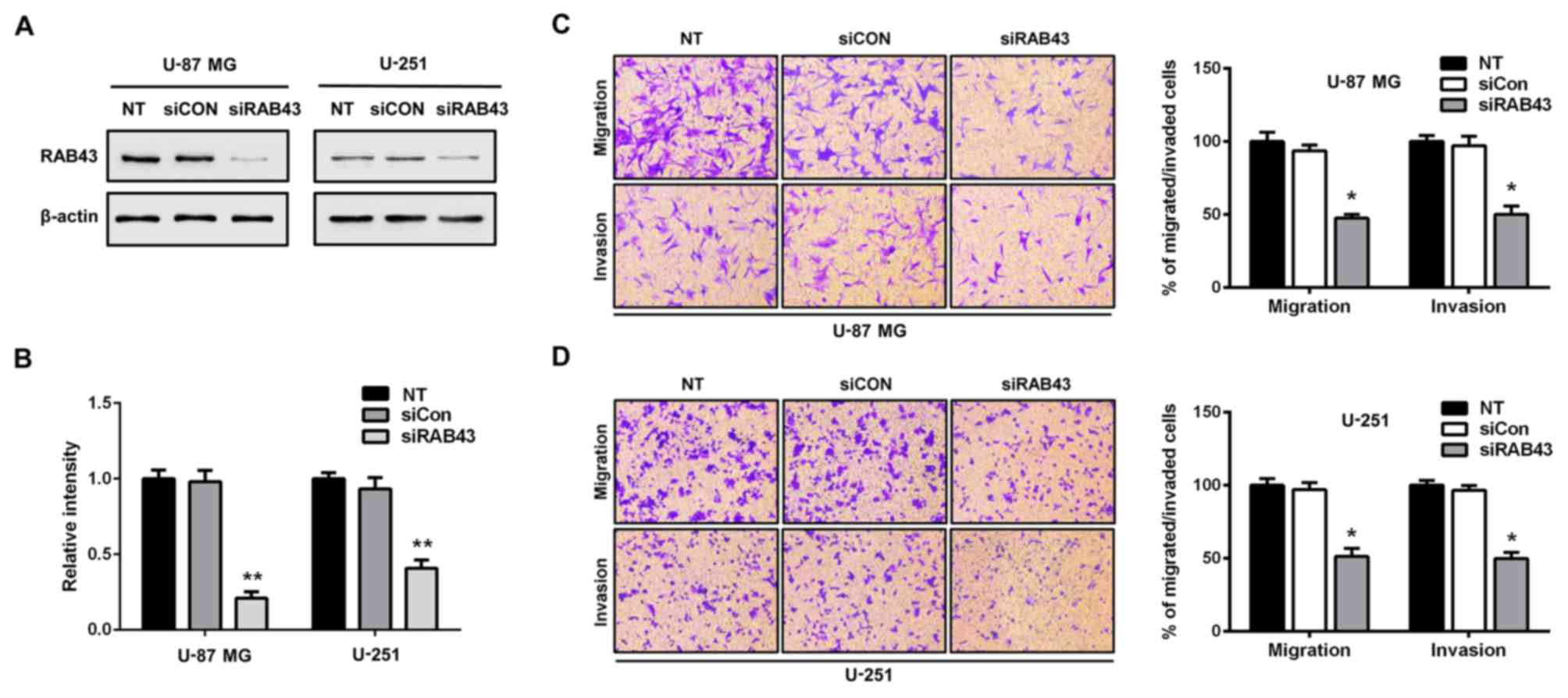
RAB43 silencing reduces glioma cell
migration and invasion
Next, we performed functional assays to determine
the influence of RAB43 on glioma cell migration and invasion. By
siRNA, RAB43 was transiently knockdown in U-87 MG (malignant glioma
cell line annotated as mesenchymal subtype) and U-251 cells
(Fig. 7A and B). Compared with the
siCON group, silencing of RAB43 significantly inhibited both the
migration and invasiveness of the U-87 MG glioma cells (p<0.05;
Fig. 7C). Similar results were
observed in the U-251 human glioma cells (p<0.05; Fig. 7D). Meanwhile, downregulation of
MMP-2 and MMP-9 levels was observed after silencing of RAB43 in the
U-87 MG and U-251 cells (Fig. 8A).
Taken together, these results indicated that suppression of RAB43
significantly inhibited the migration and invasion of glioma
cells.
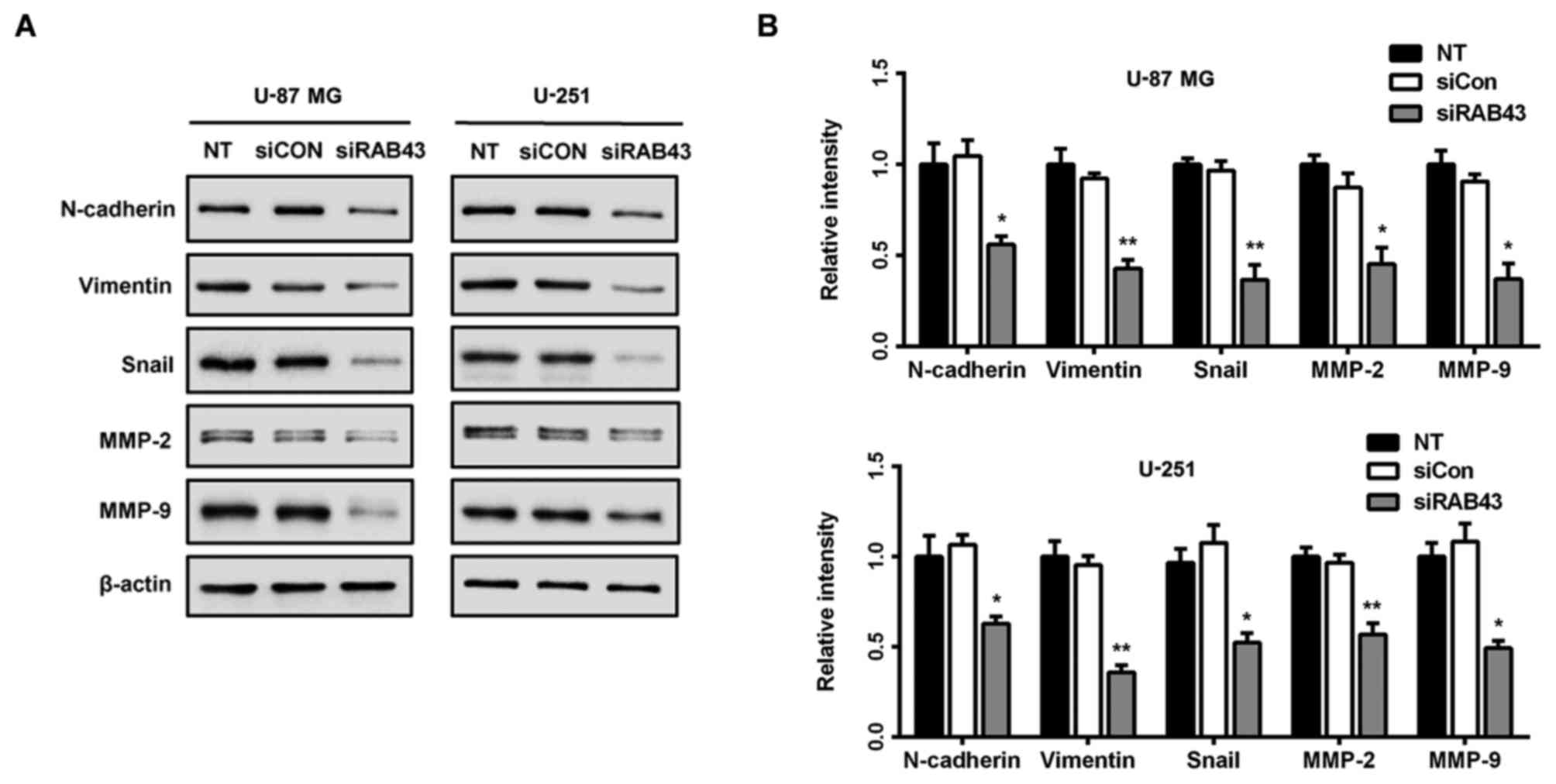
RAB43 knockdown inhibits mesenchymal
properties in glioma cells
Enhanced cell migration and invasion capabilities
are important consequences of epithelial-mesenchymal transition
(EMT), an early event in tumor metastasis (16). Therefore, we examined the expression
of molecules associated with EMT in glioma cells. As shown in
Fig. 8A and B, RAB43 knockdown
significantly suppressed expression of mesenchymal markers
(N-cadherin and vimentin), as well as EMT-related transcription
factor Snail in both the U-87 MG and U-251 cells. Thus, RAB43
expression may contribute to the invasiveness and poor prognosis in
patients by promoting EMT in glioma cells.
Discussion
Recently, several members of the RAB family have
been indicated to participate in cancer progression (17). For instance, RAB25 contributes to
tumor progression by directing the localization of
integrin-recycling vesicles and promoting the invasive ability of
tumor cells in breast and ovarian cancers (18,19).
RAB3D was reported to play a crucial role in invasion and
metastasis via the EMT process in colorectal cancer (10). As a member of the RAB family, RAB43
was identified to be located in endoplasmic reticulum and Golgi,
which plays crucial roles in protein transport and Golgi
organization (6). However, the
effect of RAB43 in tumors has not been previously reported. In the
present study, according to the mRNA microarray of CGGA, RAB43 was
highly expressed in HGGs in comparison with LGGs (Fig. 1A). Due to differences in the genetic
background between populations, we subsequently validated these
findings in 4 published cohorts. In addition, the protein level of
RAB43 was investigated by IHC from an independent group of patients
(n=52; Fig. 1B), and similar
findings were observed. Moreover, patients with high RAB43 had
worse overall survival than those with low RAB43 in both LGGs and
HGGs (Fig. 2). Collectively, these
data suggest that RAB43 may be associated with the malignant
phenotypes of gliomas and could serve as a novel prognostic
indicator in clinical practice.
Characterized by particular molecular signatures,
glioma has recently been classified into distinct molecular
subtypes, among which the mesenchymal subtype-associated genes were
more related to aggressive behavior in tumors and conferred a poor
prognosis in patients (20).
Recently, CGGA derived 3 subgroups of gliomas including G1, G2 and
G3 with differences in clinical characteristics based on the
Chinese population (5). The G1
subgroup was characterized by favorable clinical outcome, young
age, low malignant behaviors and high IDH1 mutation, while G3
groups exhibited contrasting characteristics with poor prognosis
and low rate of IDH1 mutation. Notably, consistent with the worse
outcomes of patients with high RAB43, we found a significantly
increased expression of RAB43 in mesenchymal and G3 subtypes, and
IDH1 wild-type patients. In contrast, patients with low RAB43 were
more likely to be of the proneural subtype and IDH1 mutation
patients. Additionally, RAB43 was also highly expressed in glioma
patients with unmethylated MGMT and 1p/19q non-co-deletion
(Fig. 3).
To further explore the biological relevance of the
RAB43 transcriptome, functional clustering annotation and
integration into KEGG and GO analysis were performed based on the
CGGA dataset. GO analyses revealed that RAB43-assosiated genes
showed significant enrichment mainly in biological processes
related to cell adhesion and migration, extracellular matrix
organization, as well as response to wounding (Fig. 5A; Table
II). In the KEGG analysis, the upregulated genes were enriched
in pathways related to focal adhesion, cell adhesion molecules
(CAMs), ECM-receptor interaction in cancer progression and
immune-related pathways (Fig. 5B).
Together, these data further implicate the critical role of RAB43
in invasiveness and metastasis of gliomas.
Excessive migration and invasion are hallmarks of
malignant tumors. Recent studies suggest a role of matrix
metalloproteinases (MMPs) in the process of glioma cell invasion
(21,22). Among all MMPs, MMP-2 and MMP-9 play
important roles in basement membrane type IV collagen degradation
during tumor migration and invasion (23). In the present study, we evaluated
the influence of RAB43 alteration on the migration and invasion of
glioma cells. As expected, RAB43 knockdown prominently reduced the
migratory and invasive abilities of the glioma cells (Fig. 7C and D). Meanwhile, MMP-2 and MMP-9
were also significantly reduced after RAB43 knockdown (Fig. 8), which suggest the role of RAB43 in
glioma metastasis.
It has been reported that EMT occurs during tumor
progression, leading to increased motility and invasiveness of
cancer cells. An obvious characteristic of EMT is upregulation of
mesenchymal markers such as N-cadherin, vimentin and Snail
(24–26). However, the impact of the RAB family
on EMT processes remain unclear. Hence, we evaluated whether RAB43
was involved in the regulation of EMT-specific proteins. As shown
in Fig. 8A and B, the siRAB43 group
showed significant decreases in N-cadherin, vimentin and Snail
expression. Taken together, these findings imply that RAB43
expression may contribute to metastasis and poor prognosis in
patients by promoting EMT in glioma cells. However, further
investigation is still needed to elucidate the regulatory
mechanisms of RAB43 in the EMT process.
In summary, the present study demonstrated for the
first time that RAB43 is overexpressed in glioma tissues, and
increased expression of RAB43 is associated with poor prognostic
features. In addition, high RAB43 expression is related to the
mesenchymal and G3 subtypes, as well as the wild-type IDH1 gene in
glioma patients. In vitro experiments revealed that RAB43
regulates EMT and the invasiveness of glioma cells. Altogether,
RAB43 may serve as a novel biomarker and a potential therapeutic
target for malignant gliomas.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of China (nos. 81502164, 81402060 and
81572487), the Shandong Provincial Natural Science Foundation
(BS2015YY004 and BS2014YY033), the Special Foundation for Taishan
Scholars (nos. ts20110814 and tshw201502056), the Fundamental
Research Funds of Shandong University, the Department of Science
and Technology of Shandong Province (2015GGE27101 and
2015ZDXX0801A01), the University of Bergen, The Helse Bergen,
Norway and the Norwegian Centre for International Cooperation in
Education (SIU) (UTF-2014/10047).
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brennan C: Genomic profiles of glioma.
Curr Neurol Neurosci Rep. 11:291–297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huse JT, Phillips HS and Brennan CW:
Molecular subclassification of diffuse gliomas: Seeing order in the
chaos. Glia. 59:1190–1199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sulman EP and Aldape K: The use of global
profiling in biomarker development for gliomas. Brain Pathol.
21:88–95. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan W, Zhang W, You G, Zhang J, Han L, Bao
Z, Wang Y, Liu Y, Jiang C, Kang C, et al: Molecular classification
of gliomas based on whole genome gene expression: A systematic
report of 225 samples from the Chinese Glioma Cooperative Group.
Neuro Oncol. 14:1432–1440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dejgaard SY, Murshid A, Erman A, Kizilay
O, Verbich D, Lodge R, Dejgaard K, Ly-Hartig TB, Pepperkok R,
Simpson JC, et al: Rab18 and Rab43 have key roles in ER-Golgi
trafficking. J Cell Sci. 121:2768–2781. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu N and Zhou Z: Membrane trafficking and
phagosome maturation during the clearance of apoptotic cells. Int
Rev Cell Mol Biol. 293:269–309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang R, Zhang Y, Liu S, Li C, Sun L, Bao
L, Feng J and Liu Z: Analysis of 52 Rab GTPases from channel
catfish and their involvement in immune responses after bacterial
infections. Dev Comp Immunol. 45:21–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nikoshkov A, Broliden K, Attarha S,
Sviatoha V, Hellström AC, Mints M and Andersson S: Expression
pattern of the PRDX2, RAB1A, RAB1B, RAB5A and RAB25 genes in normal
and cancer cervical tissues. Int J Oncol. 46:107–112.
2015.PubMed/NCBI
|
|
10
|
Luo Y, Ye GY, Qin SL, Mu YF, Zhang L, Qi
Y, Qiu YE, Yu MH and Zhong M: High expression of Rab3D predicts
poor prognosis and associates with tumor progression in colorectal
cancer. Int J Biochem Cell Biol. 75:53–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H, Zhao Y, Zhang C, Li M, Jiang C and
Li Y: Rab27a was identified as a prognostic biomaker by mRNA
profiling, correlated with malignant progression and subtype
preference in gliomas. PLoS One. 9:e897822014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang H and Jiang C: RAB38 confers a poor
prognosis, associated with malignant progression and subtype
preference in glioma. Oncol Rep. 30:2350–2356. 2013.PubMed/NCBI
|
|
13
|
Tortosa A, Viñolas N, Villà S, Verger E,
Gil JM, Brell M, Caral L, Pujol T, Acebes JJ, Ribalta T, et al:
Prognostic implication of clinical, radiologic, and pathologic
features in patients with anaplastic gliomas. Cancer. 97:1063–1071.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang
X, Jiang C, Kang C, Li X, Chen L, et al: Chinese Glioma Cooperative
Group (CGCG): CGCG clinical practice guidelines for the management
of adult diffuse gliomas. Cancer Lett. 375:263–273. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the Central Nervous System: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kahlert UD, Nikkhah G and Maciaczyk J:
Epithelial-to-mesenchymal(−like) transition as a relevant molecular
event in malignant gliomas. Cancer Lett. 331:131–138. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Subramani D and Alahari SK:
Integrin-mediated function of Rab GTPases in cancer progression.
Mol Cancer. 9:3122010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Caswell PT, Spence HJ, Parsons M, White
DP, Clark K, Cheng KW, Mills GB, Humphries MJ, Messent AJ, Anderson
KI, et al: Rab25 associates with alpha5beta1 integrin to promote
invasive migration in 3D microenvironments. Dev Cell. 13:496–510.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng KW, Lahad JP, Kuo WL, Lapuk A,
Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et
al: The RAB25 small GTPase determines aggressiveness of ovarian and
breast cancers. Nat Med. 10:1251–1256. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arimappamagan A, Somasundaram K,
Thennarasu K, Peddagangannagari S, Srinivasan H, Shailaja BC,
Samuel C, Patric IR, Shukla S, Thota B, et al: A fourteen gene GBM
prognostic signature identifies association of immune response
pathway and mesenchymal subtype with high risk group. PLoS One.
8:e620422013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu L, Wu J, Ying Z, Chen B, Han A, Liang
Y, Song L, Yuan J, Li J and Li M: Astrocyte elevated gene-1
upregulates matrix metalloproteinase-9 and induces human glioma
invasion. Cancer Res. 70:3750–3759. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tester AM, Ruangpanit N, Anderson RL and
Thompson EW: MMP-9 secretion and MMP-2 activation distinguish
invasive and metastatic sublines of a mouse mammary carcinoma
system showing epithelial-mesenchymal transition traits. Clin Exp
Metastasis. 18:553–560. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan LX, Liu YH, Xiang JW, Wu QN, Xu LB,
Luo XL, Zhu XL, Liu C, Xu FP, Luo DL, et al: PIK3R1 targeting by
miR-21 suppresses tumor cell migration and invasion by reducing
PI3K/AKT signaling and reversing EMT, and predicts clinical outcome
of breast cancer. Int J Oncol. 48:471–484. 2016.PubMed/NCBI
|
|
26
|
Yokoyama K, Kamata N, Fujimoto R, Tsutsumi
S, Tomonari M, Taki M, Hosokawa H and Nagayama M: Increased
invasion and matrix metalloproteinase-2 expression by Snail-induced
mesenchymal transition in squamous cell carcinomas. Int J Oncol.
22:891–898. 2003.PubMed/NCBI
|