Introduction
Gastric cancer (GC) is a malignant tumor that occurs
in the gastric mucosa of the digestive tract (1). GC has become the fourth most common
cancer in the world and ranks second as the cause of cancer death
(2). GC is one of the main diseases
in the clinic, with features such as a high incidence rate, a lack
of obvious clinical symptoms, invasion and metastasis and a low
cure rate (3).
Surgical resection is an ideal method for the
treatment of gastric cancer. A total of 90% of early gastric cancer
patients survive 5 years after the surgical resection of the
lesions (4). However, most patients
confirmed with gastric cancer are in the progressive stage due to
the lack of specific symptoms of early gastric cancer (5). Therefore, the mortality rate of
gastric cancer stays at a high level. Although the cancer genes,
tumor suppressor genes and tumor signaling pathways associated with
gastric cancer have been discovered and confirmed, its pathogenesis
is not fully clear. Thus, the search for new biological targets to
monitor and intervene in GC carcinogenesis is urgent for GC
prevention.
Circular RNA (circRNA) is a class of endogenous
non-coding RNA (ncRNA) molecules that regulate gene expression
(6). Some circRNAs can act as
microRNA (miRNA) sponges to regulate the expression levels of other
related RNAs (7,8). In other words, circRNAs can use miRNA
response elements to bind miRNAs and block the inhibitory effects
of miRNAs on the expression of their target genes (9).
circRNAs play very important roles in many diseases,
such as nervous system disorders, atherosclerosis, diabetes and
cancer (10). Although the role of
circRNAs in the occurrence of the disease has received attention,
research into the relationship between circRNAs and gastric cancer
is rare.
In the present study, we analyzed the differential
expression profiles of circRNAs in gastric cancer tissues and
adjacent tissues to explore the relationship between the circRNA
expression and GC and to provide a preliminary theoretical basis
for search of biomarkers for the early diagnosis and malignant
progression of GC.
circRNAs have important functions within living
organisms. At present, circRNAs are known to function through a
number of other molecules, such as splicing factors, RNA polymerase
II (11), nuclear small ribose
nucleoprotein (snRNP) (12), and
miRNAs (13), to regulate host gene
(host gene) linear mRNA production; these interactions can promote
or inhibit the transcription of the corresponding linear mRNA. If
the molecular mechanism is based on the circRNA regulation of host
genes as described above, we can analyze the same gene produced by
circRNA and linear mRNA in the tested sample to evaluate trends in
changes of expression to speculate on the molecular mechanism
underlying the functions of circRNAs in the organism.
Materials and methods
Human subjects: Patients and healthy
controls
The 8 GC tissues and adjacent tissue samples used in
this study were collected from the Shenzhen People's Hospital. All
patients aged from 42 to 76 years who had not been treated with
radiotherapy or chemotherapy prior to surgery were included.
Specimens were collected from March to May in 2015. The
experimental study was approved by the hospital ethics committee.
The present study obtained the consent of the patients or their
families by signing the informed consent forms.
Tissue sample collection
GC and adjacent tissues (1 cm3) obtained
after stomach cancer surgeries were immediately washed with 0.9%
NaCl (RNase-free) and quickly dipped in RNase inhibitor (Epicentre,
Madison, WI, USA) according to the manufacturers instructions.
After storage at 4°C overnight, the depressors were removed from
the biopsies and the biopsies were stored at −80°C for further
testing.
RNA sample preparation
Total RNA was extracted from the renal cortex pieces
using TRIzol (Invitrogen) according to the manufacturers
instructions. The concentration and quality of the total RNA were
measured by UV absorbance at 260 nm and 280 nm (A260/280) and
verified by gel electrophoresis.
Screening and identification of
gastric cancer-associated circRNAs
Sample labeling and array hybridization were
performed according to the manufacturers protocol (Arraystar Inc.,
Rockville, MD, USA). Briefly, total RNA was digested with RNase R
(Epicentre) to remove linear RNAs and to enrich circular RNAs.
Then, the enriched circular RNAs were amplified and transcribed
into fluorescent cRNA using a random priming method (Arraystar
Super RNA Labeling kit; Arraystar). The labeled cRNAs were purified
with the RNeasy Mini kit (Qiagen). The concentration and specific
activity of the labeled cRNAs (pmol Cy3/µg cRNA) were measured by
NanoDrop ND-1000. A total of 1 µg of each labeled cRNA was
fragmented by adding 5 µl of 10X blocking agent and 1 µl of 25X
fragmentation buffer. Then, the mixtures were heated at 60°C for 30
min. Finally, 25 µl of 2X hybridization buffer was added to dilute
the labeled cRNA. A total of 50 µl of the hybridization solution
was dispensed into the gasket slide and assembled to construct the
circRNA expression microarray slide. The slides were incubated for
17 h at 65°C in an Agilent Hybridization Oven. The hybridized
arrays were washed, fixed and scanned using the Agilent scanner
G2505C.
Screening and identification of
gastric cancer-associated mRNAs
Human whole genome oligonucleotide microarrays
representing all known genes and transcripts in the human gene were
applied in the extensive view. The sequences were collected from a
wide range of sources and were validated and optimized through
comparison to the assembly of the human genome. Sample labeling and
hybridization were performed in accordance with the Agilent
One-Color Microarray-Based Gene Expression Analysis experiment
scheme with slight modifications. First, the total RNA was
amplified from each sample and used as a Cy3-UTP marker. Second,
the labeled cRNAs were purified with the RNeasy Mini kit and the
concentration and activity were detected with the NanoDrop ND-1000.
Finally, the slides were incubated, washed, fixed and scanned using
the Agilent scanner G2505C.
Differentially expressed gene analysis
and functional analysis
We used the Agilent Feature Extraction v11.0.1.1
software graph chip to read the values and to obtain the original
data. The GeneSpring GX v12.1 software was used for quantile
normalization and subsequent data processing of the original data.
After the original data standardization, we screened for the high
quality probe (a probe that is marked as 1 for detection in at
least two samples). Two groups of samples with significantly
differentially expressed genes were identified by scatter plot
screening. The difference between the gene expression in the two
samples was validated through fold change screening. Hierarchical
clustering was performed using a script prepared by Kang Cheng
(Shanghai, China). The GO analysis and pathway analysis were
performed using the standard enrichment calculation method.
Data acquisition and analysis
Scanned images were imported into the Agilent
Feature Extraction software for raw data extraction. Quantile
normalization of the raw data and subsequent data processing were
performed using the R software package. When comparing two groups
of profile differences (i.e., disease vs. control), the ‘fold
change’ (i.e., the ratio of the group averages) was computed
between the groups for each circRNA. The statistical significance
of the difference may be conveniently estimated using the t-test.
circRNAs with fold changes <1.5 and P-values <0.05 were
selected as significantly differentially expressed genes.
Validation of differentially expressed
circRNAs by qRT-PCR
We randomly chose circRNAs with upregulated and
down-regulated expression to verify the results using qRT-PCR. The
primers are listed in Table I. The
cycle parameters for the PCR reaction were 95°C for 5 min, followed
by 40 cycles of a denaturing step at 95°C for 10 sec and an
annealing/extension step at 60°C for 60 sec. All reactions were run
in triplicate. The Rotor-Gene Real-Time Analysis software 6.0 was
used for the data analysis.
 | Table I.Real-time qRT-PCR primer list. |
Table I.
Real-time qRT-PCR primer list.
| circRNA | Primer sequences
(5–3) | Product sizes/bp | Annealing
temperature/°C |
|---|
| GAPDH(HUMAN) | F:
GGGAAACTGTGGCGTGAT |
|
| R:
GAGTGGGTGTCGCTGTTGA | 299 |
|
hsa_circRNA_400071 | F:
CCCAAGGGTTTGGTGGGAT |
|
| R:
AGAGCCCAGAGTGGGAGAAGTC | 109 |
|
hsa_circRNA_000792 | F:
AGGAAGTACAATGAAGTGCCAGT |
|
| R:
AGAATGCCCAAAGACAAAGC | 157 |
|
hsa_circRNA_000543 | F:
GAATAGATTTCAGCTTTATGC |
|
| R:
CATAACTGATCTGACTTTGTATG | 83 | 60 |
|
hsa_circRNA_001959 | F:
GTCTAGTGGAGCAGGTGGAGG |
|
| R:
AGGGCCATGACCTGGGTTA | 137 |
|
hsa_circRNA_400066 | F:
GACCCTTCCTGGCGGTTAC |
|
| R:
AGACCCACAAGCCCATTCC | 237 |
|
hsa_circRNA_001066 | F:
CTTTTTCAGACTATTTCGTGTCC |
|
| R:
CTTCTGGAAGTACCCAATATGC | 88 |
The fold changes were calculated with the
2−∆∆Ct method (14). To
reduce errors caused by the RNA concentration and transcription
efficiency, we used the housekeeping gene GAPDH (its expression
level was basically constant in the different samples) as an
internal control. The relative gene contents under the test
conditions were the gene value ratios of the sample and internal
control.
Correlation analysis of circRNAs and
mRNAs in gastric cancer
We used the Agilent circRNA and mRNA expression
profile microarray data to screen the differentially expressed
circRNA and mRNA gene expression profiling information for the
integration expression analysis. To investigate whether
differentially expressed circRNAs could perform miRNA-specific
adsorption, we evaluated the regulation of the expression of their
target gene mRNAs. We investigated the functions of circRNAs and
the mechanism underlying the occurrence of gastric cancer.
Results
Screening of the differentially
expressed circRNA
The circRNA expression profile in the GC and
adjacent tissues is shown in Fig.
1. Through the data analysis, we selected 4,458 circRNAs whose
expression levels were upregulated or downregulated in the GC
tissues.
To determine whether there was a significant
difference in circRNA expression between the two groups of samples,
we performed a statistical analysis of data from the screened
differentially expressed circRNAs. The circRNA expression levels
were normalized to the same order of magnitude prior to the
statistical analysis. circRNAs whose differences in expression were
>2-fold and had P-values <0.05 were regarded as
differentially expressed circRNAs. There were 467 differentially
expressed circRNAs, of which 214 were significantly upregulated and
253 were significantly downregulated.
qRT-PCR validation results
After the comparison and the analysis of the
differentially expressed multiple circRNAs in the microarray
experiments, we randomly verified three circRNAs
(hsa_circRNA_400071, hsa_circRNA_000792 and hsa_circRNA_000543)
with upregulated expression and three circRNAs (hsa_circRNA_001959,
hsa_circRNA_400066 and hsa_circRNA_001066) with downregulated
expression.
Using GAPDHA as the internal control, we used
qRT-PCR to verify the expression levels of the above 6 circRNAs.
The results are shown in Table II.
There were significant differences in all of the selected circRNAs
(P<0.05) with the exception of hsa_circRNA_000543 (P=0.235). The
objective circRNA validation rate was 5/6, showing that the circRNA
expression profiles were reliable.
 | Table II.Verification of the circRNA
microarray results. |
Table II.
Verification of the circRNA
microarray results.
| circRNA | Fold change |
2−∆∆Ct | P-value |
|---|
|
hsa_circRNA_400071 | 3.77 | 3.09 | 0.007 |
|
hsa_circRNA_000792 | 3.63 | 1.28 | 0.046 |
|
hsa_circRNA_000543 | 3.62 | 1.16 | 0.235 |
|
hsa_circRNA_001959 | −2.14 | 0.29 | 0.00005 |
|
hsa_circRNA_400066 | −2.33 | 1.27 | 0.025 |
|
hsa_circRNA_001066 | −4.04 | 0.67 | 0.002 |
Interaction between differentially
expressed circRNAs and miRNAs
Differentially expressed circRNAs contained
corresponding miRNA binding sites. To facilitate the investigation,
the interactions between miRNAs and circRNAs were predicted by an
in-house miRNA target prediction software (Arraystar). The
circRNA/miRNA interaction information for all circRNAs is noted in
detail. For example, in hsa_circRNA_101242, the 248th-253rd and the
602nd-607th nucleotides starting from the 5′ terminus in the
high-expression gene PAN3 were completely complementary to the
miR-21-3p seed region in the 7mer-m8 binding mode. However, the
248th-253rd nucleotides starting from the 5′ terminus were not
completely complementary to the miR-21-3p seed region (Fig. 2A1).
In hsa_circRNA_001622, the 148th-153rd nucleotides
starting from the 5′ terminus in the high-expression gene SLAMF6
gene were completely complementary to the miR-130b-5p seed region
in the 8mer binding mode (Fig.
2A2). Genes including PPP4R1, PPP4R1, TMCO3, SNX13 and LPAR1
were complementary to the miR-130b-5p seed region.
In hsa_circRNA_103122, the 609th-614th nucleotides
starting from the 5′ terminus in the low-expression gene DONSON
were completely complementary to the miR-135a-5p seed region in the
Offset 6mer binding mode (Fig.
2B1). Genes including MYH14, ATG3, PDS5A, MYO10 and STXBP5 were
complementary to the miR-135a-5p seed region.
In hsa_circRNA_101320, the 164th-169th nucleotides
starting from the 5′ terminus in the low-expression gene PRMT5 were
completely complementary to the miR-335-5p seed region in the 8mer
binding mode. Moreover, the 164th-169th nucleotides starting from
the 5′ terminus in the low-expression gene PRMT5 were completely
complementary to the miR-188-5p seed region in the 7mer-m8 binding
mode (Fig. 2B2).
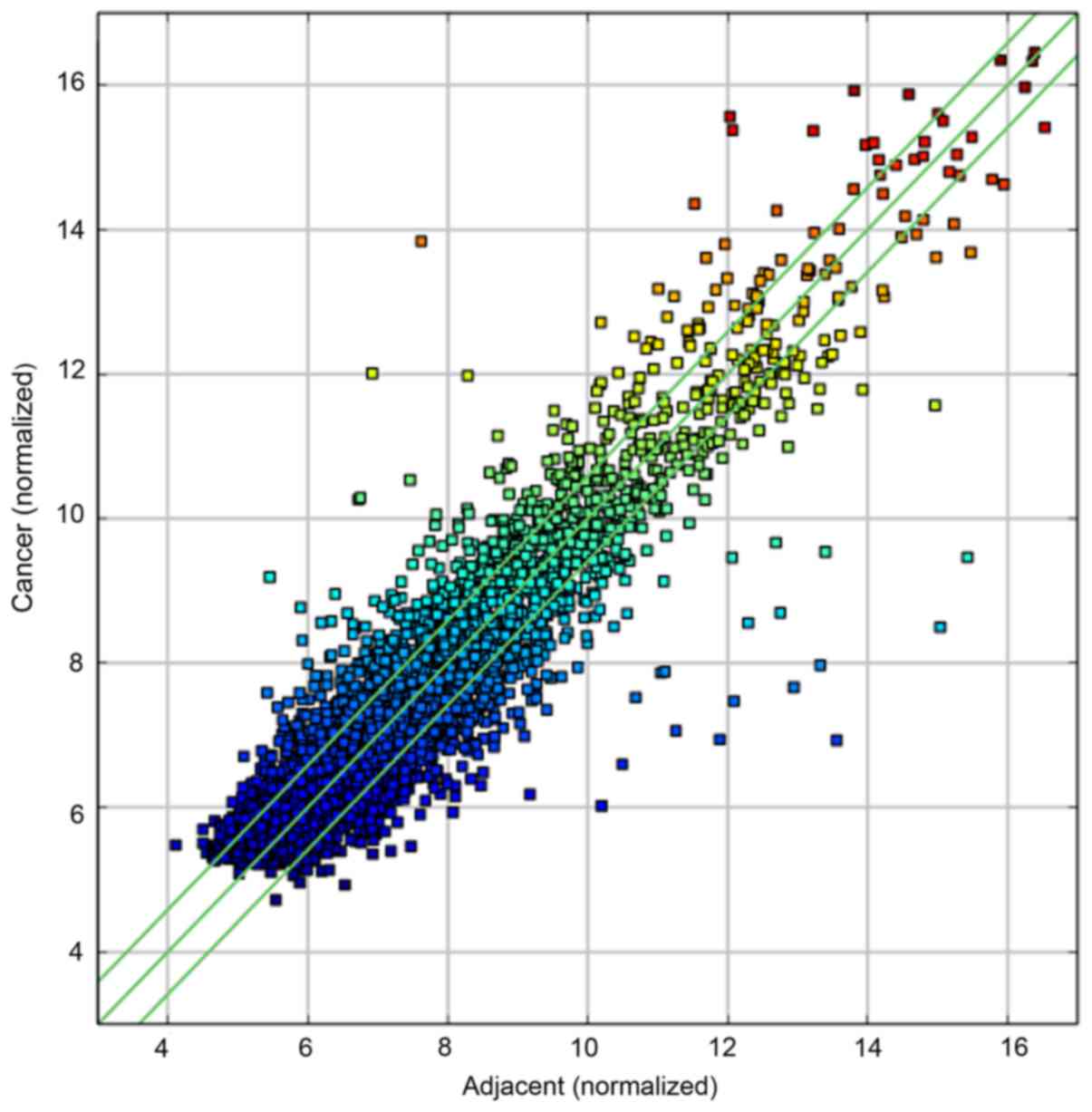
mRNA GeneChip analysis
miRNA expression profiles were constructed in
gastric cancer tissues and adjacent tissues. The data were analyzed
and 29,112 genes were found in the gastric cancer tissues and
adjacent tissues as shown in Fig.
3.
Genes with known expression levels were included in
the two groups of sample statistics. To determine whether there was
a significant difference in expression between the two groups of
samples, gene expression was normalized to the same order of
magnitude in the two groups based on the mRNA expression profiles.
The statistical analysis of the two groups of samples included fold
differences >2-fold and P<0.05, with visual differences
between gene expression. Differential expression was detected for
5,460 genes in the two groups of samples. The differential gene
expression was significantly upregulated in 239 genes, with SI
showing the maximum difference in expression. Conversely,
differential gene expression was significantly downregulated in
3,070 genes, with ATP4B showing the greatest difference in gene
expression.
qRT-PCR validation results
Through access to the relevant literature combined
with the analysis of the microarray experiments in the
differentially expressed gene multiple alignment, we selected
relative literature that reported microarray gene results for
verification. Our analysis included three upregulated genes (MYH9,
CD44 and MALAT1) and three downregulated genes (CXXC5, ARVCF and
ARIH1) compared to GAPDHA for reference. The expression results of
the six genes were analyzed by qRT-PCR for chip verification. The
results were as follows: CD44, CXXC5 and ARVCF in the experimental
group were upregulated by 2.38-, 1.89- and 0.82-fold, respectively,
whereas MYH9, MALAT1 and ARIH1 in the experimental group were
downregulated by 2.42-, 1.95- and 1.07-fold, respectively, compared
to the control group. The differences were significant (P<0.05).
Target gene expression in the microarray analysis results was
consistent with the confirmed mRNA expression levels from the
microarray gene test results, indicating that the results were
credible (Table III).
 | Table III.The verification results of
differential gene expression. |
Table III.
The verification results of
differential gene expression.
| Gene | Differential
expression |
2−∆∆Ct | P-value |
|---|
| MYH9 | −1.15 | 2.42 | 0.0001 |
| CD44 | 1.70 | 2.38 | 0.0002 |
| MALAT1 | −1.31 | 1.95 | 0.0005 |
| CXXC5 | 3.71 | 1.89 | 0.00008 |
| ARVCF | 4.25 | 0.85 | 0.041 |
| ARIH1 | −1.08 | 1.07 | 0.033 |
Differential gene bioinformatics
analysis
GO analysis is the international standard gene
function classification system. GO is used to describe biological
genes and perform expression analysis of product attributes. GO
includes three areas (biological process, cell component and
molecular function). GO classifications are calculated for selected
genes using a particular branch of the hypergeometric distribution
relationship for each gene to obtain a P-value (under normal
circumstances, the function of P<0.05 for enrichment items).
Genetic differences in the GO terms in the analysis of the
experimental results indicate enrichment. GO suggests a role based
on the differences in the gene analysis and can find enriched genes
that are differentially categorized entries based on changes in
different samples. Different genes may have related functions, such
as those associated with signal transduction and positioning.
Differentially expressed genes were identified in
the biological process (1,638 genes, including 841 upregulated
genes and 797 downregulated genes), cell component (205 genes,
including 119 upregulated genes and 86 downregulated genes) and
molecular function categories (147 genes, including 155 upregulated
genes and 92 downregulated genes) in the GO analysis. The GO
analysis results of the differentially expressed genes are shown in
Figs. 4–9.
Analysis of differentially expressed
genes in metabolic pathways
In the pathway analysis using the differentially
expressed genes, 83 genes were significantly enriched in pathways,
from which 20 were selected for the enrichment of gene pathways
analysis shown in Table IV.
 | Table IV.The main pathways. |
Table IV.
The main pathways.
| Pathway ID | Access information
description | Adjust | Count | P-value |
|---|
| hsa02010 | ABC
transporters | Upregulated | 44 | 9.85701E-06 |
| hsa03460 | Fanconi anemia
pathway | Upregulated | 53 | 0.000519872 |
| hsa05200 | Pathways in
cancer | Upregulated | 327 | 0.001215533 |
| hsa03440 | Homologous
recombination | Upregulated | 28 | 0.001408952 |
| hsa04974 | Protein digestion
and absorption | Upregulated | 88 | 0.002310888 |
| hsa05214 | Glioma | Upregulated | 65 | 0.005344975 |
| hsa05218 | Melanoma | Upregulated | 71 | 0.005613469 |
| hsa04320 | Dorso-ventral axis
formation | Upregulated | 24 | 0.006804497 |
| hsa03008 | Ribosome biogenesis
in eukaryotes | Upregulated | 85 | 0.007767791 |
| hsa04950 | Maturity onset
diabetes of the young | Upregulated | 25 | 0.008931677 |
| hsa05150 | Staphylococcus
aureus infection - Homo sapiens | Downregulated | 57 | 5.32523E-06 |
| hsa04978 | Mineral
absorption | Downregulated | 51 | 4.16404E-05 |
| hsa04971 | Gastric acid
secretion | Downregulated | 75 | 7.6987E-05 |
| hsa00830 | Retinol
metabolism | Downregulated | 64 | 0.000143095 |
| hsa04080 | Neuroactive
ligand-receptor interaction | Downregulated | 321 | 0.000149431 |
| hsa04514 | Cell adhesion
molecules (CAMs) | Downregulated | 145 | 0.000529636 |
| hsa04270 | Vascular smooth
muscle contraction | Downregulated | 131 | 0.000677474 |
| hsa04020 | Calcium signaling
pathway | Downregulated | 181 | 0.000860266 |
| hsa04672 | Intestinal immune
network for IgA production | Downregulated | 49 | 0.000923253 |
| hsa00910 | Nitrogen
metabolism | Downregulated | 17 | 0.001285622 |
Correlation analysis of circRNAs and
mRNAs in gastric cancer
Because interactions may exist between circRNAs and
miRNAs and between miRNAs and mRNAs in gastric cancer tissues and
adjacent normal tissues, significantly differentially expressed
circRNAs and mRNAs may exist in relationships.
circRNAs with a 1.5-fold difference in expression
were associated with the corresponding differential gene expression
profile. Only 69 of the differentially expressed circRNA
transcripts could be found in the corresponding gene expression
profile, as shown in Fig. 10.
A total of 42 genes were upregulated, and
significant expression was detected for the following 17 genes
(FC≥2.0, P<0.05): SLAMF6, LAPTM4A, LIN52, MYH9, C16orf72, CD44,
MALAT1, RPPH1, SPI1, AXIN1, MLXIP, SLC45A4, Cul4A, RPL18a, ATP13A1,
CNPY3 and MSI2. Additionally, 17 genes exhibited downregulated
expression, and the following 8 of these genes were significantly
expressed (FC ≤2.0, P<0.05): CXXC5, C19orf18, CCS, RABGGTB,
ARVCF, GUSBP11, FABP6 and ARIH1.
Discussion
There is a lack of specific data showing the early
stage factors that lead to the high mortality of GC. Therefore, the
search for new biological targets for the monitoring and
intervention of GC has become an urgent problem.
Differential expression of
circRNAs
circRNAs widely exist in many varied types of
biological cells (11). They are
not easily degraded by RNase and thus, are stably expressed in
human cells. Relevant research has shown that circRNA expression
levels in human cells may be >10-fold greater than the
expression levels of their linear isomers (11,15).
Some circRNAs have microRNA response elements. These
circRNAs can regulate miRNA expression through chelation, which
plays a key role in the regulation of miRNA expression (16). For example, ciRS-7 contains multiple
tandem miRNA binding sites and may act as an endogenous miRNA
sponge to inhibit the activity of its miR-7 target gene.
Kai et al (17) indicated that the high expression of
miR-21-3p might indicate its involvement in the invasion and
metastasis of esophageal cancer. In the present study, we found
differentially expressed circRNAs with corresponding miRNA binding
sites. This finding suggests that circRNAs may affect the
occurrence and development of gastric cancer and other diseases
through interactions with miRNAs.
In the present study, we found 467 differentially
expressed circRNAs, of which 214 were significantly upregulated and
253 were significantly downregulated. Most of the differentially
expressed circRNAs contained corresponding miRNA binding sites.
Due to the defects in GeneChip technology, great
differences exist in screening results. Therefore, in the present
study we used qRT-PCR to verify the differential expression of the
circRNAs. Since the number of samples was small, we could not
confirm whether there was specificity between the selected circRNAs
and the GC invasion, metastasis or apoptosis.
To clarify the specific circRNAs associated with GC
for early diagnosis and clinical treatment, we need to analyze
circRNA expression profiles in a large number of samples. We also
need to select more differentially expressed circRNAs to confirm
the relationship between their expression levels and GC at a
cellular level in animal experiments. To this end, we may be able
to clarify the biological functions of specifically expressed
circRNAs in GC.
Differentially expressed genes
Tumors are a type of genetic disease that occur as
the result of interactions between multiple genes, leading to their
generation and transfer. The deepening of the study of gene
functions has furthered our understanding of tumor cell metastasis.
On the molecular level, tumor cell metastasis can be divided into
the following stages: transformation of normal cells, hyperplasia
of tumor cells and tumor cell growth; proliferation; cell cycle
survival of tumor cells and extracellular matrix adhesion; release
of proteolytic enzymes from tumor cells into the extracellular
matrix, resulting in basement membrane hydrolysis; tumor cell
infiltration or vascular invasion into organs; escape from host
immune mechanisms and the production of new microvessels; and
proliferation in the new environment. These processes are related
to abnormal expression. Apart from the primary tumor, tumor cells
are rarely malignant and most transfers occur during the process of
gene exchange. GeneChip technology is a high-throughput method that
screens differentially expressed genes through parallel detection;
thus, this technology can be used for studies of the mechanisms
underlying gastric cancer and metastasis.
Differential expression profiles can be obtained
from the Agilent microarray. The chip includes almost all genes
found to date. Expression profile microarray data have been used to
find new possibilities and gastric cancer-related genes and have
provided a wealth of information. Experimental studies on results
in cancer tissues and adjacent organizations from the Communist
Party of China (CPC) found that 5,460 out of 29,112 mRNA genes
exhibited differential expression, including 2,390 genes that were
significantly upregulated and 3,070 genes that were significantly
downregulated. By analyzing differences in gene clustering, we can
more accurately divide gastric cancer tissues from adjacent tissue
areas. The comparative expression microarray screened 1,115
upregulated genes and 975 downregulated genes using GO analysis.
The results showed that the upregulated genes were primarily
involved in cell metabolism, nucleic acid binding activity of
transcription factors, cell generation, DNA binding and other
biological processes; these processes may stimulate the cells to
lose their normal regulation of growth at the gene level, leading
to abnormal hyperplasia and resulting in the formation of gastric
cancer. The pathway analysis showed 2,090 genes involved in 83
signaling pathways, including focal adhesion, Rap1 signaling and
cytokine receptor interactions, protein digestion and ceiling,
alcoholism, transfer RNA and the cell cycle. These pathways also
participated in asthma, basal cell carcinoma, bladder and prostate
cancer, leishmaniasis, inflammatory bowel disease (IBD), allogeneic
shift in the allograft rejection reaction and other metabolic
process diseases. In this study, we reported genetic differences in
38 genes involved in the Rap1 signaling pathway. At present, the
Rap1 signaling pathway has been associated with cancer based on
biological information (18). These
findings suggest that mRNA microarray screening of selected
differentially expressed genes may be accomplished with a variety
of methods to assess the roles involved in the occurrence of
gastric cancer.
We performed spectral mining of the mRNA expression
data to assess the relevant documents. We found that the
differentially expressed genes participated in multiple pathways
that were closely related to tumor occurrence. The PDGFRB
(platelet-derived growth factor receptor beta polypeptide) gene
encodes a platelet derivative growth factor family member and cell
surface amino acid kinase receptor. Studies have shown the
participation of PDGFR and its ligand in angiogenesis and vascular
functions, which suggests that PDGF-B indirectly regulates
endothelial cell functions and is an important factor for tumor
development (19,20). A research has shown that PDGFRb can
selectively regulate and promote colon cancer and osteosarcoma cell
growth. Ionization radiation experiments were performed to assess
non-small cell lung cancer cell survival and it was found that
PDGFR was upregulated in the cell spheroids that survived the
radiation exposure (21). Thus,
CD44 and PDGFR may be possible markers for non-small cell lung
cancer radiotherapy. GREB1 is located in the promoter of the
estrogen response element and is a feature of estrogen receptor
target genes. Its expression was inversely related to the
proto-oncogene epidermal growth factor receptor 2 (HER2) status. In
breast cancer cells, GREB1 overexpression promoted cell
proliferation and increased the clone formation ability. IL-6/STAT3
regulation by estrogen induced GREB1 transcriptional activity
(22). Upregulated expression of
the KIT gene, which is a stem cell growth factor receptor and tumor
stem cell marker, was associated with gene mutations in a variety
of tumors. Akiyama et al (23) noted that the tumor carcinogenic
signal c-kit and other tyrosine kinase receptors played very
important roles.
In gastric cancer and paracancerous tissues, mRNA
expression differences may suggest their diagnostic and treatment
values for gastric cancer. This study used expression profile
microarray technology to construct gastric cancer and adjacent
tissue mRNA expression profiles. These profiles were used to screen
differentially expressed mRNAs to identify differentially expressed
genes involved in gastric cancer. We assessed signal transduction
pathways to provide insights into direction of future studies on
the occurrence of gastric cancer and to help elucidate the
development mechanism to provide an experimental basis.
Relationship between circRNAs and
mRNAs
The expression of approximately one-third of human
genes is regulated by miRNAs. miRNAs can act through a variety of
pathways and can target multiple genes for regulation. A number of
miRNAs can also regulate target genes through different pathways
(24). An increasing number of
studies has shown that miRNAs and mRNAs exist in very complex
regulatory relationships. A miRNA regulates multiple target genes
that are enriched by one or more signaling pathways. In turn, the
miRNA acts on multiple target proteins to regulate signaling
pathways to more effectively achieve the miRNA-mRNA regulation
network. The ultimate manifestation is the regulation of the cell
phenotype and function (25,26).
miRNAs also play a regulatory role in disease by interacting with
circRNAs. Investigations of these interactions may provide a basis
for the elucidation of the disease mechanism, target intervention
and clinical diagnostic markers. CDR1as interacts with miR-7 and
acts as a miR-7 sponge based on a combination of nearly 70 binding
sites, which effectively influence miR-7 target gene activity.
CDR1as and miR-7 both exhibit high expression levels in the
midbrain development zone. Overexpression in zebrafish embryos
results in a fetal brain volume reduction; the midbrain volume can
be partially restored by injection of miR-7, suggesting that miR-7
and its interaction with CDR1as is at least partially responsible
for the biological effect. CDR1as is a cyclic miR-7 inhibitor;
thus, CDR1as can influence tumor regulation through miR-7 because
miR-7 plays a negative role in regulation. Therefore, CDR1as can
fulfill its function as a natural miRNA sponge.
We analyzed circRNA and mRNA expression profiles
from gastric cancer tissues and adjacent normal tissues and their
correlations. The results showed only 69 differences in circRNA
transcripts with corresponding gene expression profile information.
For instance, upregulated expression of the tumor
metastasis-related 42. MYH9 gene can cause changes in gastric
cancer cell invasion and migration (27). CD44 is one type of tumor stem cell
marker that is found with high expression in a variety of tumors
and contributes to tumor growth, invasion and metastasis (28). MALAT1 [also known as nuclear
enrichment of transcription factor 2 (neat2)] is a gene expression
regulator of lung cancer metastasis and a prognostic indicator for
some solid tumors (29). ARVCF
plays a role in renal cell cancer, kidney and prostate cancer.
CXXC5 was demonstrated to enhance the ability of ovarian cancer
cells to migrate and invade and to inhibit the apoptosis of ovarian
cancer cells that occurred during the analysis of its relationship
with its associated mRNA (30). We
detected differential expression in 69 circRNA transcriptional
genes and mRNAs in the gene profiles of the same genes. These
results suggest that gene expression is regulated by circRNA and
miRNA and miRNA and mRNA interactions in a differential manner.
Interactions and mutual influences are likely to have an important
impact on the occurrence and development of gastric cancer.
By analyzing circRNAs, mRNAs, and their
relationships in gastric cancer tissues and adjacent tissues, we
found that circRNAs and mRNAs had a relationship in gastric cancer
that led to a more sober understanding: circRNAs are controlled by
adsorption and miRNA interactions, thereby establishing a possible
role for circRNA-miRNA-mRNA in the regulation of gastric cancer
development.
In conclusion, this experiment in eight patients who
underwent gastric cancer resection surgery for lesions in gastric
cancer tissues and adjacent tissues as the research object used
Agilent microarray expression to construct circRNA and mRNA
differential expression profiles for the gastric cancer tissues and
adjacent tissues. The significant differences in circRNA and mRNA
expression in the association analysis suggested that the circRNAs
regulated the expression of various genes in gastric cancer. These
results contribute to our understanding of the molecular mechanism
underlying the development gastric cancer and the search for an
effective early diagnostic biomolecule marker for gastric cancer
and malignant progression on a theoretical basis. The qRT-PCR
validation of differentially expressed circRNAs suggested that RNA
and gene expression was regulated in the gastric tissues and
adjacent tissues.
To summarize, the present study draws the following
conclusions: i) differential expression of circRNAs and the
corresponding miRNAs interact through circRNA binding sites to
regulate the expression of target genes. These circRNAs are likely
to become new molecular biomarkers for gastric cancer in the
future. ii) Genes involved in cell metabolism, nucleic acid binding
activity of transcription factors, the generation of organelles and
DNA binding (biological processes) may be prompting cells to lose
their normal growth regulation at the gene level. These genes are
also involved in the Rap1 signaling pathway, cell factor
interactions, protein digestion and absorption, basal cell
carcinoma, prostate cancer, the allogeneic shift in the allograft
rejection reaction and other metabolic process diseases. The genes
may be differentially expressed through a variety of pathways and
play roles in the occurrence of gastric cancer. iii) CD44, CXXC5,
MYH9, MALAT1 and other genes are regulated through interactions
between circRNAs and miRNAs and between miRNAs and mRNAs via
different mechanisms, including direct interactions and mutual
influence. These interactions may be important influences on the
occurrence and development of gastric cancer.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park DJ, Thomas NJ, Yoon C and Yoon SS:
Vascular endothelial growth factor a inhibition in gastric cancer.
Gastric Cancer. 18:33–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Degiuli M and Calvo F: Survival of early
gastric cancer in a specialized European center. Which
lymphadenectomy is necessary? World J Surg. 30:2193–2203. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schwarz RE and Smith DD: Clinical impact
of lymphadenectomy extent in resectable gastric cancer of advanced
stage. Ann Surg Oncol. 14:317–328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maass PG, Luft FC and Bähring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valdmanis PN and Kay MA: The expanding
repertoire of circular RNAs. Mol Ther. 21:1112–1114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai Y, Sui W, Lan H, Yan Q, Huang H and
Huang Y: Comprehensive analysis of microRNA expression patterns in
renal biopsies of lupus nephritis patients. Rheumatol Int.
29:749–754. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Doberstein K, Bretz NP, Schirmer U, Fiegl
H, Blaheta R, Breunig C, Müller-Holzner E, Reimer D, Zeimet AG and
Altevogt P: miR-21-3p is a positive regulator of L1CAM in several
human carcinomas. Cancer Lett. 354:455–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fukuhara S, Sakurai A, Sano H, Yamagishi
A, Somekawa S, Takakura N, Saito Y, Kangawa K and Mochizuki N:
Cyclic AMP potentiates vascular endothelial cadherin-mediated
cell-cell contact to enhance endothelial barrier function through
an Epac-Rap1 signaling pathway. Mol Cell Biol. 25:136–146. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arai K, Sakamoto R, Kubota D and Kondo T:
Proteomic approach toward molecular backgrounds of drug resistance
of osteosarcoma cells in spheroid culture system. Proteomics.
13:2351–2360. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cho HJ, Baek KE, Kim IK, Park SM, Choi YL,
Nam IK, Park SH, Im MJ, Yoo JM, Ryu KJ, et al: Proteomics-based
strategy to delineate the molecular mechanisms of RhoGDI2-induced
metastasis and drug resistance in gastric cancer. J Proteome Res.
11:2355–2364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
PLoS One Editors, . Retraction: GREB1
functions as a growth promoter and is modulated by IL6/STAT3 in
breast cancer. PLoS One. 9:e1022872014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Akiyama H, Watanabe T, Wakabayashi K,
Nakade S, Yasui S, Sakata K, Chiba R, Spiegelhalter F, Hino A and
Maitani T: Quantitative detection system for maize sample
containing combined-trait genetically modified maize. Anal Chem.
77:7421–7428. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feigin VL and Findlay M: Advances in
subarachnoid hemorrhage. Stroke. 37:305–308. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van der Voet M, Olson JM, Kuivaniemi H,
Dudek DM, Skunca M, Ronkainen A, Niemelä M, Jääskeläinen J,
Hernesniemi J, Helin K, et al: Intracranial aneurysms in Finnish
families: Confirmation of linkage and refinement of the interval to
chromosome 19q13.3. Am J Hum Genet. 74:564–571. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Juvela S: Natural history of unruptured
intracranial aneurysms: Risks for aneurysm formation, growth, and
rupture. Acta Neurochir Suppl. 82:27–30. 2002.PubMed/NCBI
|
|
27
|
Liang S, He L, Zhao X, Miao Y, Gu Y, Guo
C, Xue Z, Dou W, Hu F, Wu K, et al: MicroRNA let-7f inhibits tumor
invasion and metastasis by targeting MYH9 in human gastric cancer.
PLoS One. 6:e184092011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Naor D, Nedvetzki S, Golan I, Melnik L and
Faitelson Y: CD44 in cancer. Crit Rev Clin Lab Sci. 39:527–579.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1 - a paradigm for long noncoding RNA function in cancer. J
Mol Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang JH, Ren Y, Zhang R, Han Y, Sheng YH,
Hou WJ and Ao HF: Effects of CXXC finger protein 5 up-regulated
expression in epithelial ovarian cancer. China Oncol. 25:260–268.
2015.(In Chinese). http://www.china-oncology.com/EN/Y2015/V25/I4/260
|