Introduction
Hepatocellular carcinoma (HCC) is a primary
malignancy of the liver with over 500,000 people affected. HCC is
now the third leading cause of cancer deaths worldwide which
attracts intensive research (1).
The occurrence and development of HCC is a complex process
involving multi-genes, multi-steps and multi-stages. The invasion
and metastasis of HCC is closely linked with HCC patient's
treatment and prognosis (2). Thus,
it is of great significance to reveal HCC specific genes and
investigate the mechanism of aberrant molecules in the occurrence
and development of HCC.
The recently identified RNA-binding motif protein 8A
(RBM8A, also known as Y14) belongs to the RNA-binding motif protein
(RBM) family (3). Scientists have
focused on this protein family for its diverse functions. These
proteins bind to RNA and tightly control gene expression, cell
cycle, apoptosis and RNA splicing (4–8). RBM8A
is a vital apoptosis regulator and loss of RBM8A leads to increased
apoptosis. Ishigaki et al (9) found that loss of RBM8A inhibited the
proliferation of HeLa and A549 cells and cells were blocked at M
phase. Importantly, they found the abnormal centrosome in cancer
cells, which is a symbolic feature of apoptosis, indicating the
loss of RBM8A directly regulate cell apoptosis. Moreover,
downregulated RBM8A led to decreases of pro-apoptotic genes such as
Bcl-Xs belonging to Bcl-2 family, Bim and Mcl1S (10,11).
These studies showed that PBM8A participated in various steps of
apoptosis. Evidence also demonstrated that RBM8A control tumor
occurrence and development via regulating mRNA splicing.
The function of RBM8A in HCC is poorly understood.
Although Zindy et al (12)
found that RBM8A was upregulated in liver cancer as a target gene
of re-establishment of extracellular matrix, metabolism and
post-transcription regulation, the function of RBM8A in HCC remain
largely unknown. Hence, we analyzed the expression level of RBM8A
in patient HCC tumor samples and evaluated the relationship of
RBM8A expression level and clinicopathological factors and
prognosis. We further explored the function of RBM8A in HCC tumor
cell migration, invasion as well as apoptosis.
Materials and methods
Patient data
A total of 105 surgical tissue samples of HCC were
collected from patients at Affiliated Tumor Hospital of Guangxi
Medical University after obtaining the subject informed consent and
institutional review board approval.
Cell lines
The human HCC cell lines were purchased from Stem
Cell Bank, Chinese Academy of Sciences. Bel-7404, and HL-7702 cells
were maintained in Dulbecco's modified Eagle's medium (DMEM).
MHCC97H, a high-metastatic human HCC, was established as reference
(13). The media were supplemented
with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen,
Carlsbad, CA, USA), penicillin (100 U/ml), and streptomycin (100
µg/ml) in a humidified atmosphere of 5% CO2 at 37°C. All
the cells were confirmed to be free from mycoplasma
contamination.
Establishment of stable cell lines in
which RBM8A was overexpressed or knocked down
The siRNA sequences were: sh1:
5′-AGAGCATTCACAAACTGAA-3′; sh2: 5′-CATCAGCGTTGACTGGTGT-3′; sh3:
5′-GCAACAGGTCTAGGGTTAAGG-3′. Lentiviral vectors encoding shRNA were
designed based on the sequences of siRNA to knock down RBM8A
expression (RBM8A-KD). These vectors were constructed by Hanyin Co.
(Shanghai, China). The recombinant lentiviruses (KD) and the
negative control (NC) lentivirus (Hanyin Co.) were prepared and
titered to 109 transfection units (TU)/ml. To obtain stable cell
lines, cells were seeded in 6-well plates and infected with virus
and polybrene the following day. Positive clones were selected with
puromycin for 14 days to establish the stable cell lines.
Additionally, the lentiviruses expressing the RBM8A sequence (OE)
and the negative control lentivirus (Vector) were constructed by
Hanyin Co. RBM8A-OE and control stable cell lines were then
established. The efficiency of RBM8A knockdown and overexpression
was confirmed by qRT-PCR.
Western blot analysis
Protein lysates (50 µg per lane) were separated by
sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and
transferred onto nitrocellulose membranes. After blocking with 5%
fat-free milk, the membranes were incubated with primary antibodies
(1:500 dilution) at 4°C overnight, followed by horseradish
peroxidase-conjugated secondary antibodies (1:3,000 dilution).
Anti-human RBM8A (Santa Cruz Biotechnology, sc-32312), anti-human
actin (Proteintech, HRP-60008), anti-human Snail (Cell Signaling
Technology, 3879), anti-human p-STAT3 (Cell Signaling Technology,
9131), anti-human STAT3 (Cell Signaling Technology, 12640),
anti-human fibronectin (Sigma-Aldrich, F3648), anti-human vimentin
(Cell Signaling Technology, 5741), anti-human Notch (Abaways,
CY52444) and donkey anti-Rabbit IgG (Cell Signaling Technology,
7074) were used in this study. Immune-reactive proteins were
visualized by enhanced chemiluminescence (ECL).
Histology and
immunohistochemistry
Histological and immunohistochemical analyses were
performed using an HRP kit (UltraTek; Scytek). Anti-human RBM8A
antibody diluted 1:50 in blocking buffer were used as primary
antibodies. The diaminobenzidine-stained specimens were visualized
using a general optical microscope with a camera (AxioCam ICc5;
Carl Zeiss). Images were processed with equivalent parameters using
ZEN Light Edition software (Carl Zeiss).
Total RNA isolation and quantitative
real-time PCR (qRT-PCR) analysis
Total RNA was isolated from liver cancer cells and
tumor tissues using TRIzol reagent according to the manufacturer's
directions (Invitrogen). cDNA was reverse transcripted from 1 mg
total RNA. qRT-PCR was performed with the SYBR Premix Ex Taq
(Takara, Dalian, China). PCR primers were as follows: F:
5′-GCGTGAGGATTATGACAGCGTG-3′, R: 5′-TTCGGTGGCTTCCTCATGGACT-3′.
The cycling conditions were as initial denaturation
at 95°C for 5 min, and then 36 cycles of denaturation at 95°C for
10 sec and annealing at 60°C for 30 sec. The relative mRNA
expression was calculated using the comparative Ct (∆∆Ct)
method.
Wound-healing assay
The in vitro migration ability of HCC cells
was assessed by wound-healing assay. Cells were plate in 6-well
plates and the monolayer was artificially scratched with 10-µl
pipette tips. The wound areas were photographed 0 and 20 h after
scratching and measured using a caliper. The wound-closure
percentages were calculated using the following formula:
[1-(current wound size/initial wound size)] ×100.
Cell invasion assay
Cells were detached and re-suspended in a serum-free
medium and seeded on the upper chamber of Matrigel-coated Transwell
inserts with a pore size of 8 µm. The culture medium containing 10%
FBS as a chemoattractant was added to the lower chamber. After 24 h
incubation, the cells on the upper surface of the insert were
gently removed with a cotton swab. Invading cells (lower surface of
the insert) were fixed with 4% paraformaldehyde (Sigma-Aldrich),
stained with crystal violet, and counted under a microscope. Five
random microscopic fields were examined for each insert.
Cell Counting Kit-8 (CCK-8) assay
The Bel7404 and BL7702 cells were seeded into
96-well culture plates (5×103 cells/well). At 24, 48, 72
and 96 h, 10 µl CCK-8 reagent (Beyotime Biotechnology) was added to
each well and incubated for 4 h. Then the absorbance values were
read at a wavelength of 450 nm using a microplate reader
(SpectraMax 250; GE Healthcare Life Sciences, Pittsburgh, PA,
USA).
Flow cytometry analysis
Cells were seeded into 6-well plates at a density of
1×106 cells/well for 24 h. Subsequently, the cells were
collected and stained with the ANXA5 (Annexin V)-PE apoptosis
detection kit (4A Biotech Co. Ltd., FXP018-100) according to the
manufacturer's instructions and analyzed by flow cytometry (FACS
Calibur, BD Biosciences, San Jose, CA, USA).
Statistical analyses
Unless stated otherwise, data are presented as mean
± SD in the figures. A Student t-test was performed to compare the
in vitro data. The Kaplan-Meier survival analysis was
generated using SPSS version 13.0 (SPSS Inc., Chicago, IL, USA).
One-way analysis of variance was initially performed to determine
whether overall statistically significant changes occurred before
using two-tailed paired or unpaired Student's t-tests. Multiple
test-adjusted P-values of <0.05 were considered statistically
significant. All statistical tests were two-sided.
Results
RBM8A is upregulated in hepatocellular
carcinoma
In order to explore the function of RBM8A in
hepatocellular carcinoma (HCC), we first detected the expression of
RBM8A in normal liver tissues, carcinoma adjacent tissues and HCC
tissues. Of note, we found that the mRNA level of RBM8A was
significantly higher in HCC (7.99±5.49) compared to carcinoma
adjacent tissues (4.66±3.78) as well as normal liver tissues
(1.64±1.53). We also noted that the mRNA level of RBM8A in
carcinoma adjacent tissue was significantly higher than normal
liver tissues (Fig. 1A). The
western blotting results showed that RBM8A protein was detectable
in all tissues and the protein level of RBM8A was consistently
higher in HCC tissues (1.86±0.36) compared to carcinoma adjacent
tissues (1.35±0.32) as well as normal liver tissues (0.95±0.35)
demonstrating that RBM8A was upregulated in HCC (Fig. 1B and Table I).
 | Table I.Quantification of RBM8A protein levels
in HCC tissues, adjacent tissues and normal non-carcinoma
tissues. |
Table I.
Quantification of RBM8A protein levels
in HCC tissues, adjacent tissues and normal non-carcinoma
tissues.
|
|
| RBM8A/β-actin |
|---|
|
|
|
|
|---|
| Tissues | Cases | Mean ± SD | P-value |
|---|
| Carcinoma | 105 | 1.86±0.36 |
|
| Adjacent | 105 | 1.35±0.32 | 0.000a |
| Non-carcinoma | 67 | 0.95±0.35 | 0.000b |
To further validate the expression differences, we
performed immunohistochemistry (IHC) against RBM8A on normal liver
tissue, carcinoma adjacent tissue and HCC tissue. As shown in
Fig. 1C, strong positive signals
were observed in HCC samples with a positive rate of 87.6%.
Statistical analysis results indicated that RBM8A IHC signals were
significantly higher in carcinoma tissues and signals in carcinoma
adjacent tissue were also higher than normal liver tissue (Table II).
 | Table II.IHC results of RBM8A in HCC tissues,
adjacent tissues and normal non-carcinoma tissues. |
Table II.
IHC results of RBM8A in HCC tissues,
adjacent tissues and normal non-carcinoma tissues.
| Tissues | Cases | Strong
staining | Medium
staining | Weak staining | Negative | Positive rate
(%) | P-value |
|---|
| Carcinoma | 105 | 18 | 46 | 28 | 13 | 87.62 |
|
| Adjacent | 105 | 3 | 28 | 33 | 41 | 60.95 | 0.00a |
| Non-carcinoma | 67 | 0 | 1 | 2 | 64 |
4.47 | 0.00b |
Relationships between RBM8A expression
levels with clinical indexes and survival
To further investigate the relationship between
RBM8A expression levels with clinical indexes at both mRNA and
protein levels, we set the relative mRNA expression level 7.99 as a
limit and divided the 105 cases into RBM8A mRNA high group and
RBM8A mRNA low group. There were 85 cases in the RBM8A mRNA high
group with IHC positive while 20 cases in the RBM8A mRNA low group.
Noteworthy, our results revealed significant differences of RBM8A
protein levels with different HBsAg levels, tumor diameters, TNM
staging and Edmondson pathological grading. We observed significant
correlations between RBM8A high group with HBsAg, Edmondson
pathological grading and TMN stages (Table III). However, no significant
differences between RBM8A mRNA levels with tumor diameter and TNM
staging. Thus, our result showed that at both mRNA and protein
level, high level of RBM8A were associated with HBsAg and Edmondson
pathological grading.
 | Table III.The strength of RBM8A expression in
HCC tissues and the relationship of clinical index. |
Table III.
The strength of RBM8A expression in
HCC tissues and the relationship of clinical index.
|
|
| RBM8A mRNA |
| RBM8A protein |
|
|---|
|
|
|
|
|
|
|
|---|
|
Characteristics | Cases | High | Low | P-value | Positive | Negative | P-value |
|---|
| Gender |
|
|
| 0.104 |
|
| 0.144 |
|
Male | 78 | 66 | 12 |
| 71 | 7 |
|
|
Female | 27 | 19 | 8 |
| 21 | 6 |
|
| Age |
|
|
| 0.079 |
|
| 0.463 |
|
≥60 | 35 | 25 | 10 |
| 29 | 6 |
|
|
<60 | 70 | 60 | 10 |
| 63 | 7 |
|
| AFP (µg/l) |
|
|
| 0.516 |
|
| 0.433 |
|
≥400 | 78 | 62 | 16 |
| 70 | 8 |
|
|
<400 | 27 | 23 | 4 |
| 22 | 5 |
|
| HBsAg |
|
|
| 0.000 |
|
| 0.000 |
|
Positive | 82 | 73 | 9 |
| 81 | 1 |
|
|
Negative | 23 | 12 | 11 |
| 11 | 12 |
|
| Diameter of tumor
(CM) |
|
|
| 0.248 |
|
| 0.013 |
|
≥10 | 28 | 20 | 8 |
| 27 | 1 |
|
|
5–10 | 50 | 41 | 9 |
| 39 | 11 |
|
|
<5 | 27 | 24 | 3 |
| 26 | 1 |
|
|
Hepatocirrhosis |
|
|
| 0.329 |
|
| 0.58 |
|
Yes | 58 | 45 | 13 |
| 54 | 4 |
|
| No | 47 | 40 | 7 |
| 38 | 9 |
|
| Portal vein
thrombosis |
|
|
| 0.128 |
|
| 0.276 |
|
Yes | 42 | 31 | 11 |
| 35 | 7 |
|
| No | 63 | 54 | 9 |
| 57 | 6 |
|
| Child-Pugh
score |
|
|
| 0.283 |
|
| 0.670 |
| A | 43 | 34 | 9 |
| 37 | 6 |
|
| B | 58 | 49 | 9 |
| 52 | 6 |
|
| C | 4 | 2 | 2 |
| 3 | 1 |
|
| Edmondsom
grades |
|
|
| 0.004 |
|
| 0.003 |
| I | 20 | 10 | 10 |
| 12 | 8 |
|
| II | 45 | 40 | 5 |
| 42 | 3 |
|
|
III | 38 | 33 | 5 |
| 36 | 2 |
|
| IV | 2 | 2 | 0 |
| 2 | 0 |
|
| TNM |
|
|
| 0.153 |
|
| 0.000 |
| I | 24 | 16 | 8 |
| 15 | 9 |
|
| II | 60 | 51 | 9 |
| 58 | 2 |
|
|
III | 21 | 18 | 3 |
| 19 | 2 |
|
| IV | 0 |
|
|
|
|
|
|
Non-parametric rank correlation analysis between
RBM8A protein expression and Edmondson pathological grading showed
that the Spearman correlation coefficient was 0.992 (P<0.001)
indicating that RBM8A protein expression levels are closely
associated with Edmondson pathological grading and stronger RBM8A
protein level indicates lower degree of differentiation (Table IV).
 | Table IV.Correlation between RBM8A protein
expression level and Edmondson pathological grades. |
Table IV.
Correlation between RBM8A protein
expression level and Edmondson pathological grades.
| Edmondson
grades | No. | RBM8A/β-actin | P-value |
|---|
| I | 20 | 1.41±0.51 |
|
| II | 45 | 1.63±0.89 | 0.004a |
| III | 38 | 2.21±0.63 | 0.001b |
| IV | 2 | 2.50±0.71 | 0.031c |
Among the 105 cases, the follow-up periods varies
from 3 days to 620 days without loss. All cases were fitted into
the analysis and the follow-up rate was 100%. We divide all cases
into RBM8A protein high group (IHC staining: positive and strong
positive, n=64) and RBM8A protein low group (IHC: staining weak
positive and negative, n=41). By applying Kaplan-Meier survival
curve, we calculated overall survival and progression-free survival
and tested the statistic difference with Log-rank test. Our results
showed that median of overall survival for RBM8A high expression
group and RBM8A low expression group were 160 and 229 days,
respectively, while median of progression-free survival for RBM8A
high expression group and RBM8A low expression group were 90 and
114 days, respectively (Fig. 2).
Both overall survival and progression-free survival in RBM8A low
expression group were significantly longer than that in RBM8A high
expression group indicating that patients with higher RBM8A
expression are expecting shorter survival time.
Knockdown of RBM8A reduces cell
migration and invasion
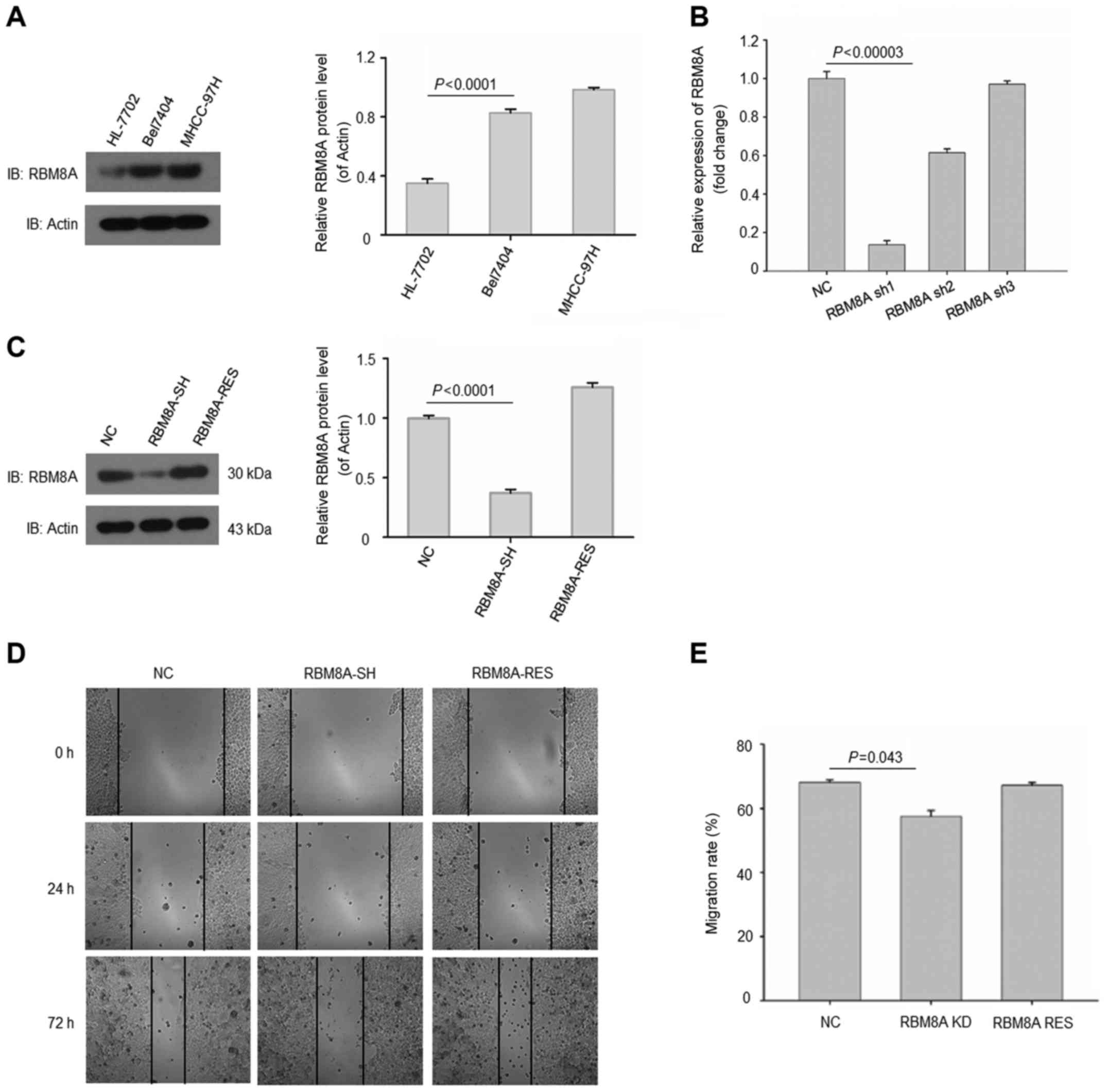
We next validated the results in human normal
hepatic immortalized cell line and HCC cell lines. As expected, we
found that RBM8A expressions were abundant in HCC cell lines
Bel-7404 and MHCC-97H (Fig. 3A). On
the contrary, RBM8A exhibited a low protein level in human hepatic
immortalized cell line HL-7702. To further elucidate the function
of RBM8A, we generated a set of shRNAs to knock down RBM8A in
Bel-7404 cell line. Our results revealed that one of the shRNAs
targeting 5′-AGAGCATTCACAAACTGAA-3′ was able to reduce the mRNA of
RBM8A by more than 80% (Fig.
3B).
Additionally, the protein level of RBM8A was also
effectively downregulated with this shRNA while RBM8A was
successfully rescued in Bel-7404 (Fig.
3C). To investigate the effect of RBM8A on cell migration and
invasion, control cells, RBM8A knockdown cells and RBM8A rescued
Bel-7404 cells were analyzed by migration assay. Our results showed
that the migration of HCC cells were significantly reduced upon
RBM8A knockdown while the RBM8A rescued group regained migration
ability (Fig. 3D and E). We next
applied gain of function experiment by overexpressing RBM8A in
HL-7702 cell line. Both mRNA and protein level of RBM8A were
significantly increased in HL-7702 RBM8A-OE cells (Fig. 4A and B). Moreover, the invasion
ability of HCCs positively associated with the expression levels of
RBM8A. (Fig. 4C and D). Thus, our
results demonstrated that RBM8A promotes cell migration and
invasion in HCC cells.
RBM8A regulates HCC cell
epithelial-mesenchymal transition
In order to elucidate the underlying mechanisms of
RBM8A-mediated HCC migration and invasion, we investigated the
epithelial-mesenchymal transition (EMT) markers and related
signaling. We found a significant reduction of Snail and
phosphorylated STAT3 in RBM8A knockdown Bel-7404 while
unphosphorylated STAT3 remained stable indicating that EMT was
impaired upon RBM8A knockdown (Fig.
5A). We noted a clear reduction of EMT markers fibronectin and
vimentin in RBM8A knockdown Bel-7404 cells which also suggested a
reduction of EMT (Fig. 5A). On the
contrary, in RBM8A overexpressed HL-7702 cells, we observed an
increase of Snail and phosphorylated STAT3 as well as fibronectin
and vimentin (Fig. 5B) compared
with control cells. Furthermore, immunofluorescence staining
validated our western blotting results (Fig. 5C). All above results support that
RBM8A is responsible for the epithelial-mesenchymal transition of
HCC cells.
RBM8A has an important function in HCC
cell survival
To further explore the role RBM8A plays in HCC cell,
we examined whether RBM8A regulate cell proliferation in HCC cells.
Noteworthy, our results displayed that knockdown of RBM8A in
Bel-7404 cells significantly reduced the population of HCC after
three days of cultivation while, ectopic expression of RBM8A in
HL-7202 cells significantly increased the HCC population (Fig. 6A and B). Since RBM8A is a vital
apoptosis regulator and loss of RBM8A leads to increased apoptosis
(16), we examined the HCC
apoptosis upon RBM8A knockdown or RBM8A overexpression.
Accordingly, our FACS results showed a marked increase of apoptotic
cells when RBM8A was knocked down in Bel-7404 (Fig. 6C). However, the number of apoptotic
cells was greatly reduced in ectopic expression of RBM8A in HL-7202
cell line (Fig. 6D).
Taken together, our results demonstrated that high
expression of RBM8A can predict poor patient prognosis and promote
tumor progression in HCC cells.
Discussion
RBM8A is an RNA binding protein with molecular
weight of 26 kDa locating at chromosome 14q21-q23. The gene codes
four transcripts and expresses broadly within the cell and shuffles
between the nuclear and cytoplasm (14). Compared to other RNA binding motif
proteins, the structure and molecular function of RBM8A is poorly
understood. The known biological function is that RBM8A could
participate in forming the exon-junction binding complex (EJC)
(15). RBM8A is the core factor of
nonsense-mediated mRNA decay (NMD) and participates in the
NMD-mediated mRNA surveillance. NMD keeps the stability of mRNA by
degrading the abnormal mRNA products with premature termination
codons and reduces the production of harmful shortened proteins
(16–18). In 2001, Noensie and Dietz proposed
the use of siRNA or drugs to inhibit NMD and in turn to identify
mutated genes and treat the tumor (19). Since it has been proposed that the
development of malignant tumor is closely related to mRNA
stability; it is possible to discover new routes to treat cancer by
interfering with the NMD of malignant tumors. As the core factor of
NMD, RBM8A may play an essential role during cell malignant
transformation.
RBM8A is found highly expressed in malignant tumors
such as primary liver cancer, pleural endotheliomas, multiple
myeloma and other malignant tumors (3,11,12,20–23).
Further analysis revealed that the function of RBM8A could be that
the knockdown of RBM8A in lung adenocarcinoma resulted in cell
cycle arrest in G2/M phase. RBM8A-Mago complex mediated the
location of centrosome which further inhibited the mitosis of tumor
cells and then induced apoptosis (9,24).
Moreover, as the core factor of EJC complex, loss of RBM8A led to
downregulation of the apoptotic genes Bcl-xS in Bcl-2 family and
Mcl1 and resulted in loss of apoptotic control and further induced
the metastasis of hepatocellular carcinoma (HCC) (2,10).
Ras/MAPK, JAK/STAT3 and NF-κB signaling pathways are confirmed to
be closely related to occurrence, development, invasion and
metastasis of liver cancer (25–27).
Loss of RBM8A led to changes of all these signaling pathways
(23,28–30):
loss of RBM8A led to reduction of MAPK protein and inhibited
Ras/MAPK signaling pathway which resulted in cell apoptosis; loss
of RBM8A resulted in inhibition of JAK/STAT3 signaling pathway and
prohibited bindings between DNA and STAT3; loss of RBM8A also led
to decrease of phosphorylation of TNF-α/STAT3 signaling pathway and
reduced the effect of NF-κB. As a common factor of these three
signaling pathways, RBM8A protein plays vital roles in inhibiting
cell growth and apoptosis. Our results also indicated that RBM8A
inhibited apoptosis and loss of RBM8A would clearly prevent
invasion and metastasis. Our data showed that EMT signaling
pathways were affected upon RBM8A demonstrating that the stability
of invasion and metastasis was disrupted due to abnormal EMT
process.
Our studies revealed from both in vivo and
in vitro level that RBM8A is significantly increased in HCC
and could be used as a diagnostic marker for HCC. One of the
mechanisms of malignant tumor occurrence is the high mutation
frequency of oncogenes. These mutations induce NMD pathway and
result in pre-inhibition of oncogene translation. The high
expression of RBM8A in HCC tissues verified that NMD pathway is
highly active in liver cancer tissues indicating that PTC mutation
is higher compared to normal tissues. Furthermore, additional study
is required to prove whether micro-metastasis exists and causes
differentially expressed RBM8A.
RBM8A expression is associated with HbsAg expression
level. Epidemiology results revealed that hepatitis B virus
infection is the most critical factor of HCC in China. Almost all
the hepatitis B virus-associated liver cancers contain hepatitis B
virus integration and the integrated virus promoted the development
of liver fibration and liver cell inflammation and in turn promoted
the occurrence of liver cancer (31,32).
Our result found that significantly high expression of RBM8A at
both mRNA and protein level in HbsAg positive group indicating that
RBM8A participates in the integration of HbsAg gene and interacts
with hepatitis B virus surface antigen and promotes the occurrence
of liver cancer.
Clinical grading and clinical TNM staging are vital
for clinical evaluation of liver cancer and provide great
significance for prognosis (33).
Our results showed that RBM8A expression is higher in late stage
liver cancer than early stage liver cancer. In histology, lower
liver cancer pathological grading corresponds to higher RBM8A
expression level. Kim et al (21), confirmed that cervical carcinoma
patients with lymphatic metastasis possessed higher RBM8A
expression than patients without lymphatic metastasis indicating
that RBM8A could serve as a molecular marker for liver cancer early
diagnosis and metastasis evaluation.
We divided clinical cases into RBM8A high expression
level group and RBM8A low expression level group based on IHC
results and followed up these cases. Our results showed that median
of overall survival are 160 and 229 days, respectively, while
median of progression-free survival for RBM8A high expression group
and RBM8A low expression group are 90 and 114 days, respectively,
indicating that RBM8A could serve as a marker for liver cancer
prognosis since RBM8A is closely related to reoccurrence, migration
and overall survival.
In summary, our results demonstrated that RBM8A is
highly expressed in HCC. RBM8A displayed significant differences in
different HBsAg expression, tumor diameter, TNM staging and
Edmondson pathological grading which guides pathological and
clinical conclusion. Additionally, we found that patients with
lower RBM8A expression level had longer OS and PFS. Besides, our
results indicate that RBM8A promotes invasion and metastasis via
EMT signaling pathway. Loss of RBM8A significantly induces
apoptosis and reduces tumor cell population demonstrating that
RBM8A could serve as a potential target for clinical treatment.
Acknowledgements
This study was supported by Self-Raised Scientific
Research Fund of the Ministry of Health of Guangxi Province (no.
Z2015605), the Regional Science Fund Project of China Natural
Science Foundation (no. 81660498), the China Postdoctoral Science
Foundation, the 60th Grant Funding of General Program for the
Postdoctoral Funding Program in the Western Region (no.
2016M602919XB).
References
|
1
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lei X, Li YF, Chen GD, Ou DP, Qiu XX, Zuo
CH and Yang LY: Ack1 overexpression promotes metastasis and
indicates poor prognosis of hepatocellular carcinoma. Oncotarget.
6:40622–40641. 2015.PubMed/NCBI
|
|
3
|
Salicioni AM, Xi M, Vanderveer LA, Balsara
B, Testa JR, Dunbrack RL Jr and Godwin AK: Identification and
structural analysis of human RBM8A and RBM8B: Two highly conserved
RNA-binding motif proteins that interact with OVCA1, a candidate
tumor suppressor. Genomics. 69:54–62. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bechara EG, Sebestyén E, Bernardis I,
Eyras E and Valcárcel J: RBM5, 6, and 10 differentially regulate
NUMB alternative splicing to control cancer cell proliferation. Mol
Cell. 52:720–733. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boman K, Segersten U, Ahlgren G, Eberhard
J, Uhlén M, Jirström K and Malmström PU: Decreased expression of
RNA-binding motif protein 3 correlates with tumour progression and
poor prognosis in urothelial bladder cancer. BMC Urol. 13:172013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ehlén A, Brennan DJ, Nodin B, O'Connor DP,
Eberhard J, Alvarado-Kristensson M, Jeffrey IB, Manjer J,
Brändstedt J, Uhlén M, et al: Expression of the RNA-binding protein
RBM3 is associated with a favourable prognosis and cisplatin
sensitivity in epithelial ovarian cancer. J Transl Med. 8:782010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fushimi K, Ray P, Kar A, Wang L,
Sutherland LC and Wu JY: Up-regulation of the proapoptotic caspase
2 splicing isoform by a candidate tumor suppressor, RBM5. Proc Natl
Acad Sci USA. 105:15708–15713. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jögi A, Brennan DJ, Rydén L, Magnusson K,
Fernö M, Stål O, Borgquist S, Uhlen M, Landberg G, Påhlman S, et
al: Nuclear expression of the RNA-binding protein RBM3 is
associated with an improved clinical outcome in breast cancer. Mod
Pathol. 22:1564–1574. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ishigaki Y, Nakamura Y, Tatsuno T,
Hashimoto M, Shimasaki T, Iwabuchi K and Tomosugi N: Depletion of
RNA-binding protein RBM8A (Y14) causes cell cycle deficiency and
apoptosis in human cells. Exp Biol Med (Maywood). 238:889–897.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Michelle L, Cloutier A, Toutant J, Shkreta
L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture
S, et al: Proteins associated with the exon junction complex also
control the alternative splicing of apoptotic regulators. Mol Cell
Biol. 32:954–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sudo H, Tsuji AB, Sugyo A, Kohda M, Sogawa
C, Yoshida C, Harada YN, Hino O and Saga T: Knockdown of COPA,
identified by loss-of-function screen, induces apoptosis and
suppresses tumor growth in mesothelioma mouse model. Genomics.
95:210–216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zindy PJ, L'Helgoualc'h A, Bonnier D, Le
Béchec A, Bourd-Boitin K, Zhang CX, Musso O, Glaise D, Troadec MB,
Loréal O, et al: Upregulation of the tumor suppressor gene menin in
hepatocellular carcinomas and its significance in fibrogenesis.
Hepatology. 44:1296–1307. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Tian B, Yang J, Zhao L, Wu X, Ye SL,
Liu YK and Tang ZY: Stepwise metastatic human hepatocellular
carcinoma cell model system with multiple metastatic potentials
established through consecutive in vivo selection and studies on
metastatic characteristics. J Cancer Res Clin Oncol. 130:460–468.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maderazo AB, Belk JP, He F and Jacobson A:
Nonsense-containing mRNAs that accumulate in the absence of a
functional nonsense-mediated mRNA decay pathway are destabilized
rapidly upon its restitution. Mol Cell Biol. 23:842–851. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chuang TW, Chang WL, Lee KM and Tarn WY:
The RNA-binding protein Y14 inhibits mRNA decapping and modulates
processing body formation. Mol Biol Cell. 24:1–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gehring NH, Neu-Yilik G, Schell T, Hentze
MW and Kulozik AE: Y14 and hUpf3b form an NMD-activating complex.
Mol Cell. 11:939–949. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hilleren P and Parker R: mRNA surveillance
in eukaryotes: Kinetic proofreading of proper translation
termination as assessed by mRNP domain organization? RNA.
5:711–719. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishigaki Y, Nakamura Y, Tatsuno T, Ma S
and Tomosugi N: Phosphorylation status of human RNA-binding protein
8A in cells and its inhibitory regulation by Magoh. Exp Biol Med
(Maywood). 240:438–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Noensie EN and Dietz HC: A strategy for
disease gene identification through nonsense-mediated mRNA decay
inhibition. Nat Biotechnol. 19:434–439. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carrasco DR, Tonon G, Huang Y, Zhang Y,
Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, et
al: High-resolution genomic profiles define distinct
clinico-pathogenetic subgroups of multiple myeloma patients. Cancer
Cell. 9:313–325. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim TJ, Choi JJ, Kim WY, Choi CH, Lee JW,
Bae DS, Son DS, Kim J, Park BK, Ahn G, et al: Gene expression
profiling for the prediction of lymph node metastasis in patients
with cervical cancer. Cancer Sci. 99:31–38. 2008.PubMed/NCBI
|
|
22
|
Knuutila S, Aalto Y, Autio K, Björkqvist
AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S,
Larramendy ML, et al: DNA copy number losses in human neoplasms. Am
J Pathol. 155:683–694. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Togi S, Shiga K, Muromoto R, Kato M, Souma
Y, Sekine Y, Kon S, Oritani K and Matsuda T: Y14 positively
regulates TNF-α-induced NF-κB transcriptional activity via
interacting RIP1 and TRADD beyond an exon junction complex protein.
J Immunol. 191:1436–1444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ishigaki Y, Nakamura Y, Tatsuno T,
Hashimoto M, Iwabuchi K and Tomosugi N: RNA-binding protein RBM8A
(Y14) and MAGOH localize to centrosome in human A549 cells.
Histochem Cell Biol. 141:101–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Arsura M and Cavin LG: Nuclear
factor-kappaB and liver carcinogenesis. Cancer Lett. 229:157–169.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Calvisi DF, Ladu S, Gorden A, Farina M,
Conner EA, Lee JS, Factor VM and Thorgeirsson SS: Ubiquitous
activation of Ras and Jak/Stat pathways in human HCC.
Gastroenterology. 130:1117–1128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Cigliano A, Delogu S, Armbruster
J, Dombrowski F, Evert M, Chen X and Calvisi DF: Functional
crosstalk between AKT/mTOR and Ras/MAPK pathways in
hepatocarcinogenesis: Implications for the treatment of human liver
cancer. Cell Cycle. 12:1999–2010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ohbayashi N, Taira N, Kawakami S, Togi S,
Sato N, Ikeda O, Kamitani S, Muromoto R, Sekine Y and Matsuda T: An
RNA biding protein, Y14 interacts with and modulates STAT3
activation. Biochem Biophys Res Commun. 372:475–479. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Roignant JY and Treisman JE: Exon junction
complex subunits are required to splice Drosophila MAP
kinase, a large heterochromatic gene. Cell. 143:238–250. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang L and Baker NE: Cell cycle
withdrawal, progression, and cell survival regulation by EGFR and
its effectors in the differentiating Drosophila eye. Dev
Cell. 4:359–369. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brechot C, Kremsdorf D, Soussan P, Pineau
P, Dejean A, Paterlini-Brechot P and Tiollais P: Hepatitis B virus
(HBV)-related hepatocellular carcinoma (HCC): Molecular mechanisms
and novel paradigms. Pathol Biol (Paris). 58:278–287. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu C, Zhou W, Wang Y and Qiao L: Hepatitis
B virus-induced hepatocellular carcinoma. Cancer Lett. 345:216–222.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan T, Zhao JJ, Bi XY, Zhao H, Huang Z, Li
ZY, Zhou JG, Li Y, Li C, Cai JQ, et al: Prognosis of hepatocellular
carcinoma: A study of 832 cases. Zhonghua Zhong Liu Za Zhi.
35:54–58. 2013.(In Chinese). PubMed/NCBI
|