Introduction
Chronic myelogenous leukemia (CML) is a
myeloproliferative disorder resulting from the neoplastic
transformation of hematopoietic stem cell (1). At present, tyrosine kinase inhibitors
are widely used for the treatment of CML. However, cancerous cells
frequently develop multidrug resistance (MDR) to chemotherapy
agents.
MDR is defined as the resistance of cancer cells to
antineoplastic agents that have distinct structures and mechanisms
of action (2). The specific
mechanisms responsible for the MDR phenotype include
pharmacokinetic alterations, tumor microenvironmental changes, or
cancer cell-specific factors, which include increased drug efflux
or decreased drug uptake, drug inactivation, drug target
modification or apoptosis evasion (3). ABCB1 (MDR1/P-glycoprotein) is
considered to be accountable for the majority of drug efflux in
human cancers. P-glycoprotein (P-gp) is a member of the superfamily
of ATP-binding cassette (ABC) transporters. P-gp has been reported
to be associated with resistance to multiple drugs in various types
of cancer, and changes in its expression or function contribute to
MDR (4–6).
MicroRNAs (miRNAs) are a class of non-coding RNAs
that are 21–25 nucleotides in length and function as regulators of
gene expression (7). The
dysregulation of miRNA expression has also been detected in CML
(8). Loss of miR-203 was detected
and overexpression of miR-203 increased sensitivity to imatinib in
BaF3-BCR/ABL (T315I) cells (9).
miR-9, as an oncogene or tumor-suppressor gene, plays an important
role in many types of cancer (10,11).
miR-9-3 was found to be downregulated in breast cancers with either
high vascular invasion or the presence of lymph node metastasis
(12). miR-9-3 could be used as a
DNA methylation target in non-small cell lung cancers (13). miR-9-3 activated the NFκB signaling
pathway in chronic lymphocytic leukemia by downregulating NF-κB1
protein (14). However, the effect
of miR-9 in MDR of CML remains unclear.
The present study was carried out to investigate the
expression levels of miR-9 in K562 and K562/ADR cells and CML
patients. In addition, the functional role of miR-9 in the drug
resistance of CML was identified. Moreover, we explored the
correlation between the expression of miR-9 and ABCB1.
Materials and methods
Cell culture
Human chronic myelogenous leukemia blast K562 cells
were cultured in RPMI-1640 medium, supplemented with 10% fetal
bovine serum (FBS) and 1% penicillin-streptomycin (all from Gibco,
Grand Island, NY, USA) at 37̊C in humidified atmosphere containing
5% CO2. Adriamycin (Sigma, St. Louis, MO, USA) was added
to parental cell cultures in stepwise increasing concentrations
from 0.001 to 1 mg/l for 6 months to develop an adriamycin
(ADR)-resistant cell line, named K562/ADR. For K562/ADR cells, 1
mg/l of adriamycin was added in daily culture and removed before
the experiments.
Primary patient samples
Sixty-one patients were recruited from July 2012 to
June 2015 at The First Affiliated Hospital of Dalian Medical
University (Dalian, China). The age of patients ranged from 17 to
70 years of age, with a median age of 43. The diagnosis of CML was
based on cytomorphology, cytochemistry, multiparameter flow
cytometry (FCM), immunology, molecular genetics and cytogenetics.
Bone marrow cells were extracted after the patients were admitted
to the hospital before treatment. Murine anti-human P-gp (UIC2
clone)-PE was purchased from BioLegend (San Diego, CA, USA). Then,
the expression of P-gp was detected by FCM. Peripheral blood
mononuclear cells (PBMCs) were isolated using Ficoll-Hypaque and
were further cultured in plastic dishes to remove adherent cells at
37̊C for 24 h. The research protocol was approved by the Ethics
Committee of Dalian Medical University. The patient clinical
characteristics are shown in Table
I.
 | Table I.Clinicopathological characteristics of
the CML patients. |
Table I.
Clinicopathological characteristics of
the CML patients.
| Patient
demographics | CML (n=61) |
|---|
| Gender |
|
| Male | 38 |
|
Female | 23 |
| Age (years) |
|
|
Median | 43 |
|
Range | 17–70 |
| Splenic
enlargement | 52 |
| Hemoglobin <100.0
g/l | 19 |
| WBC count
(109/l) |
|
|
20–100 | 36 |
|
>100 | 25 |
| Platelet count
(109/l) |
|
|
<300 | 27 |
|
>300 | 34 |
| P-gp(+) | 33 |
MicroRNA array
miRNA arrays were performed for K562 and K562/ADR
cell groups (n=3/group) by Exiqon (KangChen, China) using the
miRCURY™Hy3™/Hy5™ power labeling kit and the miRCURY™ LNA Array
(version 10.0; 757 human miRs).
Real-time PCR
Total RNA was isolated from the cell lines with the
RNeasy Mini kit, and cDNA was synthesized using QuantiTect Reverse
Transcription kit (both from Qiagen, Valencia, CA, USA) according
to the manufacturer's protocol. The expression of miR-9 was
determined using mirVana™ qRT-PCR MicroRNA Detection kit according
to the manufacturer's protocol (Ambion Inc., Austin, TX, USA) and
was normalized to U6-small nuclear RNA. qRT-PCR was performed to
detect ABCB1 mRNA using the SYBR-Green PCR kit (Takara Bio, Inc.,
Otsu, Japan). The sequences of the upstream and downstream primers
were as follows: 5′-CCCATCATTGCAATAGCAGG-3′ and
5′-GTTCAAACTTCTGCTCCTGA-3′ for ABCB1; 5′-CTCCCTCCACCTTTGACGCTG-3′
and 5′-TCCTCTTGTGCTCTTGCTGG-3′ for GAPDH. The expression level of
target genes was determined relatively to GAPDH and calculated as:
2 -(CtTarget gene - CtGAPDH).
Oligonucleotide construction
shRNA specific for ABCB1
(5′-GUUUGUCUACAGUUCGUAA-3′); mimics (5′-AUAAAGCUAGAUAACCGAAAGU-3′
and 5′-UUUCGGUUAUCUAGCUUUAUUU-3′); mimic negative control (NC)
(5′-UUCUUCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′);
antagomiR (5′-ACUUUCGGUUAUCUAGCUUUAU-3′); and control
(5′-CAGUACUUUUGUGUAGUACAA-3′) for hsa-miR-9 were obtained from
RiboBio Co. Ltd. (Guangzhou, Guangdong, China).
In vitro drug sensitivity assay
Drug resistance was evaluated by MTT assay as
previously described (15).
Briefly, cells (1×104) were plated in a 96-well plate
and incubated with different anticancer drugs at varying
concentrations (doxorubicin, 0, 10, 30,50 and 100 mg/l;
vincristine, 0, 10, 30, 50 and 100 mg/l; paclitaxel, 0, 1, 5, 20
and 40 mg/l; Sigma), respectively. After 48 h, 100 µl MTT (5 mg/l;
Sigma) was added to each well and cultured for an additional 4 h.
Then, 100 µl of dimethyl sulfoxide (DMSO) was added into each well
and the absorbance was determined at 490 nm using a microplate
reader (Model-550; Bio-Rad Laboratories, Hercules, CA, USA).
Intracellular ADR concentration
assays
Fluorescence intensity of intracellular ADR
(doxorubicin) was determined using FCM. Briefly, cells were seeded
into 6-well plates (1×106 cells/well) and cultured
overnight. After addition of ADR to a final concentration of 5
µg/ml, the cells continued to be cultured for 1 h. The cells were
then harvested. Then, the cells were washed with
phosphate-buffered-saline (PBS), and the mean fluorescence
intensity of intracellular ADR was detected using FCM. The
experiment was independently performed 3 times.
Apoptosis analysis
Cells were seeded onto a 6-well plate at a density
of ~2×105 cells/well. Following treatment with drugs
[(K562/ADR, vincristine/paclitaxel 40 µg/ml; K562,
vincristine/paclitaxel 1 µg/ml) (for 48 h)], the cells were
collected, and washed with ice-cold PBS twice. Cells were
resuspended in 100 µl binding buffer and stained with Annexin
V/FITC followed by propidium iodide (PI). Cells were then incubated
in the dark for 15 min at room temperature, and 400 µl binding
buffer was added. The cells were immediately measured by
FACSCalibur (Becton-Dickinson, San Jose, CA, USA).
Luciferase assay
A pmirGLO dual-luciferase miRNA target expression
vector was used for the 3′-UTR luciferase assays (Promega, Madison,
WI, USA). Plasmids containing wild-type pmirGLO-ABCB1-3′-UTR,
mutant pmirGLO-ABCB1-3′-UTR were synthesized. HEK 293T cells were
seeded (5×104 cells/well) in a 24-well dish and were
incubated overnight. For the 3′-UTR luciferase assay, the cells
were co-transfected with hsa-miR-9 mimics and wild-type or mutant
target sequence using Lipofectamine 2000. The lysates were
collected 48 h after the transfection, and the activities of
firefly and Renilla luciferases were measured using the
Dual-Luciferase® Reporter Assay system (Promega) and
normalized to those of Renilla luciferase activities. The
mean of the results from the cells transfected with the miR-control
was set at 1.0. Data are presented as the mean value ± SD for
triplicate experiments.
Western blot analysis
The total cell proteins were separated by SDS
polyacrylamide gel electrophoresis with 6% spacer and 10%
separation gels. The protein samples were removed onto
polyvinylidene difluoride (PVDF) membranes and blocked with 5%
powdered skimmed milk prepared with TTBS. Then, the PVDF membranes
were respectively incubated with P-gp antibody (1/1,000 dilution;
Abcam, Cambridge, UK), and then with peroxidase-conjugated
anti-rabbit IgG (1/10,000 dilution; GE Healthcare UK Ltd., Little
Chalfont, UK). The control was the GAPDH antibody (1/2500 dilution;
Bioworld, St. Louis Park, MN, USA). All bands were detected using
ECL Western Blot kit (Amersham Biosciences, Buckinghamshire, UK),
and analyzed using LabWorks™ (version 4.6; UVP; BioImaging
Systems).
In vivo antitumor activity
Four-week-old male athymic nude mice were obtained
from the Animal Facility of Dalian Medical University, and were fed
with sterilized food and water. Approximately, 1×107
cells (K562/ADR-NC and K562/ADR-miR-9) were subcutaneously injected
into the right flank of each nude mouse, respectively. Mice bearing
palpable tumors (~1 week after tumor cell inoculation) were
randomly divided into control and treatment groups (6
animals/group). The mice in groups were intratumorally injected
with 100 µl mimics or NC combined with an intraperitoneal injection
of adriamycin (7 mg/kg) or PBS 3 times/week for 3 weeks. The mice
were sacrificed and their tumors were isolated, weighed and
photographed. The tumor volume was calculated by the following
formula: Tumor volume = 1/2 (length × width2). Tumors
were removed, weighed and fixed in 4% paraformaldehyde. These
experiments were approved by the Ethics Committee of the Animal
Experiments of the Dalian Medical University.
Immunohistochemical (IHC) staining
analysis
Visible tumors were removed from the mice and
immunohistochemistry was performed on paraffin-embedded sections.
The slides were dried, deparaffinized and rehydrated. After
deparaffinization and blocking of endogenous peroxidase, the slides
were labeled overnight at 4°C with antibodies (Abcam) at a dilution
of 1:200. The following staining was performed at room temperature
for 60 min with secondary streptavidin-HRP-conjugated antibody
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Finally, the
sections were counterstained with hematoxylin and coverslipped. The
Image-Pro Plus 4.5 software (Media Cybernetics, Inc., Rockville,
MD, USA) was used to analyze the expression of proteins.
Statistical analysis
Each test was performed at least in triplicate, and
the data are expressed as means ± standard deviation (SD).
Student's t-test was used to compare the means of two groups.
P<0.05 was considered to indicate a statistically significant
result. All analyses were performed using SPSS 13.0 statistical
packages (SPSS, Inc., Chicago, IL, USA).
Results
Differential expression of miR-9 in
K562 and K562/ADR cells and CML patients
Real-time PCR was used to analyze the expression of
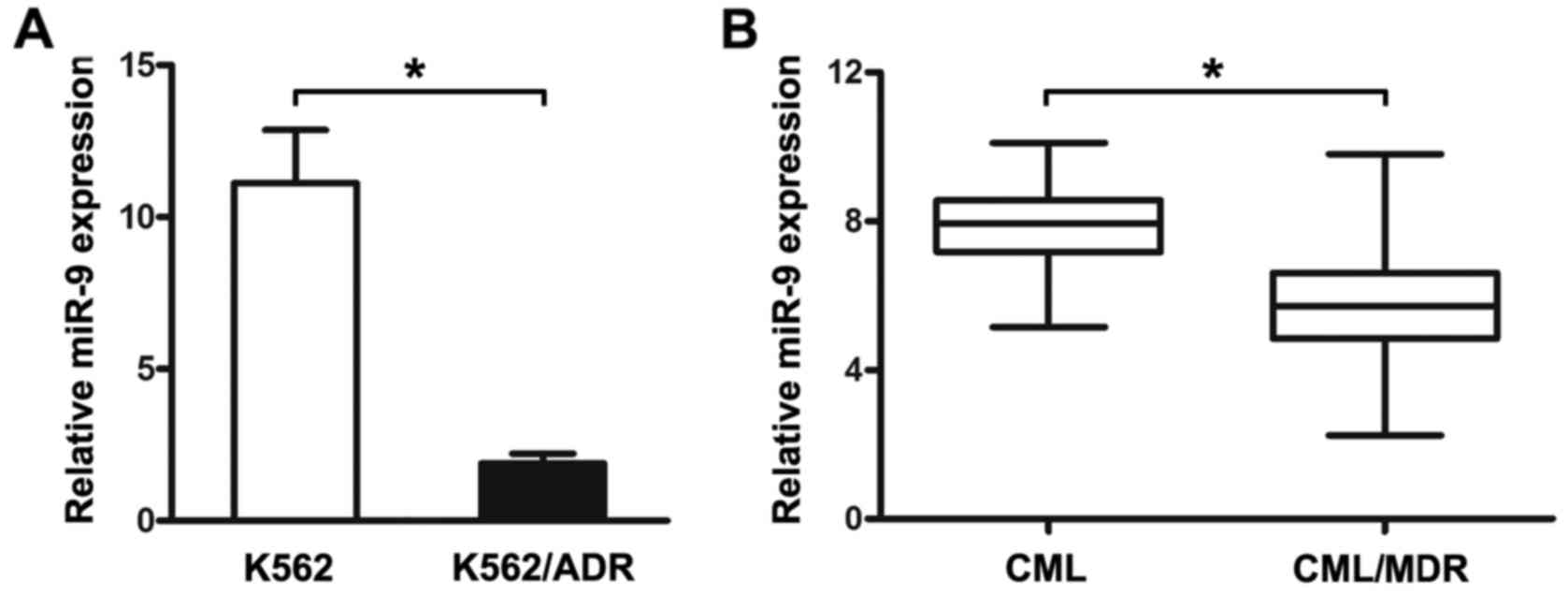
miR-9 in the K562 and K562/ADR cells and CML patients. As shown in
Fig. 1A, the expression of miR-9
was obviously increased in the K562 cells, compared with that noted
in the K562/ADR cells.
To further investigate the expression of miR-9 in
CML patients, the PBMCs isolated from CML patients were analyzed by
qRT-PCR. The PBMCs were first divided into two groups, CML without
MDR and CML/MDR. The frequency of P-gp positivity was 54.1% (33 of
61) in the CML patients. It was shown that miR-9 expression was
significantly lower in the CML/MDR patients compared to that noted
in the CML patients (Fig. 1B). The
data indicated that differential expression of miR-9 may be related
to the drug-resistance of human CML.
Inhibition of miR-9 decreases the
chemosensitivity of K562 cells in vitro
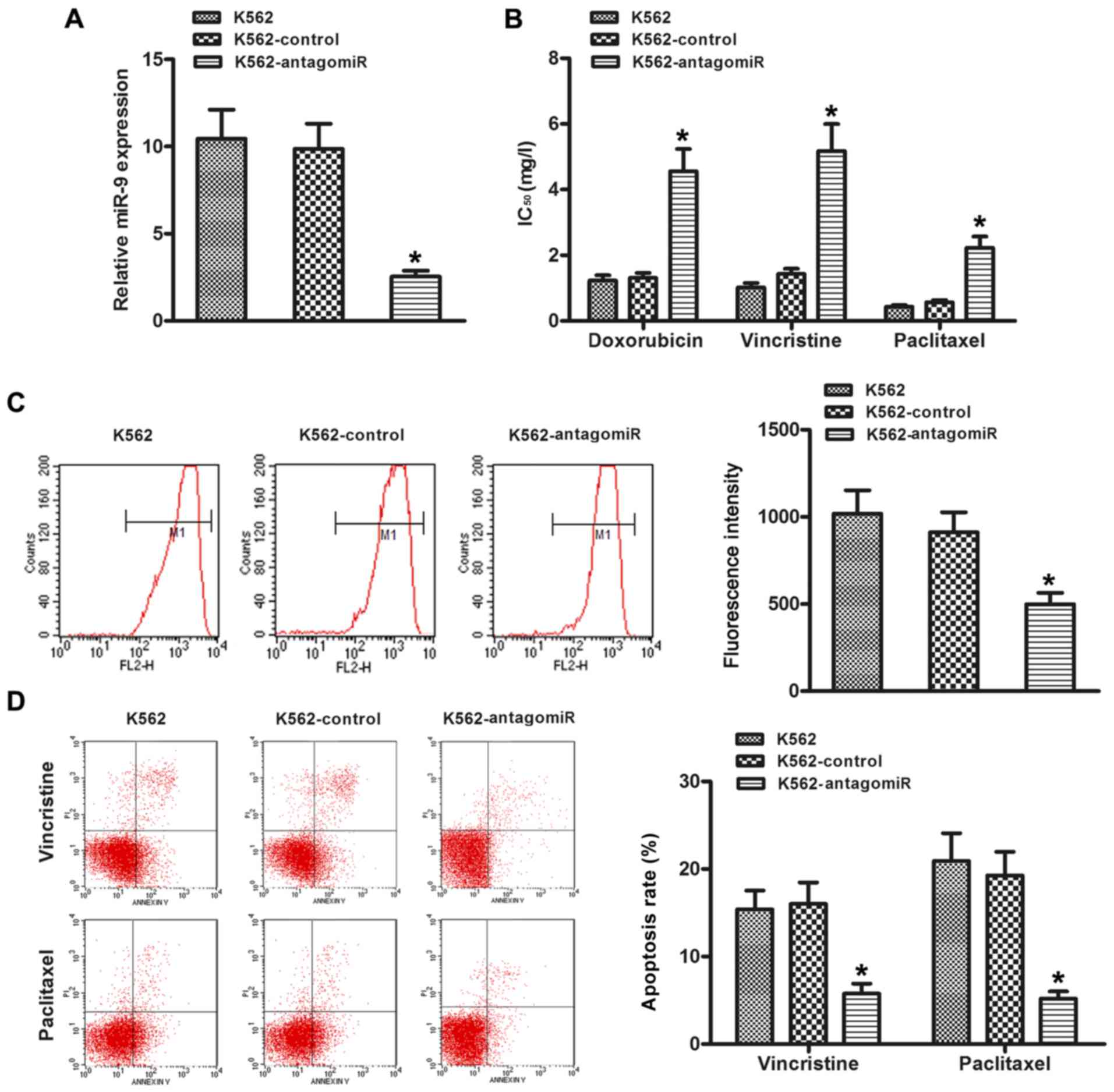
In order to investigate the effect of miR-9 on the
chemosensitivity of CML cells, the K562 cells were transfected with
miR-9 antagomiR. As shown in Fig.
2A, the expression of miR-9 was obviously decreased compared
with the control group. The IC50 values (drug
concentration that inhibits cell growth by 50%) were significantly
increased in the K562 cells transfected with the miR-9 antagomiR
(Fig. 2B), compared to the control.
Since MDR of cancer is mainly due to alterations of drug influx and
efflux, ADR was used as a probe to evaluate drug accumulation in
the cancer cells. Thus, ADR intracellular accumulation was
explored. As shown in Fig. 2C,
decreased accumulation of ADR was observed in the K562-antagomiR
cells compared with the control cells (P<0.05). Then, the role
of miR-9 downregulation in the survival of drug-resistant cells was
investigated. Annexin V assays demonstrated that silencing of miR-9
reduced the apoptosis rate of K562 cells by resistance to the
chemotherapeutic agent vincristine or paclitaxel (Fig. 2D).
Overexpression of miR-9 increases the
chemosensitivity of K562/ADR cells in vitro
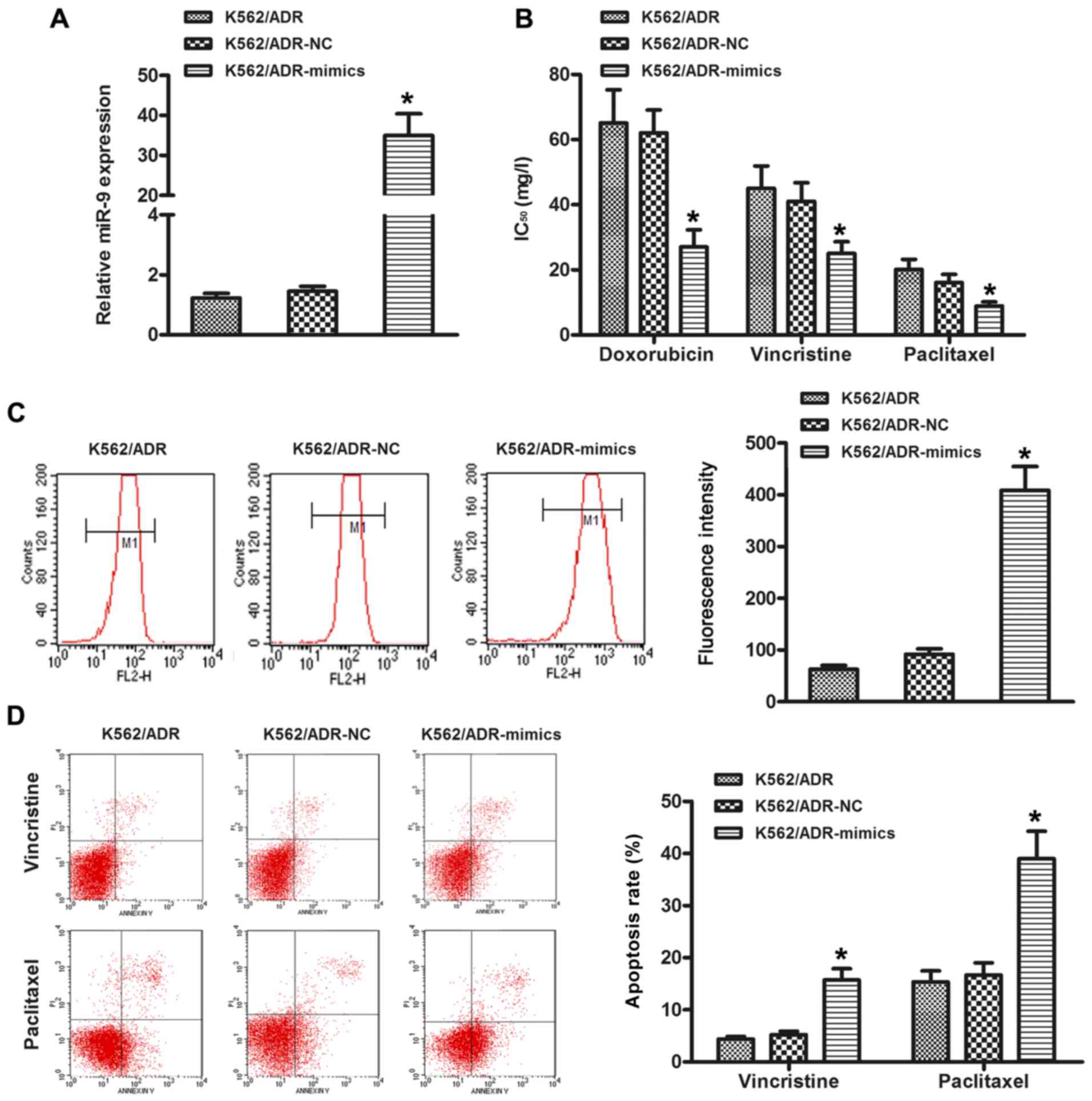
In order to further study the effect of miR-9 on
chemosensitivity, the K562/ADR cells were transfected with miR-9
mimics. As shown in Fig. 3A, the
expression level of miR-9 was increased in the K562/ADR-mimic cells
compared with the NC cells. In addition, overexpression of miR-9
sensitized cells to chemotherapy, as indicated by a decrease in the
IC50 values (Fig. 3B).
Moreover, the restoration of miR-9 increased the level of
intracellular ADR (Fig. 3C).
Meanwhile, ectopic expression of miR-9 increased the apoptosis rate
of K562/ADR cells treated with chemotherapeutic agent vincristine
or paclitaxel (Fig. 3D).
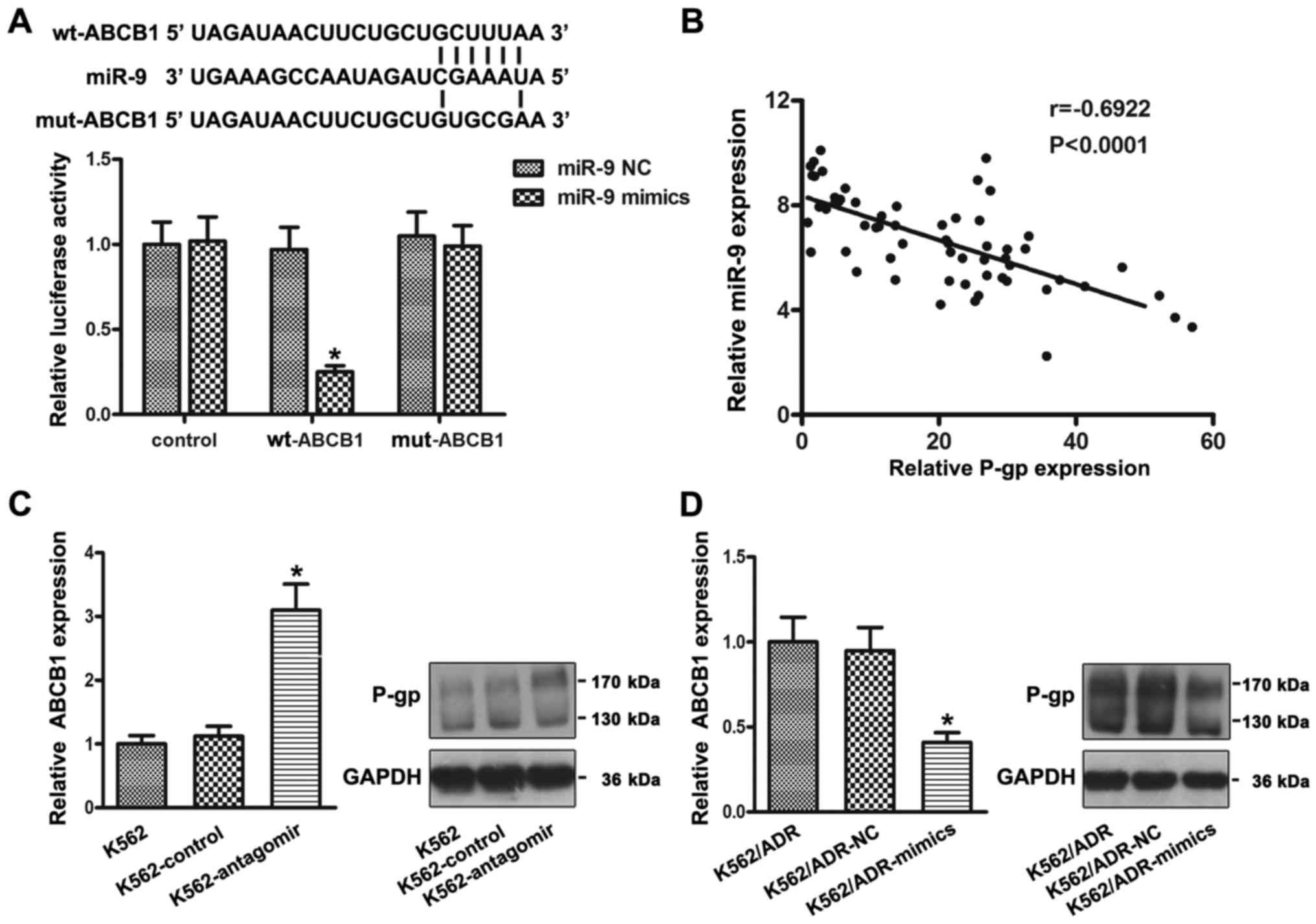
ABCB1 is a target of miR-9
Bioinformatic analyses suggested that ABCB1 is a
potential target of miR-9. To test whether ABCB1 is a direct target
of miR-9, luciferase activity assays were performed. 293T cells
were transfected with luciferase reporters carrying the
ABCB1-wt-3′-UTR or ABCB1-mut-3′-UTR, along with mimics or NC. As
shown in Fig. 4A, forced miR-9
expression decreased luciferase activity, and this suppression was
reversed by the mutation of the target sequences in the 3′-UTRs of
ABCB1. Furthermore, the expression of ABCB1 mRNA was analyzed by
RT-PCR and the expression of P-gp proteins was analyzed by western
blotting. The results showed that the ABCB1 mRNA and P-gp protein
were obviously increased after the knockdown of miR-9 in the K562
cells (Fig. 4C). Conversely, the
ABCB1 mRNA and P-gp protein were significantly decreased after the
overexpression of miR-9 in the K562/ADR cells (Fig. 4D). Moreover, P-gp expression in CML
patients was detected by FCM. As shown in Fig. 4B, the expression levels of miR-9
were negatively correlated with P-gp expression in the CML patients
(r=−0.6922, P<0.0001).
ABCB1 reverses the effect of
miR-9-mediated chemoresistance in K562 cells
To further confirm that ABCB1 is a direct target of
miR-9, we performed a rescue experiment by co-transfecting ABCB1
shRNA (vs. the shRNA control) and miR-9 antagomiR (vs. the control)
into K562 cells. Transfection of miR-9 antagomiR led to the
upregulation of P-gp protein as determined by western blot
analysis. Importantly, the upregulation of P-gp induced by miR-9
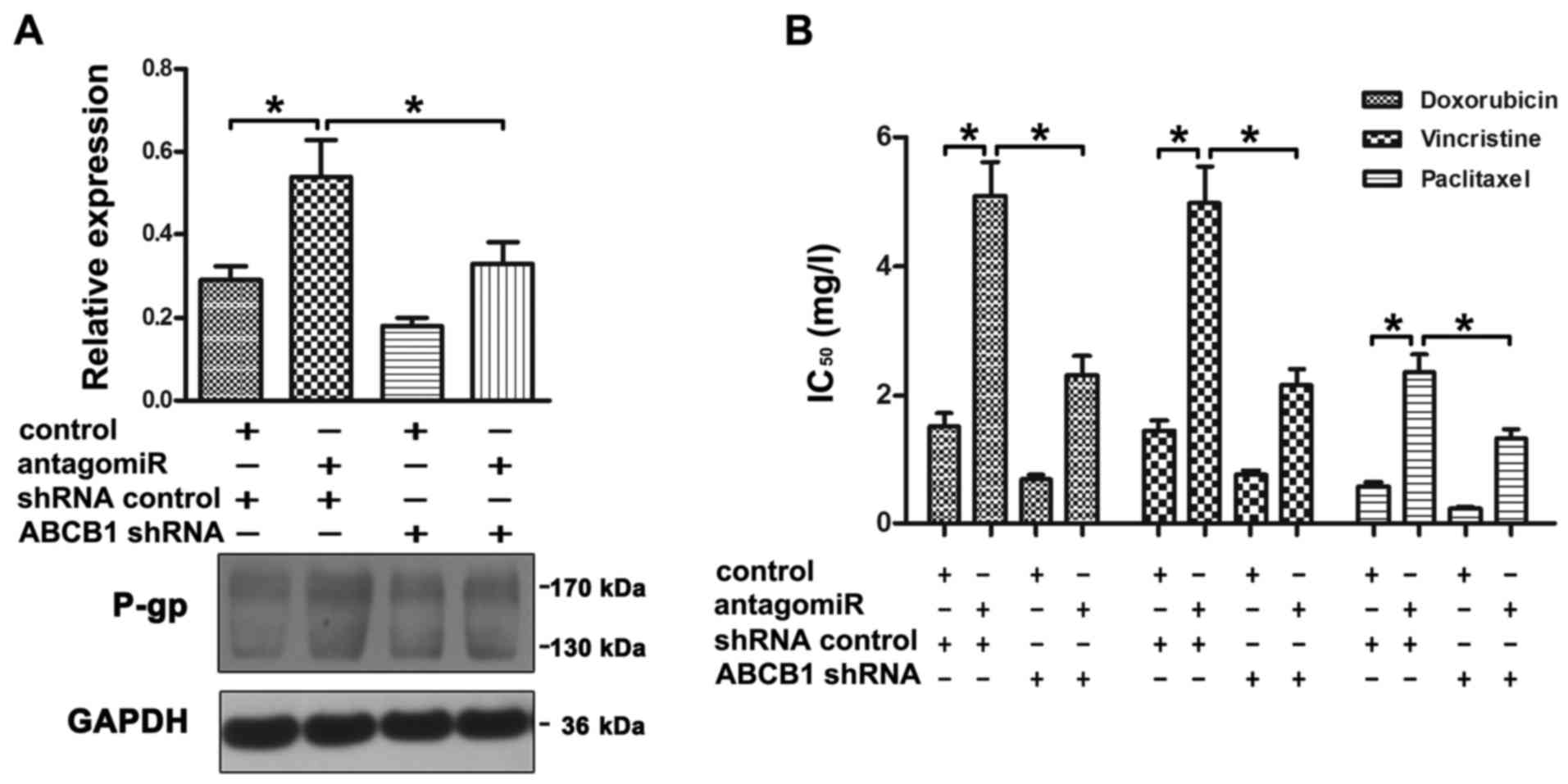
antagomiR was effectively reversed by ABCB1 shRNA (Fig. 5A). Moreover, the increase in drug
resistance induced by miR-9 antagomiR was attenuated by ABCB1 shRNA
(Fig. 5B).
Altered expression of miR-9 influences
the chemosensitivity of K562/ADR cells in vivo
Then, we investigated whether miR-9 regulates the
chemosensitivity of CML cells in vivo. As shown in Fig. 6A, overexpression of miR-9 in
K562/ADR cells resulted in a significantly lower tumor volume
compared with the cells treated with NC after 3 weeks of ADR
treatment. In addition, the tumor volume in the K562/ADR-miR-9
group was markedly decreased compared with that in the K562/ADR-NC
group. At the end of this experiment, the mice were sacrificed and
their tumors were isolated. We further analyzed the P-gp expression
pattern in the tumors by IHC staining. The expression of P-gp was
decreased in the K562/ADR-miR-9 group compared with the expression
noted in the NC group (Fig. 6B).
These results were consistent with the results in vitro.
Discussion
Multidrug resistance (MDR) and disease relapse is a
challenging clinical issue in the treatment of leukemia. In the
present study, we found that the expression of miR-9 was obviously
increased in the K562 cells and CML patients. It was shown that
miR-9 regulated the CML drug resistance both in vitro and
in vivo. In addition, we found that miR-9 and P-glycoprotein
(P-gp) had a negative correlation in the CML patients. Furthermore,
we found that ABCB1 is a direct target of miR-9.
miRNAs plays an important role in the
chemoresistance of various types of cancers, by targeting the
3′-untranslated region (3′-UTR) of a specific mRNA via base
pairing. miR-181b has been reported to be involved in MDR in
gastric cancer (15). miR-221/−222
were found to increase ER-independent growth and confer resistance
to fulvestrant by the activation of β-catenin in breast cancer
(16). It was reported that miR-214
negatively regulates phosphatase and tensin homolog (PTEN), which
is a tumor suppressor, leading to induced cisplatin resistance
(17).
Several studies have described regulatory functions
for miRNAs in the chemoresponsiveness of leukemia cells.
Overexpression of miR-370 sensitized K562 cells to
homoharringtonine (18). Moreover,
miR-17 and miR-20a have been reported to be involved in the
chemoresistance of leukemia cells (19). Additionally, ectopic expression of
miR-217 was found to sensitize dasatinib-resistant K562 cells to
dasatinib (20). More recently,
downregulation of miR-21 increased apoptosis in CML (21). However, the role of miRNAs in the
drug resistance of leukemia has not been fully addressed. In the
present study, we found that the expression of miR-9 was obviously
increased in K562 cells, compared with that in the K562/ADR cells.
Moreover, miR-9 expression was significantly lower in CML/MDR
patients compared to CML patients. Inhibition of miR-9 decreased
the chemosensitivity of K562 cells in vitro. Moreover,
overexpression of miR-9 increased the chemosensitivity of K562/ADR
cells in vitro and in vivo. Notably, the tumor
volumes in the K562/ADR-miR-9 group were markedly decreased
compared with tumor volumes in the K562/ADR-NC group. The potential
reason may be that miRNA has hundreds or thousands of mRNA targets;
a broad segment of the protein coding genome is under their
control. It has been reported that high miR-9 expression
significantly sensitized ovarian cancer cells to cisplatin in
vivo and in vitro (22),
consistent with the present study. However, another study
demonstrated that miR-9 promoted the chemoresistance of bladder
cancer cells by targeting LASS2 (23). This may suggest that miR-9 is
involved in the response to treatment. The role of miR-9 as an
oncogene or as a tumor suppressor for tumor chemoresistance depends
on the cell type.
P-gp is a membrane transporter glycoprotein. The
overexpression of P-gp in cancer cells promotes drug resistance
(6), whereas inhibition of the
expression or function of P-gp reverses drug resistance (3). miR-451 was found to be inversely
correlated with MDR1 expression in drug-resistant breast cancer
cells (24). It was also reported
that upregulated miR-27a and miR-451 stimulated MDR1 expression in
cell lines of human ovarian cancer and cervix carcinoma (25). More recently, miR-331-5p and miR-27a
were found to regulate P-gp by directly binding to its 3′-UTR
(26). In the present study, it was
confirmed that ABCB1 is a direct target of miR-9 in CML cells by
luciferase activity analysis. An altered level of miR-9 led to a
change in ABCB1 mRNA and P-gp proteins. Then, we detected the
expression of miR-9 and P-gp in CML patients. It was found that
miR-9 and P-gp had a negative correlation. Moreover, silencing of
ABCB1 reversed the drug resistance induced by the miR-9 antigomir.
Furthermore, miR-9 inhibited the expression of P-gp protein in
tumor tissues. These observations indicate that miR-9 may serve as
an MDR-specific miRNA in human CML by targeting ABCB1.
In summary, miR-9 regulated the drug resistance of
CML. miR-9 may be a potential therapeutic target for the treatment
of the MDR of CML in the future. Certainly, there are additional
target genes that contribute to the function of miR-9 in CML.
Therefore, further effort is warranted to expand the miR-9
downstream functional network.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81472014).
References
|
1
|
Sloma I, Jiang X, Eaves AC and Eaves CJ:
Insights into the stem cells of chronic myeloid leukemia. Leukemia.
24:1823–1833. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fojo A, Hamilton TC, Young RC and Ozols
RF: Multidrug resistance in ovarian cancer. Cancer. 60:(Suppl 8).
S2075–S2080. 1987. View Article : Google Scholar
|
|
3
|
Zinzi L, Capparelli E, Cantore M, Contino
M, Leopoldo M and Colabufo NA: Small and innovative molecules as
new strategy to revert MDR. Front Oncol. 4:22014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hendrikx JJ, Lagas JS, Rosing H, Schellens
JH, Beijnen JH and Schinkel AH: P-glycoprotein and cytochrome P450
3A act together in restricting the oral bioavailability of
paclitaxel. Int J Cancer. 132:2439–2447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu H, Liu Z, Tang L, Liu J, Zhou M, Xie
F, Wang Z, Wang Y, Shen S, Hu L, et al: Reversal of P-gp and
MRP1-mediated multidrug resistance by H6, a gypenoside aglycon from
Gynostemma pentaphyllum, in vincristine-resistant human oral
cancer (KB/VCR) cells. Eur J Pharmacol. 696:43–53. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsuji K, Wang YH, Takanashi M, Odajima T,
Lee GA, Sugimori H and Motoji T: Overexpression of lung
resistance-related protein and P-glycoprotein and response to
induction chemotherapy in acute myelogenous leukemia. Hematol Rep.
4:e182012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang LS, Li L, Li L, Chu S, Shiang KD, Li
M, Sun HY, Xu J, Xiao FJ, Sun G, et al: MicroRNA-486 regulates
normal erythropoiesis and enhances growth and modulates drug
response in CML progenitors. Blood. 125:1302–1313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Y, Yuan Y, Tao K, Wang X, Xiao Q, Huang
Z, Zhong L, Cao W, Wen J and Feng W: Inhibition of BCR/ABL protein
expression by miR-203 sensitizes for imatinib mesylate. PLoS One.
8:e618582013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC,
Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, et al: Aberrant
hypermethylation of miR-9 genes in gastric cancer.
Epigenetics. 6:1189–1197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Qi M, Li S, Qi T, Mei H, Huang K,
Zheng L and Tong Q: microRNA-9 targets matrix metalloproteinase 14
to inhibit invasion, metastasis, and angiogenesis of neuroblastoma
cells. Mol Cancer Ther. 11:1454–1466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heller G, Weinzierl M, Noll C, Babinsky V,
Ziegler B, Altenberger C, Minichsdorfer C, Lang G, Döme B,
End-Pfützenreuter A, et al: Genome-wide miRNA expression profiling
identifies miR-9-3 and miR-193a as targets for DNA
methylation in non-small cell lung cancers. Clin Cancer Res.
18:1619–1629. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang LQ, Kwong YL, Kho CS, Wong KF, Wong
KY, Ferracin M, Calin GA and Chim CS: Epigenetic inactivation of
miR-9 family microRNAs in chronic lymphocytic leukemia -
implications on constitutive activation of NFκB pathway. Mol
Cancer. 12:1732013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu W, Shan X, Wang T, Shu Y and Liu P:
miR-181b modulates multidrug resistance by targeting BCL2 in human
cancer cell lines. Int J Cancer. 127:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al:
MicroRNA expression profiling in human ovarian cancer:
miR-214 induces cell survival and cisplatin resistance by
targeting PTEN. Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou M, Zeng J, Wang X, Guo Q, Huang T,
Shen H, Fu Y, Wang L, Jia J and Chen C: MiR-370 sensitizes chronic
myeloid leukemia K562 cells to homoharringtonine by targeting
Forkhead box M1. J Transl Med. 11:2652013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weng H, Huang H, Dong B, Zhao P, Zhou H
and Qu L: Inhibition of miR-17 and miR-20a by oridonin triggers
apoptosis and reverses chemoresistance by derepressing BIM-S.
Cancer Res. 74:4409–4419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nishioka C, Ikezoe T, Yang J, Nobumoto A,
Tsuda M and Yokoyama A: Downregulation of miR-217 correlates with
resistance of Ph+ leukemia cells to ABL tyrosine kinase
inhibitors. Cancer Sci. 105:297–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang WZ, Pu QH, Lin XH, Liu MY, Wu LR, Wu
QQ, Chen YH, Liao FF, Zhu JY and Jin XB: Silencing of miR-21
sensitizes CML CD34+ stem/progenitor cells to
imatinib-induced apoptosis by blocking PI3K/AKT pathway. Leuk Res.
39:1117–1124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun C, Li N, Yang Z, Zhou B, He Y, Weng D,
Fang Y, Wu P, Chen P, Yang X, et al: miR-9 regulation of BRCA1 and
ovarian cancer sensitivity to cisplatin and PARP inhibition. J Natl
Cancer Inst. 105:1750–1758. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Zhang W, Zuo Y, Ding M, Ke C, Yan
R, Zhan H, Liu J and Wang J: miR-9 promotes cell proliferation and
inhibits apoptosis by targeting LASS2 in bladder cancer. Tumour
Biol. 36:9631–9640. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kovalchuk O, Filkowski J, Meservy J,
Ilnytskyy Y, Tryndyak VP, Chekhun VF and Pogribny IP: Involvement
of microRNA-451 in resistance of the MCF-7 breast cancer cells to
chemotherapeutic drug doxorubicin. Mol Cancer Ther. 7:2152–2159.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu H, Wu H, Liu X, Evans BR, Medina DJ,
Liu CG and Yang JM: Role of MicroRNA miR-27a and miR-451 in the
regulation of MDR1/P-glycoprotein expression in human cancer
cells. Biochem Pharmacol. 76:582–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng DD, Zhang H, Zhang P, Zheng YS, Zhang
XJ, Han BW, Luo XQ, Xu L, Zhou H, Qu LH, et al: Down-regulated
miR-331-5p and miR-27a are associated with chemotherapy resistance
and relapse in leukaemia. J Cell Mol Med. 15:2164–2175. 2011.
View Article : Google Scholar : PubMed/NCBI
|