Introduction
The growing incidence of cancer and associated
mortality worldwide highlights the importance of timely and
effective diagnostic methods (1).
Fluorescent probes that specifically recognize molecular targets
have proven useful in many areas of biology and medicine, and they
show potential for sensitive cancer diagnosis (2–4). Such
probes are usually created by conjugating a reporter fluorophore to
an affinity reagent, such as a monoclonal antibody, peptide or
aptamer, which binds specifically to the target of interest
(5–8). To ensure specificity and sensitivity,
the fluorophore must emit a strong signal and be photostable. It
may emit minimal fluorescence in the absence of a target, however,
in the presence of one the affinity reagent may bind to the target
with high specificity and affinity (9). A key challenge in designing
fluorescence probes is minimizing background signals in the absence
of a target.
Nucleic acid aptamers, which are short,
single-stranded RNA or DNA oligonucleotides, show substantial
promise as affinity agents (10–13).
They have several advantages over other affinity reagents,
including high affinity and specificity, facile synthesis and
modification and a structure-controlled design. Compared to other
affinity reagents, aptamers may be taken up more quickly by tissue,
penetrate more deeply into tissue, accumulate more in target tissue
and persist less in non-target organs and circulation (14,15).
These characteristics make aptamers well suited for cancer
detection.
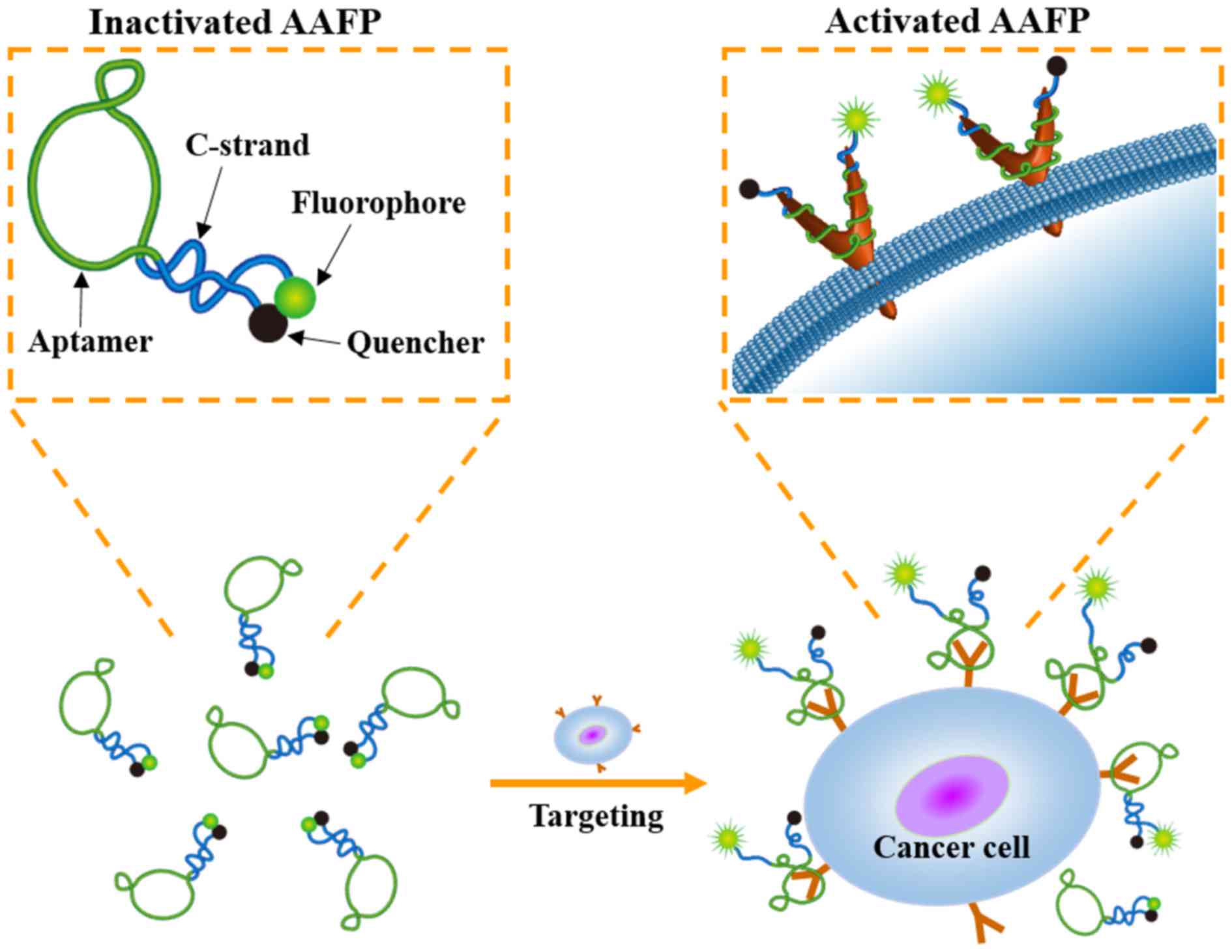
In the present study, we described the development
of an activatable aptamer-based fluorescence probe (AAFP) to detect
cancer cells and frozen cancer tissue (Fig. 1). This probe was ‘activatable’ since
it was designed to emit minimal signals in the absence of a target,
which may lead to decreased background interference than ‘always
on’ fluorescence probes (16). Our
AAFP was a short, single-stranded DNA oligonucleotide: we used the
DNA aptamer TLS11a, which binds with high affinity to cancer cells
(17), and added one short DNA
sequence (C-strand) to each end. The two C-strands were
complementary to each other, and the 5′ C-strand was conjugated to
the fluorophore FAM, while the 3′ C-strand was conjugated to the
quencher Eclipse. In the absence of a target, the two C-strands
hybridized into a hairpin structure, bringing the fluorophore and
quencher together and minimizing background fluorescence. Upon
target binding, the two C-strands separated, strongly increasing
FAM fluorescence signals.
Materials and methods
Materials
All DNA oligonucleotides were synthesized by
Shanghai Sangon Biological Engineering Technology & Services
(Shanghai, China). These oligonucleotides included the FAM-labeled
TLS11a aptamer (FAM-TLS11a), 5′-FAM-ACAGCATCCCCATGTGAACAATCGCATTGTG
ATTGTTACGGTTTCCGCCTCATGGACGTGCTG-3′; the activatable aptamer
fluorescence probe with 4 extending C-strand bases (AAFP),
5′-FAM-GGGGACAGCATCCCCAT GTGAACAATCGCATTGTGATTGTTACGGTTTCCGC
CTCATGGACGTGCTGCCCC-3′; the AAFP-3 and AAFP-5 mean probes
containing 3 and 5 extending C-strand bases respectively; a probe
with a 5-base mismatch (AAFP-mis),
5′-FAM-GGGGACAGCATCCCCATGTGAATCGAGGCAT
TGTGATTGTTACGGTTTCCGCCTCATGGACGTGCTG CCCC-3′. In all the sequences,
the extending C-strand bases are underlined and the mismatched
bases are bolded and italicized.
Cells and animals
Cell lines were obtained from the National Center
for International Research of Biological Targeting Diagnosis and
Therapy of Guangxi Medical University. Human hepatocellular
carcinoma HepG2, and human normal liver L02 cells were cultured at
37̊C in Dulbeccos modified Eagles medium (DMEM) supplemented with
10% fetal bovine serum (FBS; HyClone, Logan, UT, USA), and 100 U/ml
penicillin-streptomycin in a 5% CO2 atmosphere.
Six-week-old female BALB/c nude mice from the
Guangxi Laboratory Animal Center (Guangxi, China) were raised in
sterile conditions in a laminar flow hood. All experiments were
carried out according to the guidelines of the Federation of
European Laboratory Animal Science Associations, and all protocols
were approved by the Institutional Animal Care and Use Committee of
Guangxi Medical University.
Flow cytometry
Cells were cultured at a density of
5.0×105 cells/ml, collected by centrifugation and
resuspended in 0.5 ml of phosphate-buffered saline (PBS). In order
to investigate the ability of AAFP to bind to cancer cells, cells
were incubated with FAM-TLS11a, AAFP or AAFP-mis (250 nM) at 4̊C in
the dark for 30 min in 0.5 ml of binding buffer [PBS supplemented
with 5 mM MgCl2, 4.5 g/l glucose and 1 mg/ml bovine
serum albumin (BSA)]. Then cells were washed with PBS, suspended in
0.5 ml of binding buffer and analyzed. Fluorescent cells were
detected using flow cytometry (Beckman Coulter Epics XL; Beckman
Coulter, Inc., Brea, CA, USA), and data were analyzed using FlowJo
Software 7.6.2 (FlowJo LLC, Ashland, OR, USA).
Fluorescence spectroscopy
Cells were collected by centrifugation and suspended
in 0.5 ml of binding buffer. To assess the increase in AAFP
fluorescence upon target binding, cells were incubated with
FAM-TLS11a, AAFP or AAFP-mis (250 nM) at 4̊C in the dark for 30
min. Different lengths of the C-strand on AAFP were tested, as were
different AAFP concentrations, incubation times and incubation
temperatures. After incubation, binding reactions were analyzed by
fluorescence spectroscopy (Hitachi, F-7000; Hitachi, Tokyo, Japan)
at the wavelength range of 650–500 nm with an excitation wavelength
at 490 nm. Fluorescence spectra were also obtained from cell
suspensions or AAFP on their own in binding buffer as negative
controls.
To investigate the sensitivity of AAFP binding to
HepG2 cells, serial dilutions of cells (0–1.0×106) in
0.5 ml of binding buffer were incubated with AAFP (250 nM) at 4̊C
in the dark for 30 min. Binding reactions were analyzed by
fluorescence spectroscopy as described above.
Immunofluorescence imaging of
cells
Cells were seeded into 6-well plates and cultured
for 24 h. Cells were fixed with 4% paraformaldehyde (Sigma-Aldrich,
St. Louis, MO, USA) for 20 min, washed with PBS, then incubated
with FAM-TLS11a, AAFP or AAFP-mis in binding buffer at 4̊C in the
dark for 30 min. Cells were then washed again with PBS and stained
with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Life
Technologies, Foster City, CA, USA) for 5 min in the dark. Finally,
cells were washed and examined by fluorescence microscopy (Nikon
DS-Ri1; Nikon, Tokyo, Japan).
Cancer model and immunofluorescence
imaging of frozen cancer sections
BALB/c nude mice received a subcutaneous injection
of 5×106 HepG2 cells on their backside. Tumors were
allowed to grow for 15–20 days until reaching a diameter of 0.5–1.5
cm. Mice were sacrificed, the cancer tissue was excised, and frozen
sections 6-8-mm of thickness were prepared immediately. As a
control, normal liver tissue was obtained from the mice that did
not receive HepG2 injections.
Frozen sections were fixed with 4% paraformaldehyde
(Sigma-Aldrich) for 10 min, washed with PBS, stained with DAPI for
5 min, washed again, and were then incubated with FAM-TLS11a, AAFP
or AAFP-mis (250 nM) at 4̊C in the dark for 30 min. Finally, the
sections were washed again with PBS and observed using fluorescence
microscopy.
Statistical analyses
Each experiment was carried out in triplicate. Data
are expressed as the mean ± SD or as the median (range). All
statistical analyses were performed using GraphPad Prism 6.02
(GraphPad Software, San Diego, CA, USA). The threshold of
significance in all analyses was P<0.05.
Results
Activation of AAFP fluorescence by
target cancer cells
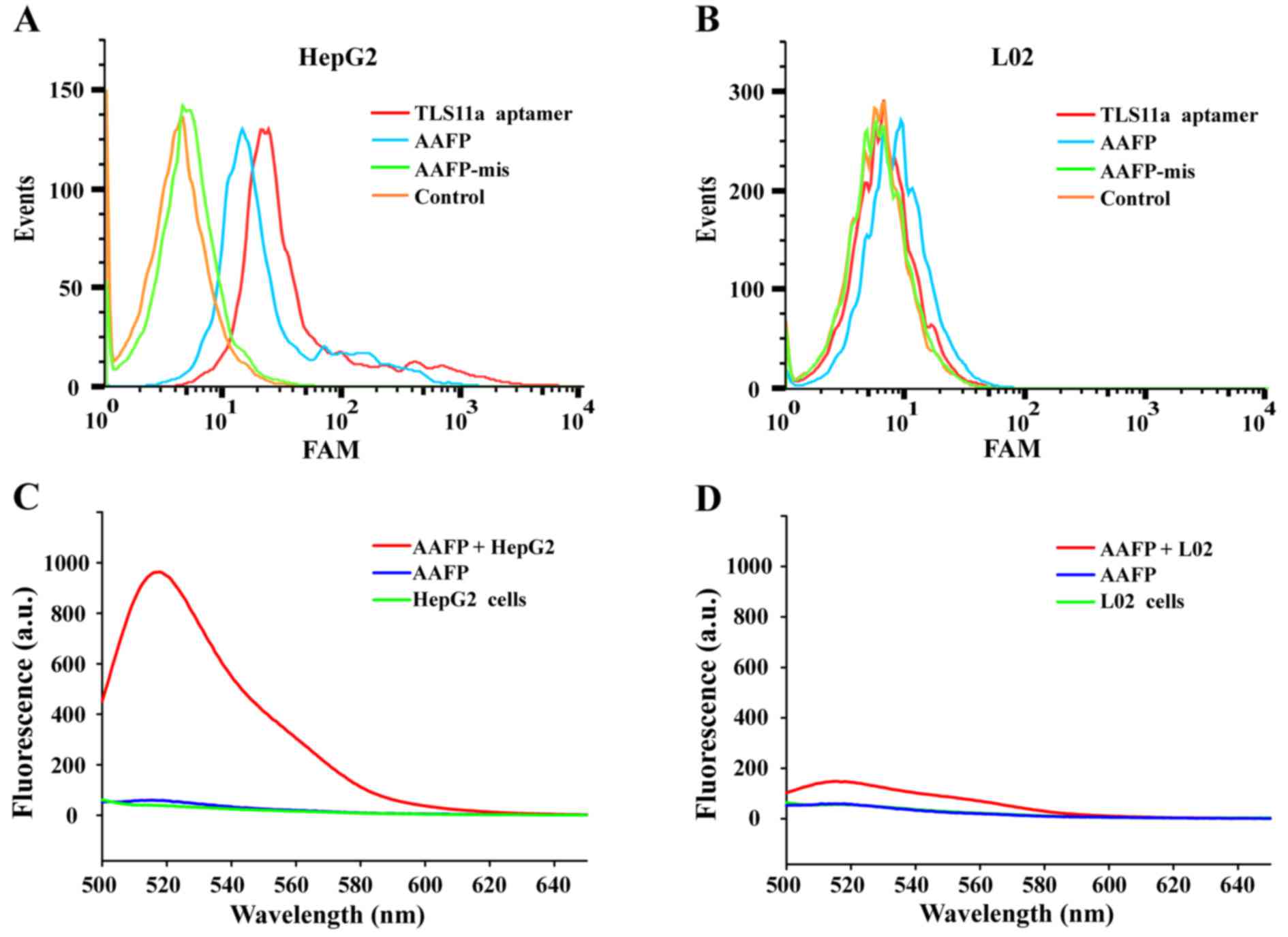
Flow cytometry was used to investigate the ability
of target cancer cells to activate AAFP fluorescence. Both
FAM-TLS11a and AAFP bound to HepG2 cells, but not to L02 cells,
while AAFP-mis did not bind to either cell line (Fig. 2A and B). FAM-TLS11a and AAFP showed
similar rates of binding to HepG2 cells, which were significantly
higher than the rate of AAFP-mis binding.
Fluorescence spectroscopy was used to analyze the
increase in FAM signals due to AAFP binding to HepG2 cells
(Fig. 2C and D). Negligible
fluorescence was detected in the cell suspensions without the probe
or in a solution of AAFP (250 nM) without cells. Incubating HepG2
cells with AAFP led to a strong fluorescence signal at 518 nm,
which was much weaker when L02 cells were incubated with AAFP.
These results indicate that AAFP fluorescence increased
specifically in response to HepG2 cancer cells.
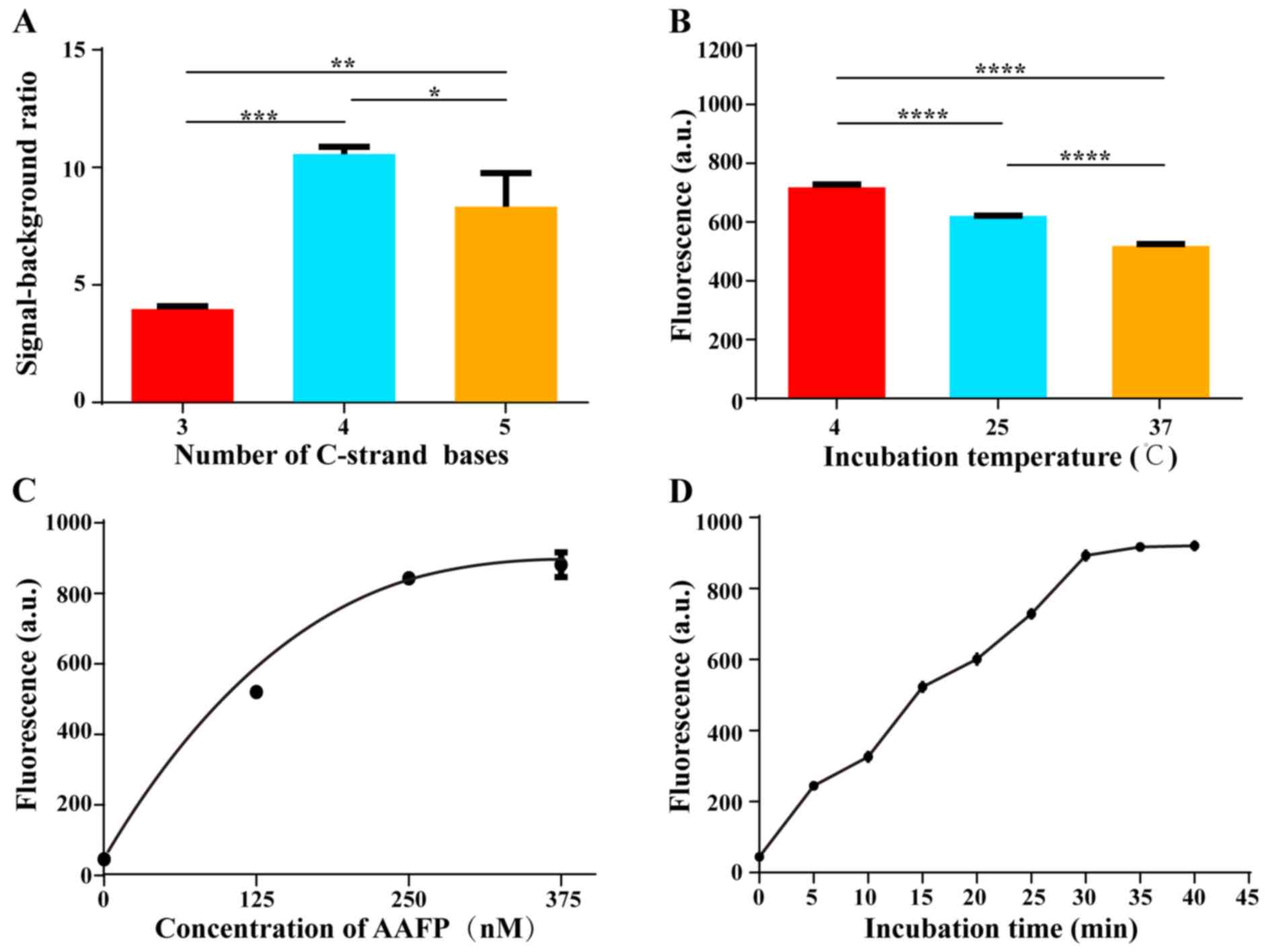
Optimization of C-strand length on
AAFP
We tested various lengths of the C-strand on AAFP,
from 3 to 5 bases, in an effort to minimize background fluorescence
while maintaining strong affinity for HepG2 cells. The highest
signal-to-background ratio was obtained when the two C-strands
contained 4 bases, corresponding to 4 base pairs in the hairpin
structure that forms in the absence of a target (Fig. 3A). This ratio gradually decreased as
the number of bases was increased above 4. We speculated that
C-strands with fewer than 4 bases are incapable of keeping the
fluorophore and quencher close together, whereas C-strands longer
than 4 bases decrease the affinity of the aptamer for target
cells.
Optimization of the detection
conditions
We investigated the optimal conditions for AAFP
binding to HepG2 cells. Cells were incubated with AAFP at 250 nM in
the dark for 30 min at 4, 25 or 37̊C, and then activation of AAFP
fluorescence was assessed using fluorescence spectroscopy (Fig. 3B). As the temperature increased,
fluorescence activation decreased, although it remained well above
the background at all temperatures tested. These results indicate
that although higher temperatures, including physiological
temperatures, slightly decrease the binding of AAFP to target
tissues in vitro, the fluorescence signal remains strong
relative to the background.
Next, we incubated HepG2 cells with varying AAFP
concentrations up to 375 nM at 4̊C for 30 min, and examined the
fluorescence signal (Fig. 3C). The
signal increased progressively up to an AAFP concentration of 250
nM, after which it almost plateaued, suggesting saturation.
Finally, we incubated HepG2 cells with AAFP at 250
nM for different lengths of time at 4̊C (Fig. 3D). The fluorescence signal increased
gradually from 0 to 30 min, after which it plateaued, suggesting
complete binding.
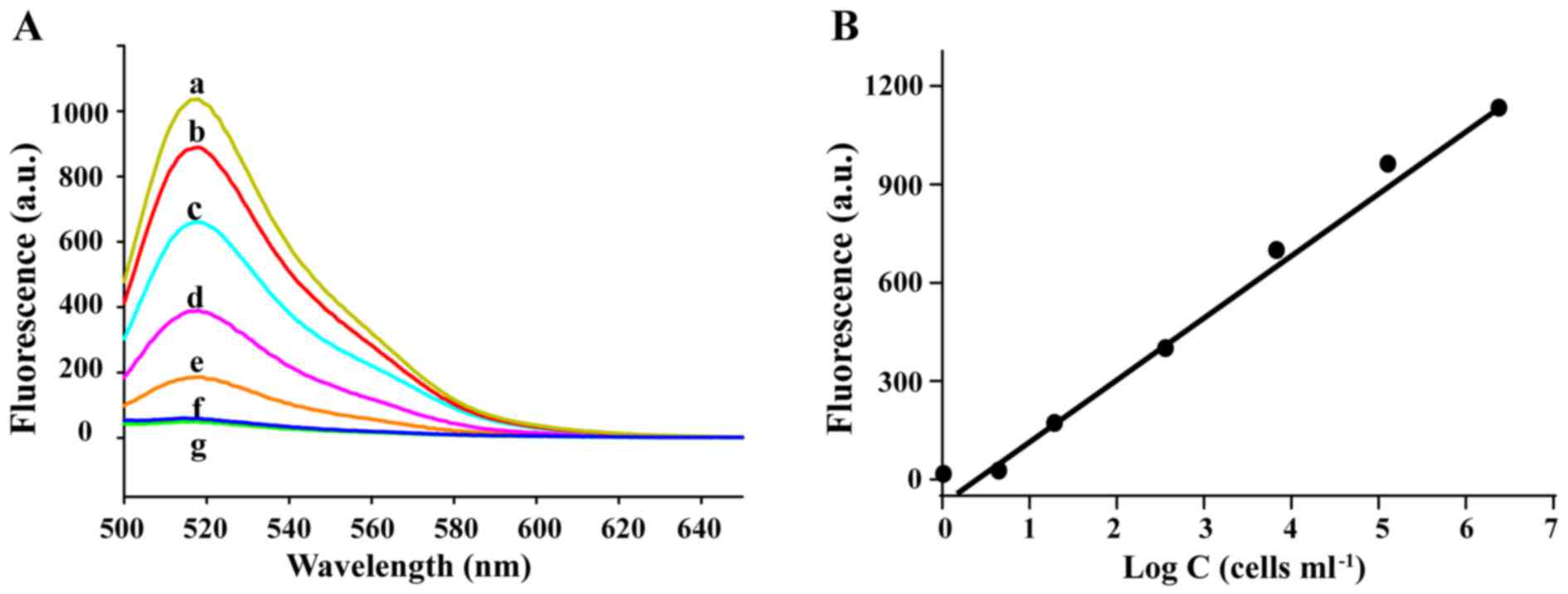
Cancer cell detection by AAFP
To assess the sensitivity of AAFP in the detection
of HepG2 cells, we incubated the probe (250 nM) with HepG2 cells at
concentrations of up to 106 cells/ml in 0.5 ml of
binding buffer, and then assessed the fluorescence signal
spectroscopically. The signal increased over the entire range of
cell concentrations assessed (Fig.
4). Regression analysis of the plot of the signal intensity and
logarithm of the cell concentration (fluorescence = 214.4×log C -
6.981) revealed that under these experimental conditions, the probe
was capable of detecting cancer cells at concentrations as low as
~100 cells/ml.
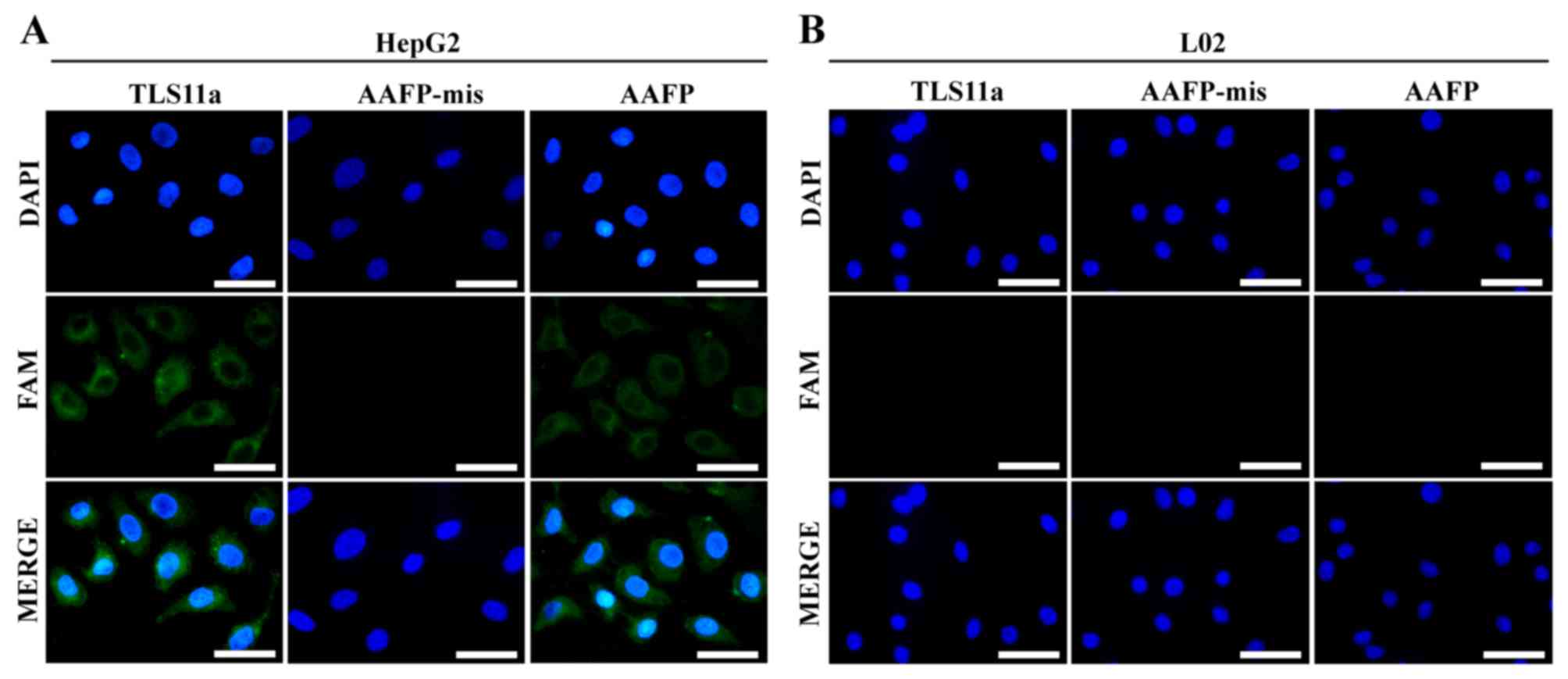
Using AAFP for fluorescence imaging of
cancer cells
To visualize directly the specificity of AAFP
binding at the cellular level, and thereby complement the flow
cytometry experiments as described in Materials and methods, we
incubated HepG2 and L02 cells with FAM-TLS11a, AAFP or AAFP-mis and
analyzed them using fluorescence microscopy. Consistent with the
flow cytometry experiments, AAFP and FAM-TLS11a were observed to
bind to HepG2 cells, but not to L02 cells, while AAFP-mis did not
bind to either cell type (Fig.
5).
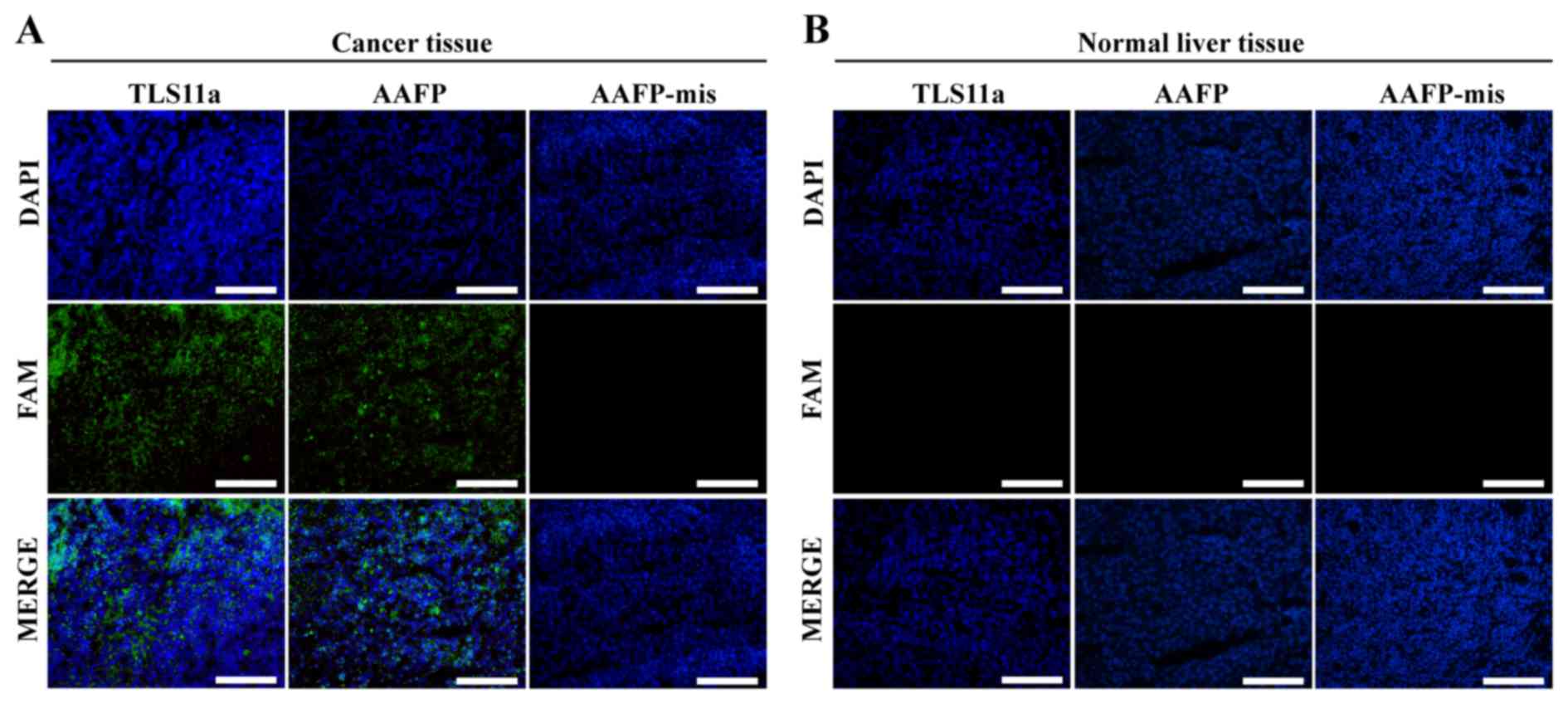
Using AAFP for fluorescence imaging of
frozen cancer tissue
We examined whether the specific AAFP binding
observed with HepG2 cells in suspension may also apply to frozen
sections of solid HepG2 tumors from a xenograft mouse model. As a
control, we also examined AAFP binding to frozen sections of normal
liver tissue from the same type of mice. Both AAFP and FAM-TLS11a
were observed to bind to sections of cancerous tissue, but not to
sections of normal tissue (Fig. 6).
AAFP-mis did not bind to either type of tissue.
Discussion
We designed an ‘activatable’ aptamer-based
fluorescence probe (AAFP) that sensitively and specifically detects
cancer cells. The probe emitted negligible fluorescence in the
absence of target cancer cells since the fluorophore and quencher,
conjugated to C-strands on either side of a cancer-specific TLS11a
aptamer, lie close together. Then, in the presence of target cancer
cells, the fluorophore and quencher separated, giving rise to a
strong FAM signal. This AAFP may be a valuable tool for the early
detection of cancer, which can facilitate timely initiation of
treatment. Using an AAFP avoids the need to conjugate a fluorophore
to biological tissue, in contrast to other types of fluorescent
probes, such as quantum dots (18,19),
carbon nanodots (20) and
fluorescent dyes (21,22). Under optimal incubation conditions,
our AAFP was able to detect cancer cells at concentrations as low
as ~100 cells/ml.
The TLS11a aptamer, although originally selected for
its binding to mouse hepatoma BNL 1ME A.7R.1 (MEAR) cells, also
binds with high affinity to human liver cancer cells. TLS11a is
likely to be a membrane protein that can be internalized into cells
(23), which may explain this dual
binding specificity. We found that TLS11a on its own or as part of
our AAFP bound efficiently to HepG2 cells, consistent with a
previous study (24). In contrast,
neither TLS11a nor AAFP bound to normal human L02 liver cells,
confirming the cancer specificity of AAFP binding. Further
confirmation came when we found that the mismatch-containing
AAFP-mis failed to bind to either HepG2 or L02 cells. These results
using flow cytometry and fluorescence spectroscopy to examine cells
in suspension were confirmed using fluorescence microscopy to
examine cells in culture.
We further confirmed the specificity of AAFP binding
using frozen sections of HepG2 tumor xenografts and normal liver
tissue. Frozen sections are often used for histopathological
examination, in part since many proteins retain their activity as
in fresh tissue sections, and since they are easier and faster to
prepare than paraffin-embedded sections (25).
‘Activatable’ probes can be superior to ‘always on’
probes since they emit much less fluorescence in the absence of a
target, thereby increasing the signal-to-background ratio (26,27).
Our experiments with fluorescence spectroscopy revealed that AAFP
emitted minimal fluorescence in the absence of HepG2 cells, likely
reflecting fluorescence resonance energy transfer between the
fluorophore and quencher lying close together in the hairpin
structure formed upon hybridization of the two C-strands. Then, in
the presence of HepG2 cells, AAFP fluorescence strongly increased,
indicating separation of the fluorophore and quencher due to
aptamer binding to targets within the cancer tissue. The high
signal-to-background ratio observed for AAFP, even at physiological
temperatures, suggests its potential for highly sensitive cancer
cell detection.
Optimization studies showed that AAFP binding
efficiency depended on the length of the C-strands, the incubation
temperature and incubation time. This suggests that studies of AAFP
or similar probes may need to screen these parameters carefully to
maximize sensitivity and specificity, and in particular to avoid
false-negative results. Our observation of maximal binding
efficiency at an AAFP concentration of 250 nM is consistent with a
previous study (28). The fact that
our AAFP bound noticeably less well at 37̊C than at 4̊C reflects
the fact that the TLS11a aptamer was selected at low temperatures
(17). This suggests that
optimization of the TLS11a aptamer sequence may improve target
binding at physiological temperatures. It also highlights the need
to design AAFP-like probes using affinity sequences selected near
in vivo temperatures.
The present study further demonstrated the power of
aptamers in the detection and imaging of tumor tissues (29). The small size of aptamers may give
them an advantage over antibodies for imaging intracellular targets
(30), however, they may be rapidly
degraded by nucleases or cleared from tissues. Chemically modifying
aptamers can render them nuclease-resistant and increase their
stability (31,32). Our results open the door to further
investigations of AAFP with different chemical modifications.
In summary, we developed a simple, highly sensitive,
specific AAFP for the detection of cancer cells and frozen cancer
tissue. Our AAFP not only holds great potential in clinical early
diagnosis, which can facilitate timely initiation of treatment, but
also suggests the possibility of replacing the TLS11a aptamer with
other sequences to target other tissue types in cancer or even
other diseases.
Acknowledgements
The present study was supported, in part, by grants
from Programs for Changjiang Scholars and Innovative Research Team
in University (no. IRT_15R13), the National Natural Scientific
Foundation of China (nos. 81430055 and 81372452), the International
Cooperation Project of the Ministry of Science and Technology of
China (no. 2015DFA31320), the Project for Innovative Research Team
in Guangxi Natural Science Foundation (2015GXNSFFA139001), and the
Project of Science and Technology of Guangxi (nos. 14125008-2-12
and 1599005-2-10).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li C, Meng Y, Wang S, Qian M, Wang J, Lu W
and Huang R: Mesoporous carbon nanospheres featured fluorescent
aptasensor for multiple diagnosis of cancer in vitro and in vivo.
ACS Nano. 9:12096–12103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kobayashi H, Longmire MR, Ogawa M, Choyke
PL and Kawamoto S: Multiplexed imaging in cancer diagnosis:
Applications and future advances. Lancet Oncol. 11:589–595. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen TT, Tian X, Liu CL, Ge J, Chu X and
Li Y: Fluorescence activation imaging of cytochrome c
released from mitochondria using aptameric nanosensor. J Am Chem
Soc. 137:982–989. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang M, Chakraborty SK, Sampath P, Rojas
JJ, Hou W, Saurabh S, Thorne SH, Bruchez MP and Waggoner AS:
Fluoromodule-based reporter/probes designed for in vivo
fluorescence imaging. J Clin Invest. 125:3915–3927. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hou Y, Zhou J, Gao Z, Sun X, Liu C,
Shangguan D, Yang W and Gao M: Protease-activated ratiometric
fluorescent probe for pH mapping of malignant tumors. ACS Nano.
9:3199–3205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sonn GA, Behesnilian AS, Jiang ZK,
Zettlitz KA, Lepin EJ, Bentolila LA, Knowles SM, Lawrence D, Wu AM
and Reiter RE: Fluorescent image-guided surgery with an
anti-prostate stem cell antigen (PSCA) diabody enables targeted
resection of mouse prostate cancer xenografts in real time. Clin
Cancer Res. 22:1403–1412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hussain T and Nguyen QT: Molecular imaging
for cancer diagnosis and surgery. Adv Drug Deliv Rev. 66:90–100.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jung YK, Woo MA, Soh HT and Park HG:
Aptamer-based cell imaging reagents capable of fluorescence
switching. Chem Commun. 50:12329–12332. 2014. View Article : Google Scholar
|
|
10
|
Lao YH, Phua KK and Leong KW: Aptamer
nanomedicine for cancer therapeutics: Barriers and potential for
translation. ACS Nano. 9:2235–2254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ellington AD and Szostak JW: In vitro
selection of RNA molecules that bind specific ligands. Nature.
346:818–822. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu Z, Lai Z, He J, Huang X, Hou X, Zhao Y
and Lu X: Research progress on cell-SELEX. Cell Commun. 2:57–66.
2015.
|
|
13
|
Tan W, Donovan MJ and Jiang J: Aptamers
from cell-based selection for bioanalytical applications. Chem Rev.
113:2842–2862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang AZ and Farokhzad OC: Current progress
of aptamer-based molecular imaging. J Nucl Med. 55:353–356. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi H, He X, Wang K, Wu X, Ye X, Guo Q,
Tan W, Qing Z, Yang X and Zhou B: Activatable aptamer probe for
contrast-enhanced in vivo cancer imaging based on cell membrane
protein-triggered conformation alteration. Proc Natl Acad Sci USA.
108:3900–3905. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meng HM, Liu H, Kuai H, Peng R, Mo L and
Zhang XB: Aptamer-integrated DNA nanostructures for biosensing,
bioimaging and cancer therapy. Chem Soc Rev. 45:2583–2602. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shangguan D, Meng L, Cao ZC, Xiao Z, Fang
X, Li Y, Cardona D, Witek RP, Liu C and Tan W: Identification of
liver cancer-specific aptamers using whole live cells. Anal Chem.
80:721–728. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Ji X, Zhang Y, Zhou G, Ke X, Wang
H, Tinnefeld P and He Z: One-pot synthesized aptamer-functionalized
CdTe:Zn2+ quantum dots for tumor-targeted fluorescence
imaging in vitro and in vivo. Anal Chem. 85:5843–5849. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu Y, Duan S, He J, Liang W, Su J, Zhu J,
Hu N, Zhao Y and Lu X: Highly sensitive detection of leukemia cells
based on aptamer and quantum dots. Oncol Rep. 36:886–892.
2016.PubMed/NCBI
|
|
20
|
Lee CH, Rajendran R, Jeong MS, Ko HY, Joo
JY, Cho S, Chang YW and Kim S: Bioimaging of targeting cancers
using aptamer-conjugated carbon nanodots. Chem Commun.
49:6543–6545. 2013. View Article : Google Scholar
|
|
21
|
Shi H, Tang Z, Kim Y, Nie H, Huang YF, He
X, Deng K, Wang K and Tan W: In vivo fluorescence imaging of tumors
using molecular aptamers generated by cell-SELEX. Chem Asian J.
5:2209–2213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan J, Yang N, Hu Z, Su J, Zhong J, Yang
Y, Yu Y, Zhu J, Xue D, Huang Y, et al: Aptamer-functionalized
fluorescent silica nanoparticles for highly sensitive detection of
leukemia cells. Nanoscale Res Lett. 11:2982016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meng L, Yang L, Zhao X, Zhang L, Zhu H,
Liu C and Tan W: Targeted delivery of chemotherapy agents using a
liver cancer-specific aptamer. PLoS One. 7:e334342012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kashefi-Kheyrabadi L, Mehrgardi MA,
Wiechec E, Turner AP and Tiwari A: Ultrasensitive detection of
human liver hepatocellular carcinoma cells using a label-free
aptasensor. Anal Chem. 86:4956–4960. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pu Y, Liu Z, Lu Y, Yuan P, Liu J, Yu B,
Wang G, Yang CJ, Liu H and Tan W: Using DNA aptamer probe for
immunostaining of cancer frozen tissues. Anal Chem. 87:1919–1924.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang Y, Huang J, Yang X, Quan K, Xie N, Ou
M, Tang J and Wang K: Aptamer-based FRET nanoflares for imaging
potassium ions in living cells. Chem Commun. 52:11386–11389. 2016.
View Article : Google Scholar
|
|
27
|
Zhang L, Cui P, Zhang B and Gao F:
Aptamer-based turn-on detection of thrombin in biological fluids
based on efficient phosphorescence energy transfer from Mn-doped
ZnS quantum dots to carbon nanodots. Chemistry. 19:9242–9250. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X,
Liu Q, Champanhac C, Teng IT, Ye M, et al: DNA aptamer selected
against pancreatic ductal adenocarcinoma for in vivo imaging and
clinical tissue recognition. Theranostics. 5:985–994. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu X, Chen J, Wu M and Zhao JX: Aptamers:
Active targeting ligands for cancer diagnosis and therapy.
Theranostics. 5:322–344. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park JY, Lee TS, Song IH, Cho YL, Chae JR,
Yun M, Kang H, Lee JH, Lim JH, Cho WG, et al: Hybridization-based
aptamer labeling using complementary oligonucleotide platform for
PET and optical imaging. Biomaterials. 100:143–151. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shangguan D, Tang Z, Mallikaratchy P, Xiao
Z and Tan W: Optimization and modifications of aptamers selected
from live cancer cell lines. ChemBioChem. 8:603–606. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shigdar S, Macdonald J, O'Connor M, Wang
T, Xiang D, Al Shamaileh H, Qiao L, Wei M, Zhou SF, Zhu Y, et al:
Aptamers as theranostic agents: Modifications, serum stability and
functionalisation. Sensors. 13:13624–13637. 2013. View Article : Google Scholar : PubMed/NCBI
|