Introduction
Keratin is an intermediate filament expressed in
epidermal cells of specific regions. The large keratin multigene
family is comprised of cytokeratins, which are differentially
expressed in various types of epithelia. Cytokeratins have been
extensively studied as breast cancer markers (1,2), and
can be divided into the acidic type I and the basic-to-neutral type
II cytokeratins. The intermediate filament network is formed
through the obligatory association of equimolar amounts of type I
and type II keratins (3). Hair
keratins are expressed in hard-keratinized structures such as the
hair and nails, and are believed to be structural proteins of the
hair or nails that are not expressed in other organs such as the
mammary gland. KRT81 is a type II hair keratin and one of the main
hair proteins that is expressed in the hair cortex (4). However, it was reported that KRT81 is
expressed in the human breast cancer cell line SKBR3 (5,6) and in
metastatic lymph nodes of breast carcinomas (7), but not in normal breast epithelial
cells. Furthermore, the expressed KRT81 was a 5′-truncated isoform
(ΔHb1), and the full size protein was not expressed (5,6).
However, its function remains unclear.
The matrix metalloproteinase (MMP) family includes
more than 20 isoforms that modulate the extracellular milieu by
degrading extracellular matrix proteins. MMPs are secreted by
fibroblasts and tumor cells and are involved in tumor invasion and
metastasis (8,9).
We previously reported that the hairless phenotype
of the Hirosaki hairless rat (HHR) is due to the deletion of basic
hair keratin genes, including KRT81 (10). HHR shows the involution by apoptosis
of the mammary gland at an early stage of lactation (11), and is resistant to mammary tumors
(12). A correlation between the
incidence of breast cancer and an observed change in the X-ray
diffraction pattern of hair from patients with breast cancer has
been reported (13). Furthermore,
X-ray diffraction of hair has the potential to provide a
non-invasive test for the presence of breast cancer (14,5).
These previous studies suggest that breast cancer
may express hair keratin, and hair keratins may have some function
in breast cells. The aim of the present study was to investigate
the expression and the function of hair keratin KRT81 in normal
breast and breast cancer cells. We investigated the expression of
KRT81 using RT-PCR, western blotting and immunohistochemical
analysis in human breast cancer cell lines, MCF7, SKBR3 and
MDA-MB231, normal human mammary epithelial cells (HMECs) and
non-neoplastic cells (MCF10A). To investigate the function of KRT81
in breast epithelial cells, we transfected MCF10A cells with
siKRT81 and analyzed gene alterations using microarrays,
Ingenuity® Pathway Analysis (IPA), and quantitative PCR
(qPCR) to assess changes in gene expression. To investigate the
effect of KRT81 on cell invasion, we performed zymography, scratch
and invasion assays using the breast cancer cell line MDA-MB-231,
which exhibits invasive properties (16). This is the first study on the
expression of hair keratin KRT81 and its function in normal breast
epithelial and cancer cells.
Materials and methods
Cell culture
HMECs were purchased from PromoCell GmbH
(Heidelberg, Germany). The MCF10A human breast epithelial cell
line, and MCF7, SKBR3 and MDA-MB-231 human breast cancer cell lines
were obtained from the American Type Culture Collection (ATCC;
Manassas, VA, USA). HMECs and MCF10A cells were routinely cultured
using a Mammary epithelial cell growth medium kit at 37̊C in 5%
CO2. MCF7, SKBR3 and MDA-MB-231 cells were routinely
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS) at 37̊C in 5% CO2.
siRNA transfection
The expression of rat KRT81 was blocked by transient
transfection with KRT81 siRNA (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA) using DharmaFECT Transfection Reagent (Thermo
Scientific, Waltham, MA, USA). Approximately 2.5×105
MCF10A or MDA-MB-231 cells were incubated with siRNA for 24–48 h
before being used for subsequent assays. For the negative control
experiments, MCF10A or MDA-MB-231 cells were transfected with
Silencer® Negative Control #1 siRNA (Applied Biosystems,
Foster City, CA, USA).
Western blot analysis
Western blotting was performed, according to the
method described by Towbin et al (17). Proteins from HMECs, MCF10A, MCF7,
SKBR3 and MDA-MB-231 cells were separated by SDS-polyacrylamide gel
electrophoresis (PAGE) (18) on
7.5% (w/v) polyacrylamide gels and electroblotted to Hybond
nitrocellulose membranes (GE Healthcare, Piscataway, NJ, USA).
Blots were probed with anti-KRT81 antibody-C-terminal (ab192689;
1:1,000; Abcam, Cambridge, MA, USA) or anti-β-actin antibody
(dilution 1:1,000; Cell Signaling Technology, Danvers, MA, USA)
followed by horseradish peroxidase-conjugated anti-guinea pig IgG
(dilution 1:2,000; Abcam) or anti-rabbit IgG (dilution 1:2,000;
Cell Signaling Technology). Signals were generated with an ECL kit
(GE Healthcare) according to the manufacturer's protocol.
Reverse transcription-polymerase chain
reaction (RT-PCR) and qPCR
Total RNA was extracted from the HMECs, MCF10A,
MCF7, SKBR3 and MDA-MB-231 cells using the RNeasy Mini kit (Qiagen,
Tokyo, Japan). cDNA was reverse-transcribed from total RNA (200 ng)
using the PrimeScript™ RT Master Mix (Takara, Tokyo, Japan). PCR
was performed with Takara LA Taq® with GC Buffer
(Takara) using specific primer pairs. PCR amplification consisted
of 30 sec at 94̊C, 30 sec at 55–60̊C and 30 sec-2 min at 72̊C for
40 cycles. Gene-specific primers were designed according to known
human sequences using the Primer3Plus software. The primers used
were as follows (5′→3′): KRT81 F-CCTGCGG ATCAGGATTTGGT
(corresponding to exon 1) and R-AAGT GGGGGATCACACAGAG
(corresponding to exon 9); GAPDH, F-AGAAGGCTGGGGCTCATTTG and
R-AGGG GCCATCCACAGTCTTC. The RT-PCR products were subjected to
electrophoresis on a 2% agarose gel and visualized with ethidium
bromide. Levels of specific mRNAs were quantified by qPCR using
SYBR-Green SuperMix (Bio-Rad Laboratories, Hercules, CA, USA).
Transcript levels were normalized to that of GAPDH cDNA. The
primers used were as follows (5′→3′): KRT81,
F-AGGCTATGTGAAGGCATTGG (corresponding to exon 8) and
R-AAGTGGGGGATCACAC AGAG (corresponding to exon 9); GAPDH,
F-AGAAGGCT GGGGCTCATTTG and R-AGGGGCCATCCACAGTCTTC; MMP9,
F-CACCTTCACTCGCGTGTAC and R-CATCTGC GTTTCCAAACCGAG. PCR specificity
was ascertained by melting curve analysis. Relative gene expression
was calculated according to the 2−ΔΔCt method.
Immunofluorescence staining
Cells were seeded onto a 4-well Slide & Chamber
(Watson, Japan) and incubated at 37̊C for 24 h, fixed immediately
in 4% paraformaldehyde, and permeabilized in 0.1% Triton X-100 in
phosphate-buffered saline (PBS) for 5 min. The slides were then
incubated with the anti-KRT81 antibody-C-terminal (ab192689;
dilution 1:100) at 4̊C overnight, followed by incubation with the
secondary antibody, Alexa Fluor 647 anti-guinea pig IgG (ab150187;
dilution 1:500) (both from Abcam) for 30 min at room temperature.
Nuclear staining and mounting were performed with Vectashield
Mounting Medium with 4,6-diamidino-2-phenylindole (DAPI) (Vector
Laboratories, Burlingame, CA, USA). Images were captured using a
fluorescence microscope FSX100 (Olympus, Tokyo, Japan).
Histological analysis and
immunohistochemistry
Breast cancer specimens, including normal areas,
were obtained from patients at the time of surgery. The archival
specimens were obtained from the Department of Pathology and
Bioscience at Hirosaki University. The sections (4-µm) were mounted
onto silane-coated slides. Immunohistochemistry was automatically
performed using the Ventana XT System Discovery® (Roche,
Basel, Switzerland). Briefly, tissue sections were treated with
protease I (Roche) at 37̊C for 16 min for antigen retrieval. The
following primary antibody was used: KRT81 polyclonal antibody
(11342–1-AP; dilution 1:25; ProteinTech, Manchester, UK), and
sections were incubated at 37̊C for 32 min. The tissue sections
were then incubated at 37̊C for 20 min with a universal secondary
antibody (Roche). The site of peroxidase binding was determined
using DISCOVERY DABMap Detection kit (Roche). Sections were then
counterstained with Hematoxylin II (Roche) for microscopic
examination. As a negative control, non-immune γ-globulin was used
instead of the antibody. The specimens were observed and
photographed using a fluorescence microscope FSX100 (Olympus). The
present study was approved by the Committee for Medical Ethics of
Hirosaki University (Hirosaki, Japan). Informed consent was
obtained from all patients prior to the beginning of the study.
Gene expression analysis by
microarray
Total RNA was extracted from the siRNA-transfected
or control MCF10A cells using the RNeasy Mini kit. One microgram of
RNA was used to produce biotin-labeled complementary RNA (cRNA).
The labeled and fragmented cRNA was subsequently hybridized to the
SurePrint G3 Human Gene Expression microarray (8×60 K version 2;
Agilent Technologies Inc., Santa Clara, CA, USA). Labeling,
hybridization, image scanning and data analysis were performed at
Bio Matrix Research Inc. (Chiba, Japan). The MCF10A microarray
dataset is available at http://www.ncbi.nlm.nih.gov/geo under accession code
GSE85236. Genes with 2-fold or greater upregulation following siRNA
transfection were analyzed using the Qiagen Ingenuity®
Pathway Analysis (IPA®) software (version 18030641). The
z-score algorithm was utilized to decrease the possibility of
false-positive results, where z ≥2 indicated that transcript
expression was significantly increased and z ≤-2 indicated that the
expression was significantly decreased.
Gelatin zymography
The MDA-MB-231 cells were grown to confluence in
Dulbeccos modified Eagles medium (DMEM) supplemented with 10% FBS.
After 24 h, the cells were transfected with siKRT81 or
Silencer® Negative Control #1 siRNA in serum-free
medium. After 48 h, the serum-free conditioned medium was harvested
by centrifugation at 1,500 rpm for 5 min. Gelatin zymography was
performed as previously described (19,20).
Scratch wound healing assay
The effects of KRT81 on cell migration were examined
using the scratch wound healing assay with KRT81-silenced and
control MDA-MB-231 cells. Briefly, 2×105 cells were
seeded onto 60-mm cell culture dishes. After cells reached ~70%
confluence as a monolayer, they were transfected with siKRT81.
After 24 h, the surface of the dishes was scratched linearly with a
200-µl pipette tip. After 24 h, images were captured using a
fluorescence microscope CKX41 (Olympus). The cells that migrated
were counted/field. Results were obtained from 3 independent
experiments with 9 fields.
Invasion assay
The invasiveness of MDA-MB-231 cells treated or not
with siKRT81 was assessed using Transwell chambers with 8-µm pore
size membrane (CytoSelect™; Cell Biolabs, Inc., San Diego, CA,
USA). Before the invasion assay, MDA-MB-231 cells were transfected
with the siKRT81 and cultured for 24 h in another culture dish. In
the upper compartment of the chamber, ~1.5×105 cells
(treated or not with siKRT81)/insert were added into the culture
medium without serum, and 500 µl of culture medium with 10% FBS
were added to the lower well of the invasion chamber. The cells
were incubated at 37̊C in 5% CO2 for 24 h. Cells were
then washed, fixed and stained with cell stain solution.
Non-migrating cells were removed from the upper surface of the
Transwell membrane with a cotton swab. Cells were counted with a
fluorescence microscope FSX100. Results were obtained from 3
independent experiments with 9 fields.
Statistical analysis
Experiments were conducted at least 3 times in
duplicate or triplicate. Results are presented as the mean ±
standard error of the mean (SEM). Statistical significance was
determined using Student's t-test. p<0.01 was considered
statistically significant.
Results
KRT81 expression in normal breast
epithelial and cancer cells
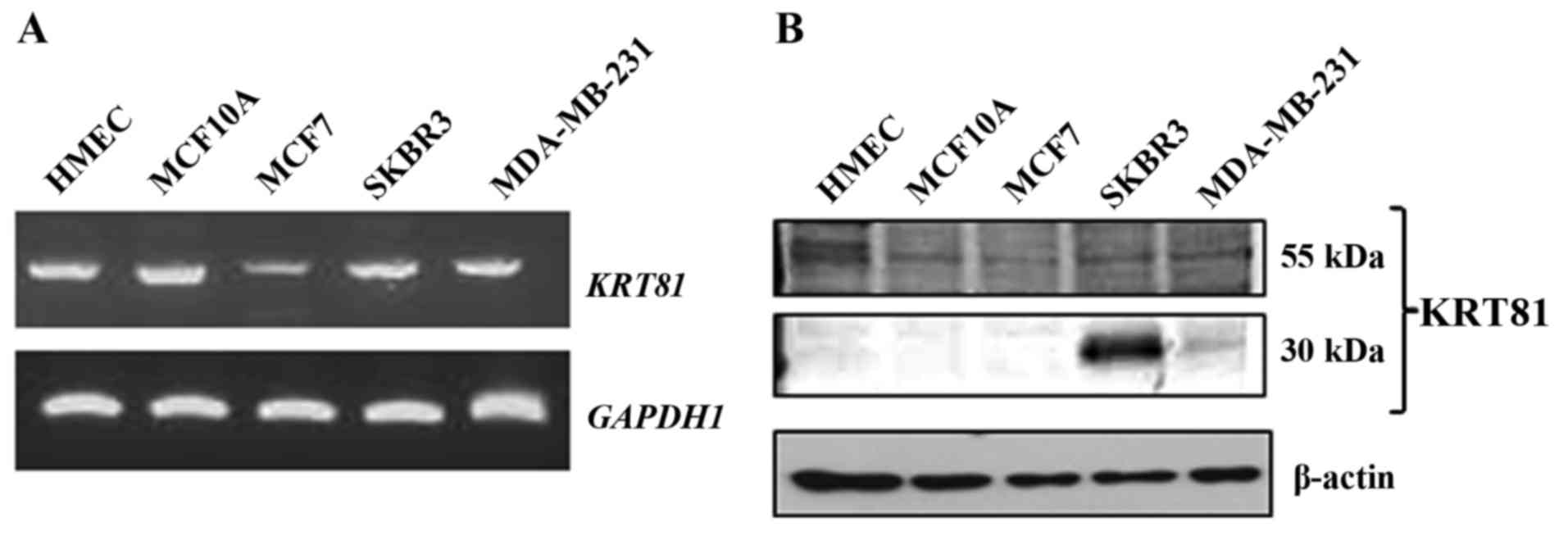
While northern blot analysis revealed that the human
truncated form of KRT81 is expressed in breast cancer SKBR3 cells,
but not in normal mammary glands (5,6), no
study has examined other breast cells by RT-PCR. Therefore, to
determine the expression of KRT81 in breast cells, RT-PCR was
performed using specific primers (forward primer located in exon 1
and reverse primer located in exon 9). Expression of KRT81,
including the 5′ region, resulting in a 1,674-bp product, was
detected in all analyzed cells (Fig.
1A). To examine whether KRT81 protein is expressed in breast
cancer cells, western blotting was performed with a KRT81 antibody.
A 55-kDa protein was detected in all breast cell lines.
Furthermore, a 30-kDa protein was detected in the SKBR3 and
MDA-MB-231 cell lines (Fig. 1B). To
examine the location of KRT81, we performed immunofluorescence
staining using an anti-KRT81 antibody. The immunofluorescence
staining revealed that KRT81 was expressed in the cytoplasm in all
breast cell lines (Fig. 2A). KRT81
expression in breast cancer tissue from patients was examined by
immunohistochemistry with an anti-KRT81 antibody. KRT81 was
expressed in the cytoplasm of ductal epithelial cells in breast
cancer and normal areas (Fig. 2B).
Staining with non-immune γ-globulin was negative. These results
indicated that the full size and truncated form of KRT81 are
expressed in human normal breast epithelial and cancer cells.
KRT81 knockdown downregulates the
expression of invasion-related genes in MCF10A cells
To analyze the function of KRT81 in mammary
epithelial cells, we transfected MCF10A cells with a KRT81
siRNA, and whole transcript profiling was performed by microarray
analysis. IPA was performed to investigate the functional
relationships between sets of genes with modified expression
levels. Table I shows that
invasion-related genes such as tumor necrosis factor (TNF),
MMP9 and Lipocalin 2 (LCN2) were downregulated in the
siKRT81-transfected cells.
 | Table I.Invasion-related genes in breast
cancer cell lines. |
Table I.
Invasion-related genes in breast
cancer cell lines.
| Gene symbol | Gene name |
Fold-changea |
|---|
| JUN | Jun
proto-oncogene | 0.48 |
| ARF6 | ADP-ribosylation
factor 6 | 0.46 |
| SP100 | SP100 nuclear
antigen | 0.45 |
| HPSE | Heparanase | 0.45 |
| MMP7 | Matrix
metallopeptidase 7 | 0.39 |
| MMP9 | Matrix
metallopeptidase 9 | 0.45 |
| MMP10 | Matrix
metallopeptidase 10 | 0.40 |
| MMP13 | Matrix
metallopeptidase 13 | 0.14 |
| HIF1A | Hypoxia inducible
factor 1 | 0.44 |
| TNF | Tumor necrosis
factor | 0.42 |
| LCN2 | Lipocalin 2 | 0.38 |
| BMP2 | Bone morphogenetic
protein 2 | 0.38 |
| PTGS2 |
Prostaglandin-endoperoxide synthase 2 | 0.29 |
| FAS | Fas (TNF receptor
superfamily, member 6) | 0.24 |
| TNFSF10 | Tumor necrosis
factor superfamily, member 10 | 0.09 |
KRT81 knockdown decreases the
migration and invasion abilities of MDA-MB-231 cells
Since MCF10A cells are non-invasive, we investigated
the effect of KRT81 on cell invasion using the invasive breast
cancer cell line MDA-MB-231. qPCR analysis revealed that KRT81
knockdown decreased KRT81 and MMP9 expression at the
mRNA level to 0.1- and 0.5-fold of that of the control cells,
respectively (p<0.01; Fig. 3A and
B). Gelatin zymography revealed that MMP9 activity was
decreased in the siKRT81-transfected cells (Fig. 3C). To investigate whether KRT81
affects migration and invasion, we performed scratch and invasion
assays using MDA-MB-231 cells. siKRT81 knockdown decreased cell
migration to 0.2-fold (p<0.01; Fig.
4A and B) and invasion to 0.5-fold (p<0.01; Fig. 4C and D). These results demonstrated
that KRT81 contributes to breast cancer cell migration and
invasion.
Discussion
Hair is produced by hair follicles. The embryonic
mammary glands and hair follicles are both derived from the ventral
ectoderm; mammary glands are skin appendages and their development
depends on a number of common fundamental developmental pathways
(21,22). Therefore, we speculated that KRT81
expressed in breast cells may have some, yet, unknown functions. In
the present study, we demonstrated the role of KRT81 in the
migration and invasion of breast cancer cells.
A previous study revealed that ΔHb1, but not the
full size KRT81, was expressed in the breast cancer cell line,
SKBR3, but not in normal breast cells using northern blotting, and
that ΔHb1 lacked the 270 amino terminal residues (5,6).
However, the present study revealed that the full size (exon 1–9)
KRT81 mRNA and the 55-kDa KRT81 protein were expressed not
only in breast cancer cells, but also in normal breast cells
(Fig. 1A and B). In western blot
analysis, the 55-kDa KRT81 protein was detected in all breast cell
lines, but the major band detected in the SKBR3 cells was ΔHb1
(30-kDa), suggesting that the amount of hair keratin proteins is
low in breast cells. Furthermore, the amount of full size KRT81 was
very low in typical breast cells, and the detection of ΔHb1 such as
in SKBR3 cells appears rare. Immunohistochemistry revealed that
KRT81 is located in the cytoplasm of breast cancer cells similar to
cytokeratins, which suggests that KRT81 may have a function related
to the cytoskeleton. However, recent studies proposed that
cytokeratins not only function in the cytoskeleton, but also
function as regulators of transcription factors and KRT17 may also
be localized in the nucleus (23,24).
Furthermore, ΔHb1 was detected not only in breast cancer, but also
in colon or nasopharyngeal carcinoma. It is thought that ΔHb1 may
inhibit some of the functions of the keratin cytoskeleton or
interfere with transcription regulation (25). Thus, KRT81 may have other functions
in breast cells.
The siRNA-mediated KRT81-knockdown in MCF10A
cells decreased the expression of invasion-related genes, including
LCN2 and matrix metalloproteinases. LCN2 promotes mammary
tumor formation and progression by upregulating matrix
metalloproteinase expression (26).
These alterations suggest inactivation of the NF-κB pathway, since
LCN2 and MMP9 expression is regulated by NF-κB
(27,28). However, microarray and IPA analysis
suggested that KRT81 knockdown did not alter the NF-κB
pathway (data not shown), except for TNF expression, which was
decreased (Table I). TNF is a known
regulator of MMPs and LCN2 (29–31).
In the present study, TNF, MMP9 and LCN2 expression was decreased
in the siKRT81-treated MCF10A cells. Additionally, the expression
of several TNF signaling-related genes, including Fas cell surface
death receptor, tumor necrosis factor superfamily member 10, and
TNF receptor superfamily member 14, was decreased (data not shown).
While the function of hair keratins has not been reported, the
functions of some keratins and TNF have been clarified. For
example, keratin 8/18 and keratin 17 interact with the TNF receptor
1 (TNFR1)-associated death domain protein (TRADD), a death adaptor
essential for TNFR1-dependent signaling. These keratins may
attenuate TNF-induced apoptosis through association with TRADD.
Furthermore, TNF regulates rat Krt23 (32) or human KRT15 (33). These studies suggest that KRT81 may
also regulate TNF-dependent functions.
Furthermore, we performed an upstream regulator
prediction analysis using IPA. The IPA analysis revealed that the
predicted upstream regulator in siKRT81-treated MCF10A cells was
the histone-lysine N-methyltransferase enzyme, enhancer of zeste 2
polycomb repressive complex 2 subunit (EZH2), as EZH2 (z-score,
−2.61) and the downstream genes of EZH2 were inhibited (Table II). Since EZH2 has been linked to
cancer invasion or growth (34),
recent studies have proposed that EZH2 may be useful as a
therapeutic target (35,36). Furthermore, it is known that EZH2
regulates the expression of KRT81, but its mechanism is unclear
(37). Since KRT81 silencing
downregulated EZH2-downstream genes such as LCN2 and
MMP9, KRT81 may be involved in EZH2 activation.
 | Table II.EZH2-regulated genes in MCF10A
cells. |
Table II.
EZH2-regulated genes in MCF10A
cells.
| Gene symbol | Gene name |
Fold-changea |
|---|
| TNF | Tumor necrosis
factor | 0.42 |
| RARRES3 | Retinoic acid
receptor responder 3 | 0.14 |
| PTGS2 |
Prostaglandin-endoperoxide synthase 2 | 0.29 |
| NCOA7 | Nuclear receptor
co-activator 7 | 0.46 |
| LCN2 | Lipocalin 2 | 0.38 |
| KRT81 | Keratin 81 | 0.15 |
| IL6 | Interleukin 6 | 0.19 |
| IL24 | Interleukin 24 | 0.40 |
| CXCL10 | C-X-C motif
chemokine ligand 10 | 0.05 |
|
C15orf48 | Chromosome 15 open
reading frame 48 | 0.20 |
In conclusion, this is the first study on KRT81 gene
expression in normal and breast cancer cells, suggesting that KRT81
is involved in breast cancer migration and invasion. Collectively,
the findings of the present study revealed that KRT81 is expressed
in clinical specimens from patients with breast cancer. Indeed, it
was reported that a KRT19 fragment in serum can be used as a marker
of liver and breast cancer (38,39),
and KRT81 microRNA is associated with the risk and survival of
patients with non-small cell lung cancer (40–42).
Therefore, KRT81 may be used as a biomarker for patients with
breast cancer once we clarify its biological significance.
Acknowledgements
The present study was supported in part by the JSPS
KAKENHI (grant no. 20770096) and a Hirosaki University
Institutional Research Grant for Young Scientists.
Glossary
Abbreviations
Abbreviations:
|
KRT81
|
keratin 81
|
|
MMP
|
matrix metallopeptidase
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
SDS-PAGE
|
sodium dodecyl sulfate-polyacrylamide
gel electrophoresis
|
References
|
1
|
Charafe-Jauffret E, Ginestier C, Monville
F, Finetti P, Adélaïde J, Cervera N, Fekairi S, Xerri L, Jacquemier
J, Birnbaum D, et al: Gene expression profiling of breast cell
lines identifies potential new basal markers. Oncogene.
25:2273–2284. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheang MC, Voduc D, Bajdik C, Leung S,
McKinney S, Chia SK, Perou CM and Nielsen TO: Basal-like breast
cancer defined by five biomarkers has superior prognostic value
than triple-negative phenotype. Clin Cancer Res. 14:1368–1376.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Langbein L, Rogers MA, Winter H, Praetzel
S and Schweizer J: The catalog of human hair keratins. II.
Expression of the six type II members in the hair follicle and the
combined catalog of human type I and II keratins. J Biol Chem.
276:35123–35132. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Langbein L and Schweizer J: Keratins of
the human hair follicle. Int Rev Cytol. 243:1–78. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boulay A, Régnier CH, Anglard P, Stoll I,
Tomasetto C and Rio MC: Transcription regulation and protein
subcellular localization of the truncated basic hair keratin
hHb1-DeltaN in human breast cancer cells. J Biol Chem.
276:22954–22964. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Régnier CH, Boulay A, Asch PH, Wendling C,
Chenard MP, Tomasetto C and Rio MC: Expression of a truncated form
of hHb1 hair keratin in human breast carcinomas. Br J Cancer.
78:1640–1644. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomasetto C, Régnier C, Moog-Lutz C,
Mattei MG, Chenard MP, Lidereau R, Basset P and Rio MC:
Identification of four novel human genes amplified and
overexpressed in breast carcinoma and localized to the q11-q21.3
region of chromosome 17. Genomics. 28:367–376. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Figueira RC, Gomes LR, Neto JS, Silva FC,
Silva ID and Sogayar MC: Correlation between MMPs and their
inhibitors in breast cancer tumor tissue specimens and in cell
lines with different metastatic potential. BMC Cancer. 9:202009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren F, Tang R, Zhang X, Madushi WM, Luo D,
Dang Y, Li Z, Wei K and Chen G: Overexpression of MMP family
members functions as prognostic biomarker for breast cancer
patients: A systematic review and meta-analysis. PLoS One.
10:e01355442015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nanashima N, Akita M, Yamada T, Shimizu T,
Nakano H, Fan Y and Tsuchida S: The hairless phenotype of the
Hirosaki hairless rat is due to the deletion of an 80-kb genomic
DNA containing five basic keratin genes. J Biol Chem.
283:16868–16875. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nanashima N, Asano J, Hayakari M, Nakamura
T, Nakano H, Yamada T, Shimizu T, Akita M, Fan Y and Tsuchida S:
Nuclear localization of STAT5A modified with O-linked
N-acetylglucosamine and early involution in the mammary
gland of Hirosaki hairless rat. J Biol Chem. 280:43010–43016. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nanashima N, Yamada T, Shimizu T and
Tsuchida S: Deletion of phospholipase A2 group IVc
induces apoptosis in rat mammary tumour cells by the nuclear
factor-κB/lipocalin 2 pathway. Biochem J. 469:315–324. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
James V, Kearsley J, Irving T, Amemiya Y
and Cookson D: Using hair to screen for breast cancer. Nature.
398:33–34. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Corino GL and French PW: Diagnosis of
breast cancer by X-ray diffraction of hair. Int J Cancer.
122:847–856. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Corino GL, French PW, Lee M, Ajaj MM,
Haklani J, Mistry DA, Phan K and Yuile PG: Characterization of a
test for invasive breast cancer using X-ray diffraction of
hair-results of a clinical trial. Breast Cancer. 3:83–90.
2009.PubMed/NCBI
|
|
16
|
Cailleau R, Young R, Olivé M and Reeves WJ
Jr: Breast tumor cell lines from pleural effusions. J Natl Cancer
Inst. 53:661–674. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: Procedure and some applications. Proc Natl
Acad Sci USA. 76:4350–4354. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miyazaki K, Hattori Y, Umenishi F,
Yasumitsu H and Umeda M: Purification and characterization of
extracellular matrix-degrading metalloproteinase, matrin (pump-1),
secreted from human rectal carcinoma cell line. Cancer Res.
50:7758–7764. 1990.PubMed/NCBI
|
|
20
|
Umenishi F, Yasumitsu H, Ashida Y, Yamauti
J, Umeda M and Miyazaki K: Purification and properties of
extracellular matrix-degrading metallo-proteinase overproduced by
Rous sarcoma virus-transformed rat liver cell line, and its
identification as transin. J Biochem. 108:537–543. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Michno K, Boras-Granic K, Mill P, Hui CC
and Hamel PA: Shh expression is required for embryonic hair
follicle but not mammary gland development. Dev Biol. 264:153–165.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mikkola ML and Millar SE: The mammary bud
as a skin appendage: Unique and shared aspects of development. J
Mammary Gland Biol Neoplasia. 11:187–203. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Escobar-Hoyos LF, Shah R, Roa-Peña L,
Vanner EA, Najafian N, Banach A, Nielsen E, Al-Khalil R, Akalin A,
Talmage D, et al: Keratin-17 promotes p27KIP1 nuclear
export and degradation and offers potential prognostic utility.
Cancer Res. 75:3650–3662. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hobbs RP, Jacob JT and Coulombe PA:
Keratins are going nuclear. Dev Cell. 38:227–233. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nishikawa J, Kiss C, Imai S, Takada K,
Okita K, Klein G and Szekely L: Upregulation of the truncated basic
hair keratin 1(hHb1-DeltaN) in carcinoma cells by Epstein-Barr
virus (EBV). Int J Cancer. 107:597–602. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang J, Bielenberg DR, Rodig SJ, Doiron R,
Clifton MC, Kung AL, Strong RK, Zurakowski D and Moses MA:
Lipocalin 2 promotes breast cancer progression. Proc Natl Acad Sci
USA. 106:3913–3918. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kang H, Lee M, Choi KC, Shin DM, Ko J and
Jang SW: N-(4-hydroxyphenyl)retinamide inhibits breast
cancer cell invasion through suppressing NF-KB activation and
inhibiting matrix metalloproteinase-9 expression. J Cell Biochem.
113:2845–2855. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li SH, Hawthorne VS, Neal CL, Sanghera S,
Xu J, Yang J, Guo H, Steeg PS and Yu D: Upregulation of neutrophil
gelatinase-associated lipocalin by ErbB2 through nuclear
factor-kappaB activation. Cancer Res. 69:9163–9168. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arena A, Stassi G, Iannello D, Gazzara D,
Calapai M, Bisignano C, Bolignano D, Lacquaniti A and Buemi M: Both
IL-1β and TNF-α regulate NGAL expression in
polymorphonuclear granulocytes of chronic hemodialysis patients.
Mediators Inflamm. 2010:6139372010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee SJ, Park SS, Cho YH, Park K, Kim EJ,
Jung KH, Kim SK, Kim WJ and Moon SK: Activation of matrix
metalloproteinase-9 by TNF-α in human urinary bladder cancer HT1376
cells: The role of MAP kinase signaling pathways. Oncol Rep.
19:1007–1013. 2008.PubMed/NCBI
|
|
31
|
Moon SK, Cha BY and Kim CH: ERK1/2
mediates TNF-alpha-induced matrix metalloproteinase-9 expression in
human vascular smooth muscle cells via the regulation of NF-kappaB
and AP-1: Involvement of the ras dependent pathway. J Cell Physiol.
198:417–427. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zer C, Sachs G and Shin JM: Identification
of genomic targets downstream of p38 mitogen-activated protein
kinase pathway mediating tumor necrosis factor-alpha signaling.
Physiol Genomics. 31:343–351. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Banno T, Gazel A and Blumenberg M: Effects
of tumor necrosis factor-alpha (TNF alpha) in epidermal
keratinocytes revealed using global transcriptional profiling. J
Biol Chem. 279:32633–32642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yoo KH and Hennighausen L: EZH2
methyltransferase and H3K27 methylation in breast cancer. Int J
Biol Sci. 8:59–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Italiano A: Role of the EZH2 histone
methyltransferase as a therapeutic target in cancer. Pharmacol
Ther. 165:26–31. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim KH and Roberts CW: Targeting EZH2 in
cancer. Nat Med. 22:128–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee ST, Li Z, Wu Z, Aau M, Guan P,
Karuturi RK, Liou YC and Yu Q: Context-specific regulation of NF-κB
target gene expression by EZH2 in breast cancers. Mol Cell.
43:798–810. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nakata B, Takashima T, Ogawa Y, Ishikawa T
and Hirakawa K: Serum CYFRA 21–1 (cytokeratin-19 fragments) is a
useful tumour marker for detecting disease relapse and assessing
treatment efficacy in breast cancer. Br J Cancer. 91:873–878.
2004.PubMed/NCBI
|
|
39
|
Uenishi T, Kubo S, Hirohashi K, Tanaka H,
Shuto T, Yamamoto T and Nishiguchi S: Cytokeratin-19 fragments in
serum (CYFRA 21–1) as a marker in primary liver cancer. Br J
Cancer. 88:1894–1899. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Campayo M, Navarro A, Viñolas N, Tejero R,
Muñoz C, Diaz T, Marrades R, Cabanas ML, Gimferrer JM, Gascon P, et
al: A dual role for KRT81: A miR-SNP associated with recurrence in
non-small-cell lung cancer and a novel marker of squamous cell lung
carcinoma. PLoS One. 6:e225092011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee SY, Choi JE, Jeon HS, Hong MJ, Choi
YY, Kang HG, Yoo SS, Lee EB, Jeong JY, Lee WK, et al: A genetic
variation in microRNA target site of KRT81 gene is
associated with survival in early-stage non-small-cell lung cancer.
Ann Oncol. 26:1142–1148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Robles AI and Ryan BM: KRT81
miR-SNP rs3660 is associated with risk and survival of NSCLC. Ann
Oncol. 27:360–361. 2016. View Article : Google Scholar : PubMed/NCBI
|