Introduction
Endometrial cancer (EC) is the fourth most common
malignancy among postmenopausal females in the developed world, and
its incidence is surpassed only by lung, colorectal and breast
cancer (1–3). Although, EC accounts for 74,000 deaths
each year worldwide, the majority of affected women have a good
prognosis, as abnormal vaginal bleeding begins at an early stage of
the disease, and thus, the 5-year survival rate is 80–82% (4). Atypical endometrial hyperplasia (AEH)
is a type of pre-cancerous lesion which is a significant risk
factor for the development or co-existence of EC. However,
traditional surgical resection, the standard method for treating EC
and AEH is a total hysterectomy, such as a bilateral oophorectomy,
which results in a total loss of fertility. As a result, this
method of treatment is unacceptable to many women diagnosed with EC
or AEH (5,6). Molecular-targeted therapies for EC,
such as microRNA (miRNA)-based therapies, have received increased
attention, even though the specific molecular events which lead to
EC development remain unclear. However, various studies have
investigated the molecular changes which lead to AEH (7–9).
Research regarding the functions of miRNAs may increase our
understanding of disease pathogenesis and particularly the
pathogenesis of cancer.
miRNAs consist of a family of single-stranded, 22
nucleotide, non-coding, evolutionarily conserved RNAs, which
regulate gene degradation or translational suppression at the
post-transcriptional level by binding to the 3′-untranslated region
(3′-UTR) of mRNAs (10,11). The relevance of miRNAs in cancers
such as EC is associated with their ability to regulate gene
expression and various cellular processes, such as cell
proliferation, differentiation, apoptosis, epigenetic dysfunction
and carcinogenesis. These regulatory abilities suggest that miRNA
expression may be important when developing a prognosis and
treatment strategy for EC patients (12–14).
Numerous studies have shown the importance of miRNAs, and miRNA
profiling analyses have revealed significant variations in miRNA
expression across different cancer subtypes and stages of
carcinogenesis. Such findings suggest that miRNAs play important
roles in the initiation and progression of human malignancies
(15–17).
The exact biological functions of miRNAs in EC
remain unclear. In the present study, we profiled miRNA expression
in cases of human EC, and focused on the relationships between
certain miRNAs that exhibited aberrant expression and the presence
of EC, in order to explore the effect of those miRNAs on cellular
functions and regulatory mechanisms. AN3CA and HEC-1-A cell lines
were used in the present study, since HEC-1-A and AN3CA are both
human EC cell lines that have been commonly used in EC research
in vitro.
Materials and methods
Patient characteristics and sample
collection
Between October 2015 and May 2016, 45 consecutive
patients (15 with EC, 15 with AEH and 15 healthy donors) at The
First Affiliated Hospital of Guangxi Medical University were
recruited to participate in the present study. The EC patients were
aged between 40 and 82 years, and had undergone a hysterectomy,
bilateral salpingo-oophorectomy, pelvic and/or para-aortic
lymphadenectomy or peritoneal washing for cytology.
Frozen fresh tissue sections and respective blood
samples were obtained from 15 patients with EC, 15 patients with
AEH, and 15 subjects with a normal endometrium. The samples were
used in a microarray assay and also analyzed by quantitative
real-time PCR. No patient had a history of adjuvant or neoadjuvant
therapy prior to surgery. The study protocol was approved by the
Ethics Committee of The First Affiliated Hospital of Guangxi
Medical University, and each enrolled subject provided their signed
informed consent for participation.
Profiling of miRNA expression
An Agilent Human miRNA Microarray kit (release 16.0;
Agilent Technologies, Santa Clara, CA, USA) containing probes for
1,205 human miRNAs and 144 human viral miRNAs was used for miRNA
expression profiling. The miRNA assays were performed according to
the manufacturer's instructions. In brief, 100 ng of total RNA from
each sample was dephosphorylated and then ligated with pCp-Cy3 dye.
The labeled RNA was purified using a Micro Bio-SpinMicro Bio-Spin
66 column (Bio-Rad, Hercules, CA, USA), and then added to the miRNA
array, which contained a hybridization buffer. After 20 h of
incubation at 55°C, the array slides were washed and scanned, and
the images were analyzed using Feature Extraction 10.7.3.1 software
(Agilent Technologies). Data quality was evaluated using an Agilent
microRNA Spike-In kit, and all samples satisfied the Spike-In QC
criteria (LabelingSpike-InSignal >2.5 and HybSpike-InSignal
>2.5). The relevant miRNA microarray data is available in the
NCBI Gene Expression Omnibus (GSE70574).
Real-time polymerase chain
reaction
Total RNA was extracted using UNIQ-10 columns and a
TRIzol Total RNA Isolation kit (Sangon, Shanghai, China). Cloned
AMV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) was used
to reverse transcribe 1 µg samples of total RNA in a reaction
volume of 20 µl. Two microliters of cDNA were used for real-time
PCR that was performed using a Takara Ex Taq RT-PCR version 2.1 kit
(Takara, Shiga, Japan). Real-time quantification of mature miRNAs
was performed using an Applied Biosystems 7500 Sequence Detection
system (Applied Biosystems, Foster City, CA, USA). Each 20 µl PCR
reaction mixture contained 1 µl of RT product (1:5 dilution), 0.5
µl of universal reverse primer, 0.5 µl of sense primer, and 10 µl
of mix buffer (DBI Bestar® SybrGreen qPCR mastermix).
The reaction mixtures were incubated in a 96-well optical plate at
94°C for 2 min, followed by 40 cycles of 94°C for 20 sec, 58°C for
20 sec, and 72°C for 20 sec. All reactions were run in triplicate.
The gene-specific miRNA primers are listed in Table I.
 | Table I.Primer sequences used for miRNA
expression analysis. |
Table I.
Primer sequences used for miRNA
expression analysis.
| Gene |
| Sequence (5′-3′) |
|---|
| U6 | F |
CTCGCTTCGGCAGCACA |
| U6 | R |
AACGCTTCACGAATTTGCGT |
| miR-1202 | RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCCCC |
| miR-1202 | F |
ACACTCCAGCTGGGGTGCCAGCTGCAGTGGG |
| miR-5787 | RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCTCC |
| miR-5787 | F |
ACACTCCAGCTGGGGGGCTGGGGCGCGGGG |
| miR-6749-5p | RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCTCCC |
| miR-6749-5p | F |
ACACTCCAGCTGGGTCGGGCCTGGGGTTGGG |
| miR-196a-5p | RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCAAC |
| miR-196a-5p | F |
ACACTCCAGCTGGGTAGGTAGTTTCATGTTG |
| miR-338-3p | RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAACAA |
| miR-338-3p | F |
ACACTCCAGCTGGGTCCAGCATCAGTGATTTT |
| miR-449a | RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCAGC |
| miR-449a | F |
ACACTCCAGCTGGGTGGCAGTGTATTGTTAGC |
| ALL | R |
CTCAACTGGTGTCGTGGA |
Cell line and culture conditions
Human endometrial cancer cell line AN3CA and HEC-1-A
were purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA), and maintained in Dulbeccos modified Eagles
medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml
penicillin, and 100 U/ml streptomycin in a humidified atmosphere of
5% CO2 at 37°C. The medium was changed every 2 or 3 days
based on the recommended culture condition. All cells were
harvested by centrifugation before being washed with
phosphate-buffered saline (PBS) and subjected to total protein or
RNA extraction.
Cells were cultured to 60–70% confluence, and then
re-suspended in serum-free DMEM at a concentration of
105 cells/ml. Six-well plates were inoculated with 2 ml
of cell suspension in each well, and 3 replicate wells were created
for each experimental group. miR-1202 inhibitors and miR-196a
mimics purchased from GenePharma (Shanghai, China) were diluted to
5 nM concentrations with 0.25 ml serum-free DMEM for use in
transfection studies. Lipofectamine 2000 (Invitrogen) transfection
reagent (5 µl) was diluted with 0.25 ml serum-free DMEM. Next, the
diluted transfection reagent was added to the diluted mimics, mixed
gently, and incubated for 20 min at room temperature. The cell
suspension was refreshed with new medium, and then added to the
mixture of Lipofectamine 2000 and the mimics aforementioned; after
which, the total mixture was incubated at 37°C in a 5%
CO2 atmosphere for 6 h. Subsequently, the medium in each
well was replaced with normal serum-containing medium and incubated
for 48 h prior to use in the following experiments.
Flow cytometric assay
After transfection for 72 h, the apoptotic cells
were quantified using an Annexin V/propidium iodide (PI) apoptosis
kit (MultiSciences, Hangzhou, China) prior to performing cell
apoptosis and cell cycle analyses. The AN3CA or HEC-1-A cells were
collected, rinsed with PBS, and then resuspended in 200 µl of
binding buffer containing 5 µl Annexin V (10 µg/ml) for 10 min in
the dark. After being incubated with 10 µl of PI (20 µg/ml), the
cells were immediately analyzed by flow cytometry (Beckman Coulter
Epics XL; Beckman Coulter, Brea, CA, USA). Data acquisition and
analysis were performed using CellQuest software (Becton-Dickinson,
Franklin Lakes, NJ, USA).
Cell migration and invasion
assays
Cell migration and invasion assays were performed in
24-well, Matrigel-coated invasion chambers. For this assay,
AN3CA/HEC-1-A (non-transfected), AN3CA/HEC-1-A-NC and
AN3CA/HEC-1-A-miR-1202 inhibitor/miR-196a mimic cells were plated
at a density of 1.0×105 cells/well in wells containing
0.5 ml of serum-free medium and polycarbonate filters (8-µm pore
size; Costar Inc., Milpitas, CA, USA). The outer chambers were
filled with 0.5 ml of the medium supplemented with 10% FBS. After
24 h, the cells were fixed in methanol and stained with crystal
violet. Subsequently, the top surface of the membrane was gently
scrubbed with a cotton bud, and the cells that had invaded through
the membrane filters were counted. The invasion inhibition rate (%)
was calculated as [(A - B)/A) × 100]; where A and B are the
percentages of invading cells for the miRNA inhibitor group and
normal control (NC) group, respectively. Each experiment was
performed in triplicate.
Statistical analysis
The miRNA array data were processed by quantile
normalization, followed by a log2 transformation. The spots called
‘absent’ by the Agilent Feature Extraction software were discarded.
The unpaired Mann-Whitney test was used to identify significant
differences in expressed miRNA between LNM-positive and -negative
CRC samples. The Benjamini-Hochberg false discovery rate (FDR)
method was used for multiple comparison corrections. miRNAs with an
FDR <0.1 and log fold-change >3 were considered as
potentially important and included in further independent
replication experiments performed for validation purposes. All data
were analyzed using GeneSpring 12.6 software (Agilent
Technologies).
All other data were analyzed using SPSS Statistics
for Windows, version 17.0. (SPSS, Inc., Chicago, IL, USA) and the
presence of a normal data distribution was assessed by the
Kolmogorov-Smirnov test. miRNA expression results for the 3 groups
of tissue are presented as the mean ± standard deviation.
Differences between groups were evaluated using one-way ANOVA for
3-group comparisons and t-tests for 2-group comparisons.
Results
Differentially expressed miRNAs are
found in endometrial adenocarcinoma and AEH tissues when compared
with normal endometrial tissue
We performed miRNA analyses to examine the global
miRNA expression profiles in tissue samples obtained from 15 EC
patients, 15 patients with AEH, and 15 healthy donors. A paired
comparison analysis identified 34 miRNAs that were expressed at
significantly different levels in the 3 groups. Among these
differentially expressed miRNAs, 14 were upregulated and 20 were
downregulated in EC patients when compared with their expression
levels in the AEH group (data not shown). The healthy donor group
served as a negative control group. A correlation analysis of
hsa-miRNA expression in the tissue samples indicated that all of
the hsa-miRNAs were expressed at significantly different levels in
the 3 groups (Fig. 1).
Verification and selection of
differentially expressed miRNAs in tissue specimens
To confirm the results obtained from the miRNA
microarray assay, the expression levels of 12 miRNAs which had been
analyzed by microarray were further analyzed by real-time PCR. Our
results revealed that when compared with their expression levels in
the healthy group, hsa-miR-5787, hsa-miR-6749-5p and hsa-miR-1202
were expressed at successively increased levels in tissue and blood
samples (Fig. 2B, D and F), while
levels of hsa-miR-338-3p, hsa-miR-449a, hsa-miR-196a were
successively downregulated in tissue and blood samples,
respectively, from patients of the AEH and EC group (Fig. 2A, C and E). Thus, the real-time PCR
results were consistent with the microarray assay results. Among
these various candidates, we selected miR-1202 and miR-196a as our
target mRNAs for further experiments.
Effects of miR-1202 and miR-196a on
cell apoptosis
An Annexin V-FITC/PI staining analysis indicated
that AN3CA cells transfected with miR-1202 inhibitor (Fig. 3A and B) or miR-196a mimics (Fig. 3C and D), respectively, for 72 h had
significantly increased levels of apoptosis and necrosis, as
indicated by the presence of a prominent sub-G1 peak (apoptotic
cells). The rates of apoptosis and necrosis were significantly
increased to 60% after 72 h of treatment, and were higher than in
either of the two NC groups. Similarly, we performed the same
assessment in the HEC-1-A cells (Fig.
3E-H). Treatment with miR-1202 inhibitor (Fig. 3E and F) and miR-196a mimics
(Fig. 3G and H), respectively, for
72 h resulted in a significant increase in apoptosis and necrosis
in the HEC-1-A cells shown as a significant sub-G1 peak (apoptotic
cells).
Effects of miR-1202 and miR-196a on
cell cycle distribution
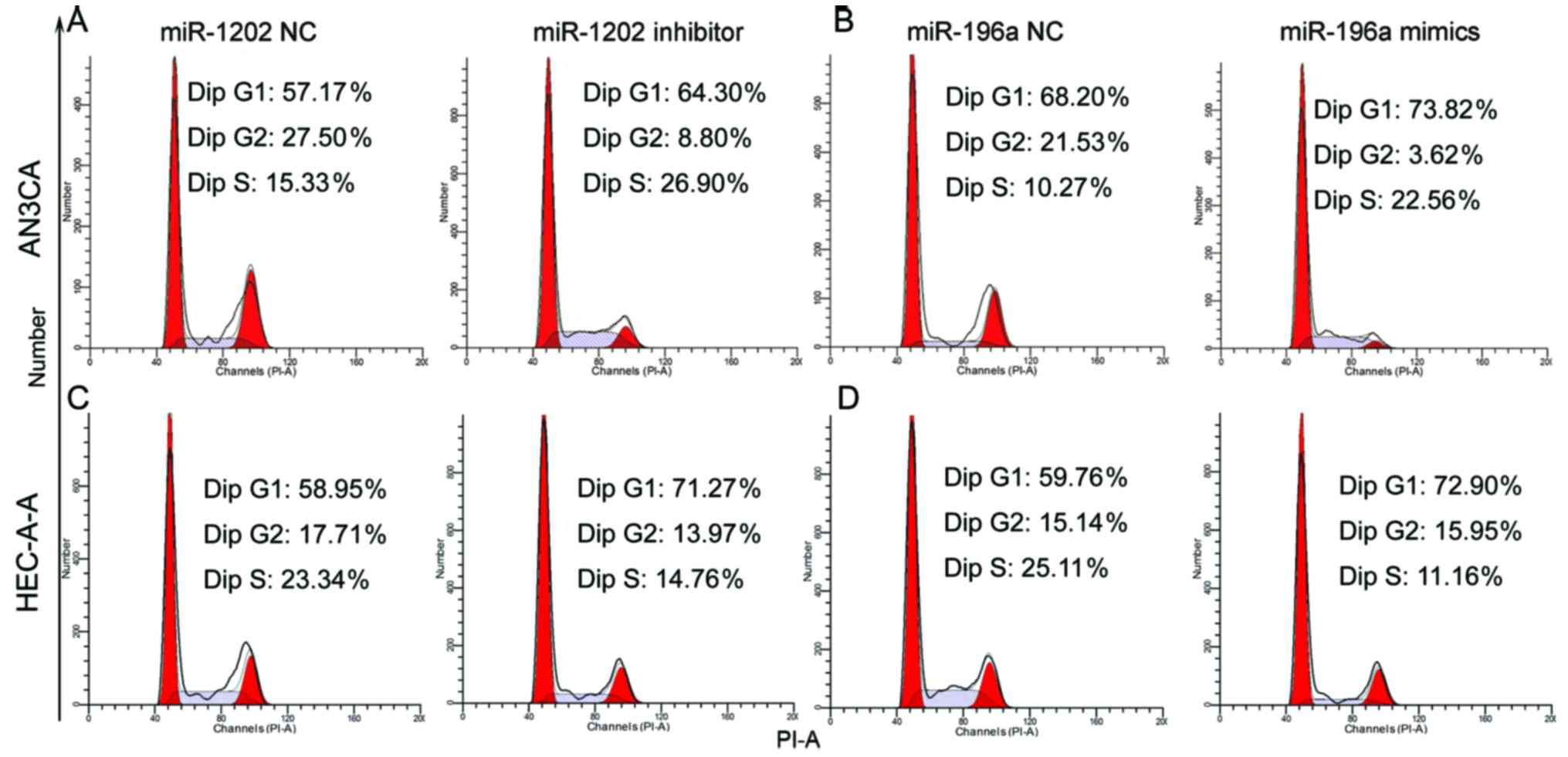
We performed flow cytometric assays to assess the
effect of miR-1202 silencing or miR-196a overexpression on the
AN3CA and HEC-1-A cell cycle. After transfection with miR-1202
inhibitor (Fig. 4A and C) or
miR-196a mimics (Fig. 4B and D) for
72 h, the percentage of G0/G1 phase cells was significantly
increased, while the percentage of cells in the G2/M phases
decreased.
Effects of miR-1202 and miR-196a on
cell migration and invasion abilities
We investigated the roles of miR-1202 and miR-196a
in mediating EC cell migration and invasion. AN3CA cells that had
been transfected with miR-1202 inhibitor or miR-196a mimics were
evaluated for their migration and invasion abilities. AN3CA cells
transfected with scrambled miRNA inhibitors or mimics were used as
NC groups. The results revealed that AN3CA and HEC-1-A cells
transfected with miR-1202 inhibitor (Figs. 5A and B, and 6A and B) or miR-196a mimics (Figs. 5C and D, and 6C and D) displayed a significantly
decreased migratory ability when compared with the NC groups
(P<0.01). In parallel, AN3CA and HEC-1-A cells transfected with
miR-1202 inhibitor (Figs. 5E and F,
and 6E and F) or miR-196a mimics
(Figs. 5G and H, and 6G and H) also displayed a significantly
decreased invasion ability (P<0.01).
Discussion
Microarray panels have been described as a new and
powerful methodology for identifying miRNA expression patterns that
may distinguish between samples of carcinoma and non-carcinoma
tissue (18,19). However, no study has described the
miRNA microarray profile of endometrial cancer (EC) tissue, and
particularly tissue in which EC is accompanied by AEH. In the
present study, we identified some global changes in miRNA
expression that characterize the difference between human malignant
and non-malignant tissue samples. The consistency of our findings
was supported by our use of TaqMan qRT-PCR methodology to validate
12 of the most significant differentially expressed miRNAs in an
extended series of human tissue samples. Finally, highly expressed
miR-1202 and lowly expressed miR-196a were selected as our research
candidates for further investigation. Next, we explored the effects
of these two miRNAs on cellular functions in vitro, and
found that miR-1202 may protect AN3CA cells against apoptosis and
increase their S phase arrest, while miR-196a may reverse these
effects. Collectively, our findings indicate the importance of
miR-1202 and miR-196a in the pathogenesis of EC, and contribute to
the understanding of the miRNA-driven pathways related to EC.
In agreement with our results, a previous study
revealed that miR-1202 was strongly correlated with a shorter
overall survival time among patients with adrenocortical carcinoma
(ACC) (20). Moreover, other
studies have shown that high levels of miR-1202 in human brain
tissue may play an important role in the pathophysiology of
depression, suggesting miR-1202 as a potential target for novel
anti-depressant agents (21,22).
Additionally, overexpression of miR-1202 may be associated with
lymph node metastasis (23). In the
present study, we demonstrated that miR-1202 expression was
significantly higher in EC patients than in AEH patients and a
healthy donor group, suggesting that miR-1202 inhibitor may exert a
protective effect in EC. We also demonstrated that miR-196a
expression levels were significantly lower in specimens of EC
tissue than in specimens of endometrial tissue obtained from AEH
patients or normal control subjects. This supports previous studies
that suggested miR-196a as a newly discovered promising biomarker
for tumor progression (24–27). However, its dysregulation in
endometrioid endometrial carcinoma (EEC), revealed that miR-196a
may also work synergistically with other miRNAs thought to be
involved in various diseases (28).
In cervical cancer (26) and head
and neck squamous cell carcinoma (29), miR-196a was overexpressed in the
absence of HOXC8/HOXB9 expression, which suggests its role as an
oncomiR. Regarding our research candidates, miR-1202 and miR-196a,
further investigation may be warranted to explore the regulation
mechanism even if numerous studies have revealed that both of them
may negatively regulate target genes through binding to the 3′UTR
of their target mRNAs, such as SOX11/12 (30), MAP3K1 (31), PCDH17 (32) and TP53 (33).
The present study has several limitations that
should be mentioned. First, the study was performed using native
human tissues, and false-positive results may exist due to the
limited sample size. Thus, only miRNAs that revealed a significant
aberrant change were included in the present study. Second, the
molecular mechanism by which alterations in miRNAs cause EC or AEH
has not identified, and requires further bioinformatic analysis and
validation testing. A better understanding of the bio-functional
significance of broad and often subtle variations in miRNA levels
which occur during development of EC would allow specific-mRNA
molecules to be identified for manipulation in cases of EC, and
thus, lead to new therapeutic interventions that are based on
rational target selection.
In conclusion, the present study identified the
miRNA profile signature in 3 different cohorts: EC and AEH
patients, and healthy donors. This pattern of aberrantly expressed
miRNAs may contribute to our understanding of EC and/or AEH. Our
findings suggest that silencing of miR-1202 or overexpression of
miR-196a may increase cell apoptosis and G1 phase arrest in EC
cells with lower migration and invasion abilities. Additionally,
the miRNA expression signature described in the present study can
aid in developing new molecular-based therapies for EC.
Acknowledgements
The present study was funded by the Natural Science
Foundation of GuangXi (no. 2013GXNSFAA01956).
References
|
1
|
Braun MM, Overbeek-Wager EA and Grumbo RJ:
Diagnosis and management of endometrial cancer. Am Fam Physician.
93:468–474. 2016.PubMed/NCBI
|
|
2
|
Prat J, Gallardo A, Cuatrecasas M and
Catasús L: Endometrial carcinoma: Pathology and genetics.
Pathology. 39:72–87. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Uharcek P: Prognostic factors in
endometrial carcinoma. J Obstet Gynaecol Res. 34:776–783. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rossouw JE, Anderson GL, Prentice RL,
LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SA,
Howard BV, Johnson KC, et al: Writing Group for the Women's Health
Initiative Investigators: Risks and benefits of estrogen plus
progestin in healthy postmenopausal women: Principal results From
the Women's Health Initiative randomized controlled trial. JAMA.
288:321–333. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Corrado G, Baiocco E, Carosi M and Vizza
E: Progression of conservatively treated endometrial complex
atypical hyperplasia in a young woman: A case report. Fertil
Steril. 90:2006.e5–2006.e8. 2008. View Article : Google Scholar
|
|
6
|
Ofinran O and Balega J: The value of
magnetic resonance imaging in investigating complex atypical
hyperplasia of the endometrium. Minerva Ginecol. 68:400–404.
2016.PubMed/NCBI
|
|
7
|
Jurcevic S, Olsson B and Klinga-Levan K:
MicroRNA expression in human endometrial adenocarcinoma. Cancer
Cell Int. 14:882014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang S, Jia Y, Liu X, Winters C, Wang X,
Zhang Y, Devor EJ, Hovey AM, Reyes HD, Xiao X, et al: Systematic
dissection of the mechanisms underlying progesterone receptor
downregulation in endometrial cancer. Oncotarget. 5:9783–9797.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Torres A, Torres K, Wdowiak P, Paszkowski
T and Maciejewski R: Selection and validation of endogenous
controls for microRNA expression studies in endometrioid
endometrial cancer tissues. Gynecol Oncol. 130:588–594. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Santulli G: MicroRNAs and endothelial
(Dys) function. J Cell Physiol. 231:1638–1644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Widodo Djati MS and Rifa'i M: Role of
MicroRNAs in carcinogenesis that potential for biomarker of
endometrial cancer. Ann Med Surg. 7:9–13. 2016. View Article : Google Scholar
|
|
13
|
Boren T, Xiong Y, Hakam A, Wenham R, Apte
S, Wei Z, Kamath S, Chen DT, Dressman H and Lancaster JM: MicroRNAs
and their target messenger RNAs associated with endometrial
carcinogenesis. Gynecol Oncol. 110:206–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu YY, Wu HJ, Ma HD, Xu LP, Huo Y and Yin
LR: MicroRNA-503 suppresses proliferation and cell-cycle
progression of endometrioid endometrial cancer by negatively
regulating cyclin D1. FEBS J. 280:3768–3779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kalia M: Biomarkers for personalized
oncology: Recent advances and future challenges. Metabolism. 64
Suppl 1:S16–S21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee TS, Jeon HW, Kim YB, Kim YA, Kim MA
and Kang SB: Aberrant microRNA expression in endometrial carcinoma
using formalin-fixed paraffin-embedded (FFPE) tissues. PLoS One.
8:e814212013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luna-Aguirre CM, de la Luz Martinez-Fierro
M, Mar-Aguilar F, Garza-Veloz I, Treviño-Alvarado V, Rojas-Martinez
A, Jaime-Perez JC, Malagon-Santiago GI, Gutierrez-Aguirre CH,
Gonzalez-Llano O, et al: Circulating microRNA expression profile in
B-cell acute lymphoblastic leukemia. Cancer Biomark. 15:299–310.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pellatt DF, Stevens JR, Wolff RK, Mullany
LE, Herrick JS and Samowitz W: Slattery ml: Expression profiles of
miRNA subsets distinguish human colorectal carcinoma and normal
colonic mucosa. Clin Transl Gastroenterol. 7:e1522016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Plieskatt JL, Rinaldi G, Feng Y, Levine
PH, Easley S, Martinez E, Hashmi S, Sadeghi N, Brindley PJ, Bethony
JM, et al: Methods and matrices: Approaches to identifying miRNAs
for nasopharyngeal carcinoma. J Transl Med. 12:32014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Özata DM, Caramuta S, Velázquez-Fernández
D, Akçakaya P, Xie H, Höög A, Zedenius J, Bäckdahl M, Larsson C and
Lui WO: The role of microRNA deregulation in the pathogenesis of
adrenocortical carcinoma. Endocr Relat Cancer. 18:643–655. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rucker JJ and McGuffin P: Chipping away at
major depressive disorder. Genome Biol. 15:4212014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lopez JP, Lim R, Cruceanu C, Crapper L,
Fasano C, Labonte B, Maussion G, Yang JP, Yerko V, Vigneault E, et
al: miR-1202 is a primate-specific and brain-enriched microRNA
involved in major depression and antidepressant treatment. Nat Med.
20:764–768. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Zhang H, Zhang P, Li J, Shan Z and
Teng W: Upregulation of miR-2861 and miR-451 expression in
papillary thyroid carcinoma with lymph node metastasis. Med Oncol.
30:5772013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen ZF, Ma LL and Xue HB: Common
polymorphisms of the microRNA genes (miR-146a and miR-196a-2) and
gastric cancer risk: An updated meta-analysis. Genet Mol Res.
14:8589–8601. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Jin L, Chen D, Liu J, Su Z, Yang S,
Gui Y, Mao X, Nie G and Lai Y: Tumor suppressive miR-196a is
associated with cellular migration, proliferation and apoptosis in
renal cell carcinoma. Mol Med Rep. 14:560–566. 2016.PubMed/NCBI
|
|
26
|
Liu P, Xin F and Ma CF: Clinical
significance of serum miR-196a in cervical intraepithelial
neoplasia and cervical cancer. Genet Mol Res. 14:17995–18002. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan Y, Fan J, Huang L, Ye M, Huang Z, Wang
Y, Li Q and Huang J: Increased expression of microRNA-196a predicts
poor prognosis in human ovarian carcinoma. Int J Clin Exp Pathol.
8:4132–4137. 2015.PubMed/NCBI
|
|
28
|
Xiong H, Li Q, Liu S, Wang F, Xiong Z,
Chen J, Chen H, Yang Y, Tan X, Luo Q, et al: Integrated microRNA
and mRNA transcriptome sequencing reveals the potential roles of
miRNAs in stage I endometrioid endometrial carcinoma. PLoS One.
9:e1101632014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Darda L, Hakami F, Morgan R, Murdoch C,
Lambert DW and Hunter KD: The role of HOXB9 and miR-196a in head
and neck squamous cell carcinoma. PLoS One. 10:e01222852015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roisman A, Huamán Garaicoa F, Metrebian F,
Narbaitz M, Kohan D, García Rivello H, Fernandez I, Pavlovsky A,
Pavlovsky M, Hernández L, et al: SOXC and MiR17-92 gene expression
profiling defines two subgroups with different clinical outcome in
mantle cell lymphoma. Genes Chromosomes Cancer. 55:531–540. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu C, Wang S, Zhu S, Wang H, Gu J, Gui Z,
Jing J, Hou X and Shao Y: MAP3K1-targeting therapeutic artificial
miRNA suppresses the growth and invasion of breast cancer in vivo
and in vitro. Springerplus. 5:112016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Yao X, Qin C, Luo P and Zhang J:
Investigation of the molecular mechanisms underlying metastasis in
prostate cancer by gene expression profiling. Exp Ther Med.
12:925–932. 2016.PubMed/NCBI
|
|
33
|
Zhu M, Xu Z, Wang K, Wang N and Li Y:
microRNA and gene networks in human pancreatic cancer. Oncol Lett.
6:1133–1139. 2013.PubMed/NCBI
|