Introduction
In China, gastric cancer (GC) is the second most
common malignancy (1). In 2012,
405,000 new cases were estimated to occur worldwide, which
accounted for 42.5% of the total cases (2). Due to a lack of early detection and
efficient screening in China, the majority of patients are often
diagnosed with distant metastases, leading to a poor 5-year
survival rate (3,4). Therefore, investigation of the
molecular pathogenesis of GC and the development of effective
diagnostic biomarkers are critical for GC patients.
miRNAs are small non-coding RNAs which play
significant roles in the initiation and progression of various
types of cancer (5–7). Thus, investigation of the mechanisms
underlying the regulation of GC by miRNAs is important. miR-15b
belongs to the miR-15 family, which is located on chromosome 3
(8). miR-15b-5p is a mature miRNA
spliced from the 5′-end of pre-miR-15b. Aberrant miR-15b expression
has been reported in a large number of cancers, and contributes to
increased proliferation, reduced apoptosis, tumor metastasis,
recurrence and poor patient prognosis (9–12).
However, to the best of our knowledge, little has been reported
regarding the role of miR-15b-5p in GC.
The present study revealed the expression profile of
miRNAs in GC tissues and miR-15b-5p was chosen for further study.
The expression pattern of miR-15b-5p was detected in GC cell lines,
tissues and plasma samples. Moreover, progestin and adipoQ receptor
family member 3 (PAQR3) was found to be a direct target of
miR-15b-5p. Furthermore, the mechanism of miR-15b-5p in regulating
GC metastasis was analyzed.
Materials and methods
GC tissues and cell lines
Forty pairs of GC and adjacent non-tumor tissues,
100 GC patient blood samples and 100 blood samples from healthy
subjects were obtained from the Affiliated Jiangning Hospital of
Nanjing Medical University (Nanjing, China). There were no
significant differences in sex, age and accompanying diseases
between the groups. All the tissue specimens were confirmed by two
pathologists and frozen in liquid nitrogen until RNA extraction.
The blood samples from the healthy subjects were collected from
individuals who were undergoing regular physical check-ups at our
hospital. The present study was approved by the Ethics Committee of
Nanjing Jiangning Hospital and consent forms were obtained from all
of the patients.
Four GC cell lines (AGS, BGC-823, SGC-7901 and
MGC-803), a GES-1 immortalized human gastric epithelial mucosa cell
line and the HeLa cell line were purchased from the Cell Bank of
the Chinese Academy of Sciences (Shanghai, China). Cells were grown
in RPMI-1640 medium (Invitrogen Life Technologies, Carlsbad, CA,
USA), supplemented with 10% fetal bovine serum (FBS) and 100 U/ml
penicillin/streptomycin (both from Invitrogen, Carlsbad, CA, USA)
at 37°C with 5% CO2 in an incubator (Thermo Fisher
Scientific, Waltham, MA, USA).
miRNA expression profiling
analysis
Three pairs of GC and adjacent non-tumor tissues
were adopted to analyze the miRNA expression profile by miRNA
microarray at KangChen Bio-tech (Shanghai, China). The miRNA
microarray analysis was performed as previously described (13).
Plasma preparation
Peripheral venous blood (2 ml) from patients or
healthy individuals was collected in a tube containing EDTA and
centrifuged at 1,800 × g for 5 min to isolate the plasma. Then, the
plasma was stored at −80°C until RNA extraction.
qRT-PCR
TRIzol and TRIzol LS reagents (both from Invitrogen)
were respectively used to extract RNA from tissues and cells and
that from plasma. The RNA concentration was quantitated on a
protein nucleic acid spectrophotometer (BioDrop, Cambridge, UK).
Complementary DNA was obtained using a reverse-transcription
PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio, Inc.,
Otsu, Japan) according to the manufacturer's instructions. The
qRT-PCR reaction was performed on an ABI StepOne Plus System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative
expression of miR-15b-5p was calculated using the 2−ΔΔCt
method (14), and U6 served as the
internal control. qRT-PCR primer set for miR-15b-5p
(miRQ0000417-1-1) and U6 (MQP-0202) were purchased from RiboBio
(Guangzhou, China). The primer sequences were as follows: PAQR3
forward, 5′-CTCAAGGACAACCCGTACATCAC-3′ and reverse,
5′-AAACTTTTGATACACAGCCTGGAC-3′; RECK forward,
5′-TGGCAAGAGTTTGATCGCTT-3′ and reverse,
5′-ATGGCTCCTTGATCTGACTGTG-3′; SMAD7 forward,
5′-ATGCTGTGCCTTCCTCCGCTG-3′ and reverse,
5′-CCACGCACCAGTGTGACCGA-3′; β-actin forward,
5′-GATCATTGCTCCTCCTGAGC-3′ and reverse,
5′-ACTCCTGCTTGCTGATCCAC-3′.
Cell transfection
BGC-823 and SGC-7901 cell lines were used for cell
transfection. Cells (3×105) were seeded into a 6-well
culture plate (Corning, Corning, Inc., Corning, NY, USA) and
transiently transfected with miRNA using Lipofectamine®
2000 (Invitrogen) after reaching 70–90% confluency. At 6 h after
transfection, the medium was replaced with fresh medium. A final
concentration of 50 nM miR-15b-5p mimic and negative control
(RiboBio), 2.5 ng of PAQR3 expression vector and control vector
(SunShineBio, Nanjing, China) were used for transfection.
Cell proliferation assay
Cell proliferation ability was measured by Cell
Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies,
Kumamoto, Japan). Cells (3×103) were planted into a
96-well plate (Corning) and transfected as described above. At 24,
48 and 72 h after transfection, the medium was replaced with a
mixture of 100 µl of fresh medium and 10 µl of CCK-8. After a 2-h
incubation, the optical density (OD) value was measured on a
microplate reader (iMark; Bio-Rad Laboratories, Inc., Hercules, CA,
USA) at 450 nm.
Wound healing assay
Cells were transfected as described above, and
starved for 12 h before scratching. Pipette tips (20 µl) were used
to generate a wound. At 0 and 72 h after the scratch, images of the
wound areas were captured under an inverted microscope (IX51;
Olympus Co., Tokyo, Japan) at a magnification of ×200.
Transwell assay
Twenty-four hours after transfection, the cells were
starved for 8 h, trypsinized (Gibco, Grand Island, NY, USA) and
suspended in serum-free medium at a concentration of
3×106/ml. Complete RPMI-1640 medium (600 µl) was added
into the lower well and 100 µl of cell suspension was added into
the upper chamber which was coated with Matrigel (BD Biosciences,
San Jose, CA, USA). After incubating for 48 h, the upper chamber
(Corning) was wiped with a swab to remove cells on the upper
surface, and then stained with crystal violet (Beyotime Institute
of Biotechnology, Shanghai, China) for 20 min. After washing with
distilled water, the cells that had invaded to the lower surface
were counted and captured under a microscope (Olympus) at a
magnification of ×200. Five random and different vision fields were
selected to count the invasive cell number.
Western blotting
Cells were lysed using radioimmunoprecipitation
assay (RIPA) lysis buffer (Beyotime Institute of Biotechnology) on
ice for 5 min, and were then centrifugalized at 12,000 × g for 5
min at 4°C to extract the protein. The concentration of the protein
was determined by the BCA method (BCA protein assay kit; Beyotime
Institute of Biotechnology). Proteins (30 µg) were separated on 10%
SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF)
membrane (EMD Millipore, Billerica, MA, USA). The membranes were
blocked with 5% non-fat milk for 1 h, and then incubated in the
primary antibodies at 4°C overnight, including β-actin (A5441,
mouse monoclonal, 1:8,000; Sigma-Aldrich, St. Louis, MO, USA),
E-cadherin (ab76055, mouse monoclonal; 1:500; Abcam, Cambridge,
UK), vimentin (sc-6260, mouse monoclonal, 1:500), PAQR3 (sc-161992,
goat polyclonal, 1:500) (both from Santa Cruz Biotechnology, Santa
Cruz, CA, USA). After being washed in Tris-buffered solution with
Tween-20 (TBST), the membranes were incubated in the secondary
antibody (ZB-2306, rabbit anti-goat and ZB-2305, goat anti-mouse
1:10,000; ZSGB-Bio, Ltd., Beijing, China) for 2 h. The protein
bands were detected using an enhanced chemiluminescence (ECL) kit
(Pierce, Rockford, IL, USA) on a FluorChem E System (ProteinSimple,
San Jose, CA, USA).
Dual-luciferase assay
The wild-type (WT) and mutant (Mut) binding
sequences of miR-15b-5p on the 3 untranslated region (3′UTR) of
PAQR3 were synthesized (SunShineBio) and cloned into the
pMIR-reportor vector (Invitrogen), which were respectively named
pMIR-report-PAQR3-3′UTR-WT and pMIR-report-PAQR3-3′UTR-Mut. HeLa
cells (1×105) were seeded into a 24-well plate (Corning)
and cultured overnight until reaching 70–90% confluency.
pMIR-report β-galactosidase reporter plasmid (500 ng),
pMIR-report-PAQR3-3′UTR-WT (500 ng) or pMIR-report-PAQR3-3′UTR-Mut
(500 ng), miR-15b-5p mimic (50 nM) or negative control (50 nM) were
co-transfected using Lipofectamine 2000. Cells were collected at 48
h post-transfection, and luciferase activities were measured using
the Dual-Luciferase Reporter Assay System (Promega, Madison, WI,
USA) on a TD-20/20 Luminometer (Turner BioSystems, Sunnyvale, CA,
USA) according to the manufacturer's instructions.
Statistical analysis
The data are expressed as the mean ± SD from at
least three independent experiments. Student's t-test (SPSS
Statistics 17; SPSS, Inc., Chicago, IL, USA) was adopted to
determine the statistical significance between the groups. A
P-value of <0.05 was considered to indicate a statistically
significant result.
Results
miRNA expression profile in GC
tissues
Three GC and paired non-tumor tissues were
respectively pooled, and subjected to microarray analysis. The heat
map is shown in Fig. 1A. Eighty-two
miRNAs were upregulated (ratio of tumor to non-tumor tissue ≥1.5),
while 179 were significantly downregulated (ratio of tumor to
non-tumor tissue <1.5), among which, miR-15b-5p was upregulated
7.6-fold and selected for further research.
miR-15b-5p is upregulated in GC cell
lines and tissues
In order to verify the miRNA array results,
miR-15b-5p expression was first detected in GC cell lines. The
results showed that miR-15b-5p expression levels were higher in 4
GC cell lines (SGC-7901, BGC-823, MGC-803 and AGS) than that in the
normal gastric epithelium GES-1 cells (Fig. 1B). In 40 pairs of tumor and matched
non-tumor tissue, miR-15b-5p was upregulated in 72.5% (27/40) of
the cases (Fig. 1C). miR-15b-5p
expression levels were much higher in the tumor tissues than that
in the non-tumor tissues (Fig. 1D).
In addition, the expression pattern of miR-15b-5p and
clinicopathological characteristics of the GC patients were
compared, and the data showed that miR-15b-5p expression was
correlated with the degree of tumor invasion and lymph node
metastasis (Table I).
 | Table I.Relationship between miR-15b-5p
expression and clinicopathological characteristics of the GC
patients. |
Table I.
Relationship between miR-15b-5p
expression and clinicopathological characteristics of the GC
patients.
|
| Relative miR-15b-5p
expression |
|
|---|
| Patient
characteristics | Low (n=13) | High (n=27) | Total (n=40) | P-value |
|---|
| Sex |
|
|
| 0.890 |
| Male | 8 | 16 | 24 |
|
|
Female | 5 | 11 | 16 |
|
| Age (years) |
|
|
| 0.768 |
| ≤60 | 3 | 9 | 12 |
|
|
>60 | 10 | 18 | 28 |
|
| Lauren's
classification |
|
|
| 0.305 |
| Diffuse
type | 7 | 19 | 26 |
|
|
Intestinal type | 6 | 8 | 14 |
|
| Differentiation |
|
|
| 0.720 |
| Poor | 8 | 15 | 23 |
|
|
Moderate/well | 5 | 12 | 17 |
|
| Degree of
invasion |
|
|
| 0.029a |
| Tunica
mucosa | 8 | 7 | 15 |
|
| Mucous
layer outside | 5 | 20 | 25 |
|
| Lymph node
metastasis |
|
|
| 0.042a |
| Yes | 4 | 19 | 23 |
|
| No | 9 | 8 | 17 |
|
Plasma miR-15b-5p level is correlated
with distant metastasis of GC
To evaluate the diagnostic value of plasma
miR-15b-5p in GC metastasis, the expression levels of plasma
miR-15b-5p were first examined in 100 GC patients and 100
cancer-free healthy control subjects. The results showed that
miR-15b-5p expression levels in the GC group were higher than those
in the control group (p=0.011; Fig.
2A). Next, GC plasma specimens were categorized into a distant
metastasis group (n=40) and a non-metastasis group (n=60) according
to the diagnosis. The data indicated that the distant metastasis
group had a higher miR-15b-5p level than that of the non-metastasis
group (p=0.018; Fig. 2B).
miR-15b-5p promotes GC metastasis
miR-15b-5p mimics or negative control were
introduced into BGC-823 and SGC-7901 cells to observe the
biological function of miR-15b-5p in GC cells. After transient
transfection, enforced expression levels of miR-15b-5p were
detected in the two GC cell lines (Fig.
3A). The effect of miR-15b-5p on gastric cell proliferation was
evaluated by CCK-8 assay. Data showed that when compared with the
control cells, exogenous expression of miR-15b-5p in the BGC-823
and SGC-7901 cells caused significant increases in cell growth
rates (Fig. 3B). To determine the
influence of miR-15b-5p on cell migration, a wound healing assay
was employed. The results indicated that miR-15b-5p overexpression
promoted the migratory ability of both GC cell lines (Fig. 3C). The invasive capacity of
miR-15b-5p was detected by Transwell assay. Enhanced miR-15b-5p
expression increased the number of cells that invaded to the lower
surface of the membrane (Fig. 3D).
The levels of epithelial marker E-cadherin and mesenchymal marker
vimentin were further examined to confirm whether miR-15b-5p
participates in the epithelial-mesenchymal transition (EMT)
process. The results showed that, when compared with the control
cells, the expression of E-cadherin was decreased while the
expression of vimentin was increased in the
miR-15b-5p-overexpressing GC cells (Fig. 3E).
PAQR3 is a target of miR-15b-5p in
GC
Online software programs (TargetScan and miRDB) were
used to predict the target of miR-15b-5p, and numerous genes were
found to contain the binding sequence of miR-15b-5p. Among these
candidates, PAQR3, RECK and SMAD7 were identified, as their
functions were related to cancer metastasis. The qRT-PCR results
showed that miR-15b-5p overexpression did not have marked effects
on RECK and SMAD7 expression, while PAQR3 was significantly
decreased in the miR-15b-5p-overexpressing BGC-823 and SGC-7901
cells (Fig. 4A and B). Luciferase
reporter assay was performed to confirm the result. Wild-type and
mutant binding sequences of miR-15b-5p on 3′UTR of PAQR3 were
amplified (Fig. 4C), and the
results showed that miR-15b-5p decreased the luciferase activity of
pMIR-report-PAQR3-3′UTR-WT but not of pMIR-report-PAQR3-3′UTR-Mut
(Fig. 4D). Collectively, these data
indicated that PAQR3 is a direct target of miR-15b-5p.
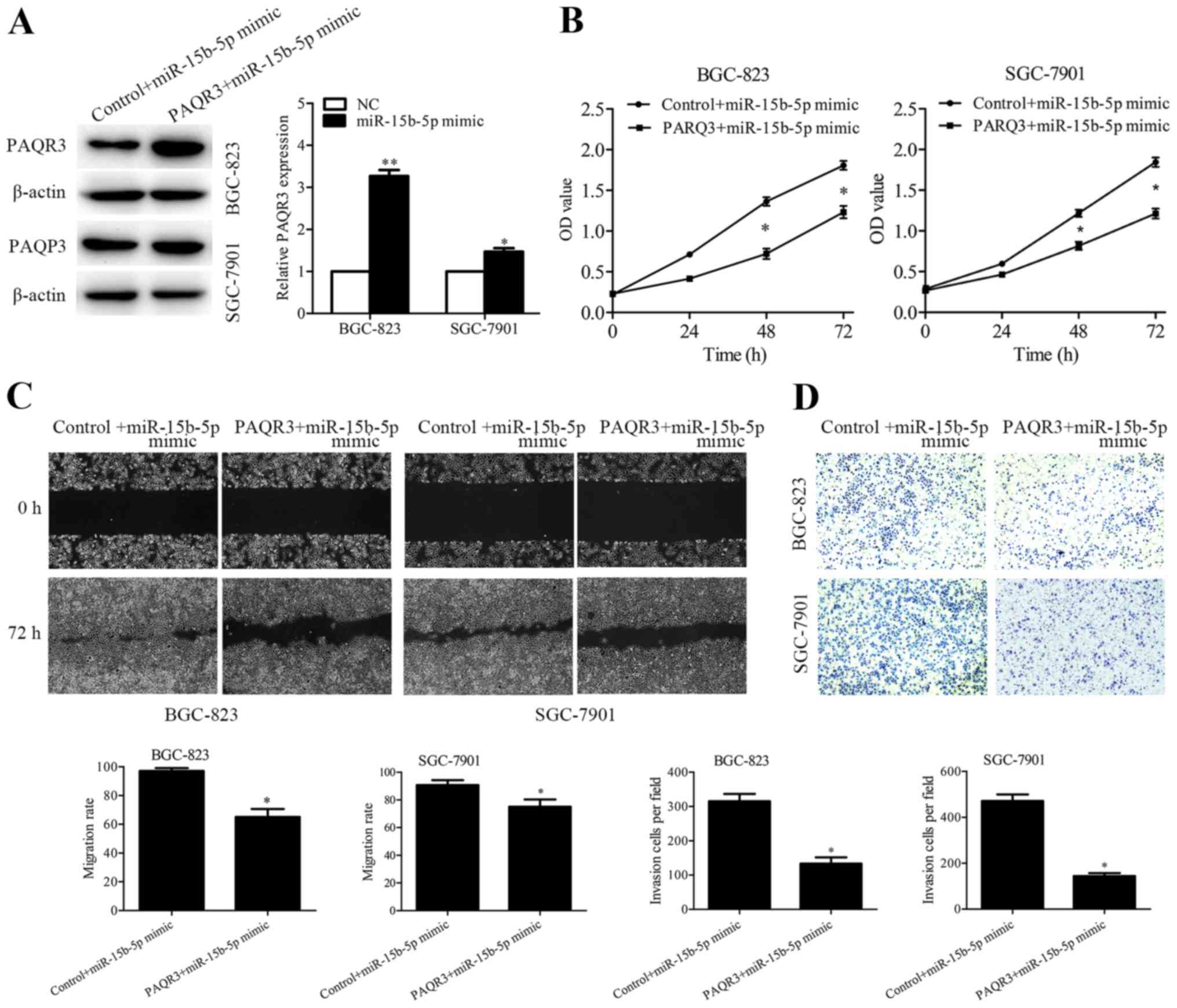
Overexpression of PAQR3 partially
rescues miR-15b-5p-induced cell proliferation, migration and
invasion
Rescue experiments were performed to verify whether
miR-15b-5p regulates GC metastasis by targeting PAQR3. PAQR3
expression vector and control vector were transfected into the
miR-15b-5p-overexpressing BGC-823 and SGC-7901 cells, and the
efficiency was detected by western blotting (Fig. 5A). In addition, CCK-8, wound healing
and Transwell assays were performed, and the results showed that
overexpression of PAQR3 partially abrogated the enhanced cell
proliferation (Fig. 5B), migration
(Fig. 5C) and invasion (Fig. 5D) induced by miR-15b-5p
overexpression.
Discussion
Previous studies have shown that miR-15b modulates
multidrug resistance (15) and
induces apoptosis (16) in gastric
cancer (GC). However, studies concerning miR-15b-5p in GC
metastasis are limited.
In the present study, the microarray analysis in GC
tissues displayed numerous abnormally expressed miRNAs. Among
these, miR-15b-5p was chosen for further research. The qRT-PCR
results showed that miR-15b-5p was markedly increased in both GC
cell lines and tissues. Moreover, high miR-15b-5p expression was
found to be correlated with lymph node metastasis and degree of
tumor invasion.
Recent studies have revealed that miRNAs can be
potential markers of cancer for their stability in tissues and
blood samples (17,18). Chen et al found that
circulating miR-15b in serum could be a potential marker for
detecting hepatocellular carcinoma (19). Given that miR-15b-5p was correlated
with lymph node metastasis and degree of tumor invasion, we aimed
to ascertain whether the plasma miR-15b-5p level could be a
biomarker of GC metastasis. The results showed that the plasma
levels of miR-15b-5p in GC patients were higher than those in the
healthy control, and patients with distant metastasis showed a
higher miR-15b-5p plasma level than the non-metastasis patients.
The results implied that high plasma miR-15b-5p may serve as a
metastatic marker of GC. Due to the small size of the GC plasma
samples, the results should be validated in a larger sample
size.
In GC cell lines, the effects of miR-15b-5p on GC
metastasis were studied by performing gain-of-function assays. We
found that miR-15b-5p overexpression enhanced cell proliferation,
migration and invasion. EMT is a critical process in tumor
metastasis (20,21). Thus, we assessed whether miR-15b-5p
could facilitate EMT. The expression of EMT markers was detected
and the results revealed that upregulation of miR-15b-5p promoted
the EMT process. These results indicated that miR-15b-5p
participates in GC metastasis partly by promoting cell
proliferation, migration, invasion and EMT.
Since miRNAs exert their function by downregulating
target genes (22), the targets of
miR-15b-5p in GC cells were investigated. Three
metastasis-associated genes (PAQR3, RECK and SMAD7) were identified
as potential miR-15b-5p targets by online software programs and the
expression levels of those selected genes were validated in
miR-15b-5p-overexpressing GC cells. Overexpression of miR-15b-5p in
GC cells inhibited the expression of PAQR3, but not RECK and SMAD7.
Luciferase assay indicated that PAQR3 is a direct target of
miR-15b-5p. Furthermore, we found that overexpression of PAQR3
partially rescued miR-15b-5p-induced cell proliferation, migration
and invasion. PAQR3 is a tumor-suppressor gene (23) that suppresses the proliferation,
migration, tumorigenicity, EMT and metastasis in different types of
cancers (24–27). In the present study, we found that
miR-15b-5p promoted GC cell metastasis by targeting PAQR3.
In summary, the present study demonstrated that
miR-15b-5p is upregulated in GC cell lines, GC patient tissues and
plasma. A high plasma miR-15b-5p level may act as a biomarker for
GC metastasis. miR-15b-5p overexpression enhances GC metastasis by
regulating PAQR3 expression.
Acknowledgements
The present study was supported by the Science and
Technology Development Plan of Jiangning District (grant no.
2014Dk04), and the Nanjing Health Bureau Research Project (grant
no. YKK14194).
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang H, Zhang H, Deng P, Liu C, Li D, Jie
H, Zhang H, Zhou Z and Zhao YL: Tissue metabolic profiling of human
gastric cancer assessed by 1H NMR. BMC Cancer. 16:3712016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jung J, Jung Y, Bang EJ, Cho SI, Jang YJ,
Kwak JM, Ryu DH, Park S and Hwang GS: Noninvasive diagnosis and
evaluation of curative surgery for gastric cancer using NMR-based
metabolomic profiling. Ann Surg Oncol. 21 Suppl 4:S736–S742. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao X, Wang Y, Deng R, Zhang H, Dou J,
Yuan H, Hou G, Du Y, Chen Q and Yu J: miR186 suppresses prostate
cancer progression by targeting Twist1. Oncotarget. 7:33136–33151.
2016.PubMed/NCBI
|
|
6
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen ZL, Wang B, Jiang KW, Ye CX, Cheng C,
Yan YC, Zhang JZ, Yang Y, Gao ZD, Ye YJ, et al: Downregulation of
miR-199b is associated with distant metastasis in colorectal cancer
via activation of SIRT1 and inhibition of CREB/KISS1 signaling.
Oncotarget. 7:35092–35105. 2016.PubMed/NCBI
|
|
8
|
MacLean JA II, King ML, Okuda H and
Hayashi K: WNT7A regulation by miR-15b in ovarian cancer. PLoS One.
11:e01561092016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li J, Chen Y, Guo X, Zhou L, Jia Z, Tang
Y, Lin L, Liu W and Ren C: Inhibition of miR-15b decreases cell
migration and metastasis in colorectal cancer. Tumour Biol.
37:8765–8773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Huang F, Wang J, Peng L and Luo
H: MiR-15b mediates liver cancer cells proliferation through
targeting BCL-2. Int J Clin Exp Pathol. 8:15677–15683.
2015.PubMed/NCBI
|
|
11
|
Sun G, Yan S, Shi L, Wan Z, Jiang N, Li M
and Guo J: Decreased expression of miR-15b in human gliomas is
associated with poor prognosis. Cancer Biother Radiopharm.
30:169–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang WL, Zhang JH, Wu XZ, Yan T and Lv W:
miR-15b promotes epithelial-mesenchymal transition by inhibiting
SMURF2 in pancreatic cancer. Int J Oncol. 47:1043–1053.
2015.PubMed/NCBI
|
|
13
|
Zhou RP, Chen G, Shen ZL and Pan LQ:
Cinobufacin suppresses cell proliferation via miR-494 in BGC-823
gastric cancer cells. Asian Pac J Cancer Prev. 15:1241–1245. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun H, Meng X, Han J, Zhang Z, Wang B, Bai
X and Zhang X: Anti-cancer activity of DHA on gastric cancer - an
in vitro and in vivo study. Tumour Biol. 34:3791–3800. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang X, Du L, Wang L, Li J, Liu Y, Zheng
G, Qu A, Zhang X, Pan H, Yang Y, et al: Serum microRNA expression
signatures identified from genome-wide microRNA profiling serve as
novel noninvasive biomarkers for diagnosis and recurrence of
bladder cancer. Int J Cancer. 136:854–862. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Chen J, Liu Y, Li S and Huang P:
Plasma miR-15b-5p, miR-338-5p, and miR-764 as biomarkers for
hepatocellular carcinoma. Med Sci Monit. 21:1864–1871. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai J, Qian C, Su M, Chen M and Chen J:
Gastrokine-2 suppresses epithelial mesenchymal transition through
PI3K/AKT/GSK3beta signaling in gastric cancer. Tumour Biol.
37:12403–12410. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Du J, Zheng J, Liu J, Xu R, Shen
T, Zhu Y, Chang J, Wang H, Zhang Z, et al: EGF-reduced Wnt5a
transcription induces epithelial-mesenchymal transition via
Arf6-ERK signaling in gastric cancer cells. Oncotarget.
6:7244–7261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu X, Li Z, Chan MT and Wu WK: PAQR3: A
novel tumor suppressor gene. Am J Cancer Res. 5:2562–2568.
2015.PubMed/NCBI
|
|
24
|
Ling ZQ, Guo W, Lu XX, Zhu X, Hong LL,
Wang Z, Wang Z and Chen Y: A Golgi-specific protein PAQR3 is
closely associated with the progression, metastasis and prognosis
of human gastric cancers. Ann Oncol. 25:1363–1372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Z, Ling ZQ, Guo W, Lu XX, Pan Y, Wang Z
and Chen Y: PAQR3 expression is downregulated in human breast
cancers and correlated with HER2 expression. Oncotarget.
6:12357–12368. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang W, Guo W, You X, Pan Y, Dong Z, Jia
G, Yang C and Chen Y: PAQR3 suppresses the proliferation, migration
and tumorigenicity of human prostate cancer cells. Oncotarget. Jun
3–2016.(Epub ahead of print). doi: 10.18632/oncotarget.9807.
|
|
27
|
Guo W, You X, Xu D, Zhang Y, Wang Z, Man
K, Wang Z and Chen Y: PAQR3 enhances Twist1 degradation to suppress
epithelial-mesenchymal transition and metastasis of gastric cancer
cells. Carcinogenesis. 37:397–407. 2016. View Article : Google Scholar : PubMed/NCBI
|