Introduction
Lung cancer is the leading cause of cancer-related
deaths worldwide (1,2). Non-small cell lung cancer (NSCLC)
accounts for ~85% of lung cancer. Despite improvement in cancer
treatment, the 5-year survival rate remains less than 10%. The
5-year survival rate is estimated to be 55–80% with an earlier lung
cancer diagnosis or surgery at an early stage (3). Preferential understanding of how NSCLC
develops and progresses plays an important role in early detection
and prevention as well as targeted treatment of NSCLC.
Previous studies have shown that DNA repair pathways
commonly exhibit functional overlap to ensure genomic stability;
therefore, challenging the perception that distinctive lesions are
repaired by different mechanisms in the mammalian genome. This
finding is especially true for oxidative damaged DNA, such as
oxidized bases (4), which might be
caused by oxidant exposure or ionizing radiation, but could also
result from normal cellular metabolism. Oxidized bases are
mutagenic and cytotoxic, and several previous studies have shown
that oxidized bases may contribute to neurodegeneration, aging and
cancer (5). Base excision repair
(BER) is an important DNA repair pathway that is responsible for
the repair of DNA base damage and single strand breaks caused by
X-rays, oxygen radicals or alkylating agents (6).
OGG1, a DNA repair glycosylase that localizes to
both the nucleus and mitochondria, is the main enzyme responsible
for the identification and excision of 8-oxoG lesions, which
produces G:C to T:A transversions (7–9). OGG1
is one of the components of the BER pathway. The human OGG1
(hOGG1) gene is found on chromosome 3p26.2, which is one of
the most frequent genomic deletion regions that contains some
potential tumor suppressor genes in various types of tumors, such
as NSCLCs (10). Previous studies
suggested that hOGG1 plays a role in several disease pathways,
including various cancers (11–13).
Despite such functional importance, how hOGG1 is regulated
at the transcriptional level in NSCLC remains largely unclear,
particularly via DNA methylation changes.
It is well known that epigenetic regulation, such as
DNA methylation, can alter gene expression (14–16).
DNA methylation frequently occurs in CpG islands, which are
frequently found in the 5′-untranslated regions (5′-UTR) of genes
(17). DNA methylation changes at
site-specific CpGs may play a crucial role in cancer progression,
including hypermethylation of tumor suppressor genes and
hypomethylation of oncogenes (14,18).
Our previous studies have shown that DNA methylation underlies
inactivation of the CpG island methylator phenotype (CIMP) and that
TSGs on 3p might be a frequent epigenetic event that confers an
increased risk of developing NSCLC (10,19).
Moreover, we previously showed that a methylated +58 CpG site in
the DCN 5′-UTR was associated with reduced DCN mRNA
expression in highly metastatic NSCLC cells (20). Hence, we hypothesize that
site-specific CpG methylation affects hOGG1 mRNA expression
levels in NSCLC.
Materials and methods
Tissue samples
Seventy-seven paired NSCLC tissues and adjacent
non-cancerous tissues were obtained after informed consent from
patients in the First Affiliated Hospital of Soochow University
between 2009 and 2013. Blood specimens were obtained after informed
consent from 30 randomly selected NSCLC and paired non-cancerous
lung patients. Blood was isolated by centrifugation at 3,500 rpm
for 20 min after blood sampling (10). The Revised International System for
Staging Lung Cancer was used to determine histological and
pathological diagnostics for NSCLC patients. No chemotherapy or
radiotherapy was given to patients with NSCLC before tissue
sampling. Tissue samples were stored at −80°C after being
snap-frozen. The present study was approved by the Ethics Committee
of the First Affiliated Hospital of Soochow University.
Cell culture and drug treatment
Human lung carcinoma cell lines (A549, H1650, H460,
SPC-A-1, 95C, 95D, H226 and SK-MES-1) were purchased from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China), and
human bronchial epithelial (HBE) cells from Shanghai Bogoo
Biotechnology, Co., Ltd. (Shanghai, China). Cell lines were seeded
and grown in RPMI-1640 medium (HyClone Laboratories, Inc., Logan,
UT, USA), with the exception of SK-MES-1, which was seeded in MEM
medium (HyClone Laboratories), with 10% heat-inactivated fetal
bovine serum (FBS; Gibco, Carlsbad, CA, USA) and L-glutamine and
antibiotics (Invitrogen, Carlsbad, CA, USA) in a humidified
incubator containing 5% CO2 at 37°C. Treatment with
5-aza-2′-deoxycitidine (5-Aza; Sigma-Aldrich, St. Louis, MO, USA)
was used to demethylate cells in culture according to the
previously described treatment protocol (10).
Quantitative determination of human
8-oxoguanine DNA glycosydase (hOGG1) concentrations and evaluation
of DNA damage in serum
The human 8-oxoguanine DNA glycosydase (hOGG1) ELISA
kit (cat. no. CSB-E12686h; Wuhan, Hubei, China) was used to
quantitatively determine hOGG1 concentrations in serum. Briefly,
standard controls and samples (100 µl) were added to each well and
covered with a provided adhesive strip; cells were incubated at
37°C for 2 h. The standard and sample results were recorded in a
plate layout that was provided by the manufacturer. Next, each well
received 100 µl of biotin-antibody (1x), and the plate was covered
with a new adhesive strip. Cells were incubated at 37°C for 1 h. If
the biotin-antibody (1x) appeared cloudy, it was warmed to room
temperature and mixed gently until the solution appeared uniform.
Next, each well received 100 µl of HRP-avidin (1x), and the plate
was covered with a new adhesive strip. Cells were incubated at 37°C
for 1 h. The aspirate was washed five times, and 90 µl of TMB
substrate was added to each well. Cells were incubated at 37°C for
15–30 min. Cell plates were protected from light. A total of 50 µl
of stop solution was added to each well, thoroughly mixed and hOGG1
was detected using a microplate reader.
8-OHdG, 8-hydroxyguanine and its 2′-deoxynucleoside
equivalent, 8-hydroxy-2′-deoxyguanosine (8-OHdG) are common
byproducts of DNA damage. During the repair of damaged DNA in
vivo by exonucleases, 8-hydroxy-2′-deoxyguanosine (8-OHdG) is
excreted (21,22); therefore, we used an OxiSelect
Oxidative DNA Damage ELISA kit (8-OHdG Quantitation; Cell Biolabs,
Inc., San Diego, CA, USA) to evaluate the level of DNA damage in
serum according to the manufacturers instructions (23–25).
The extracted DNA was dissolved in water to reach a concentration
of 1–5 mg/ml. The DNA was converted to single-stranded DNA by
incubating at 95°C for 5 min, then promptly chilling on ice. The
denatured DNA samples were digested to nucleosides by incubating
with 5–20 units of nuclease P1 at 37°C for 2 h in 20 mM sodium
acetate (pH 5.2). DNA samples were treated with 5–10 units of
alkaline phosphatase at 37°C for 1 h in 100 mM Tris (pH 7.5),
followed by centrifugation for 5 min at 6,000 × g. Subsequently,
the supernatant was used for the 8-OHdG enzyme-linked immunosorbent
assay (ELISA).
RNA extraction, cDNA synthesis and
quantitative real-time PCR (qRT-PCR)
Total RNA of cells and tissues were extracted by
adding 1.0 ml RNAiso Plus (Takara Bio, Osaka, Japan) according to
the manufacturers protocol. The RNA concentration was measured
using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA)
and synthesis of cDNA was performed using Reverse Transcriptase
M-MLV (Takara Bio) with reverse transcriptase. The sequences of
qRT-PCR for hOGG1 and GAPDH were as follows:
hOGG1, forward, 5′-ATCGTACTCTAGCCTCCACTCC-3′ and reverse,
5′-GTCAGTGTCCATACTTGATCCGC-3′; GAPDH, forward,
5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-TGCACCACCAACTGCTTAGC-3′.
qRT-PCR was performed using SYBR Premix ExTaq™ (Takara Bio)
according to the manufacturers instructions on an ABI StepOnePlus
Real-Time PCR system (Applied Biosystems, Foster City, CA, USA).
The PCR program was 50°C for 2 min, 95°C for 10 min, followed by 45
cycles of 95°C for 15 sec and 60°C for 1 min.
Quantitative methylation analysis of
DNA was performed using MassARRAY EpiTYPER assays
Quantitative methylation analysis of DNA was
performed using MassARRAY EpiTYPER assays (Sequenom, Inc., San
Diego, CA, USA) according to the protocol recommended by the
manufacturer (26). This system
uses matrix-assisted laser desorption/ionization time-of-flight
(MALDI-TOF) mass spectrometry in combination with RNA base-specific
cleavage (Mass Cleave). After bisulfite modification, genomic DNA
was amplified using MassArray primers. PCR products were introduced
to a T7 promoter sequence by the Beijing Bio-Miao Biotechnology,
Co., Ltd. (Beijing, China). Next, RNA products were transcribed
in vitro using T-base-specific cleavage, in which small RNA
fragments were obtained. The molecular weight of each fragment was
detected by flight mass spectrometry (MALDI-TOF) and methylation
levels were analyzed using EpiTyper software. PCR amplification
bias was controlled for by using DNA methylation standards (0, 20,
40, 60, 80 and 100%) and data was normalized by correction
algorithms based on an R statistical computing environment.
Construction of luciferase reporter
plasmids, transient transfection and luciferase assay
To construct a plasmid containing the hOGG1
promoter, we used pGL3 basic vector (Promega, Madison, WI, USA).
Briefly, an 88-bp fragment containing the predicted Sp1 target site
(positions +322–327) was chosen for the luciferase assay. The
wild-type and mutated fragment was directly synthesized (Genewiz,
Suzhou, China) and subcloned into the pGL3 basic vector to generate
pGL3-wild-type (WT: tggtccttgtctgggCGgggtctttgggCGtCGaCGaggcctggt
tctggg taggCGgggctactaCGgggCGgtgcctgctgtggaa) and pGL3-mutant
plasmid (Mut: tggtccttgtctgggCGgggtctttgggCGtCG
aCGaggcctggttctgggtaggCGgggctactaCTggAATgtgcctgctgt ggaa).
Subsequently, A549 and SPC-A1 cells were plated in a 24-well plate
and cotransfected with wild-type plasmid, mutated plasmid, or
pRL-TK plasmid using Lipofectamine 2000 (Life Technologies,
Carlsbad, CA, USA). After 48 h, cells were collected, and
luciferase activities were measured by the Dual-Luciferase reporter
assay kit (Promega). Each experiment was performed in
triplicate.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was carried out as previously described
(20). Briefly, immunoprecipitation
was performed using 5 µg anti-Sp1 antibody (Cell Signaling
Technology, Beverly, MA, USA). Purified ChIP DNA was subjected to
PCR, using primers specific for the hOGG1 promoter region
(positions +247 to +398) encompassing the putative Sp1-binding
site. Specific ChIP primers used for PCR were as follows: forward,
5-TAAGGGTCGTG GTCCTTGTC-3 and reverse, 5-TGGAGGCTAGAGTACGA
TGC-3.
Results
Serum levels of hOGG1 are decreased
and 8-OHdG levels are increased in NSCLC samples
hOGG1 gene encodes a DNA glycosylase that
catalyzes the excision and removal of 8-OH-dG adducts (27). A previous report has shown a
decrease of hOGG1 in the brain of Alzheimers patients (28), which caused the accumulation of
8-oxoG in the mitochondrial DNA of neurons and calpain-dependent
neuronal loss (12). In addition,
an association between the hOGG1 gene and lung cancer risk
has been reported (27,29). Here, we detected the level of hOGG1
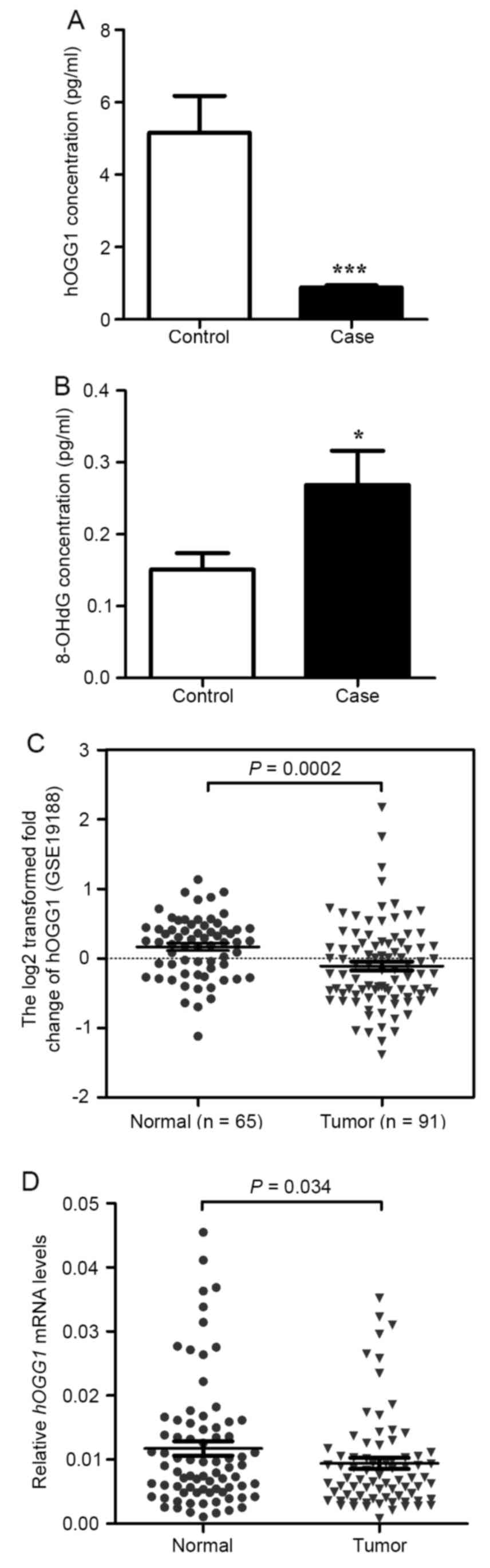
and 8-OH-dG using ELISA assays. As illustrated in Fig. 1A, the level of hOGG1 was lower in
NSCLC serum than in paired normal serum. Furthermore, we detected
8-OH-dG and found that the level of 8-OH-dG was higher in NSCLC
serum than in paired normal serum (Fig.
1B).
hOGG1 mRNA expression is downregulated
in NSCLC tissues
hOGG1 mRNA levels were significantly lower in
NSCLC tissues compared with adjacent non-cancerous lung tissues
(P=0.034; Fig. 1D). No significant
differences were observed in hOGG1 mRNA levels between NSCLC
tissues classified by various clinicopathological characteristics
(Table I). Moreover, a public data
set (GSE19188) containing 91 NSCLC tissues and 65 normal lung
tissues showed that hOGG1 mRNA expression was downregulated
in human NSCLC tissues (P=0.002; Fig.
1C).
 | Table I.Demographic and clinical
characteristics of NSCLC patients and the association with hOGG1
mRNA expression in tumor tissue specimens. |
Table I.
Demographic and clinical
characteristics of NSCLC patients and the association with hOGG1
mRNA expression in tumor tissue specimens.
|
Characteristics | No. of cases
(%) | hOGG1
expression | P-value |
|---|
| Age (years) |
|
≤65 | 36 (46.8) | 0.0098±0.0011 | 0.6832 |
|
>65 | 41 (53.2) | 0.0091±0.0013 |
|
| Sex |
|
Male | 52 (67.5) | 0.0092±0.0011 | 0.7653 |
|
Female | 25 (32.5) | 0.0098±0.0014 |
|
| Histology |
|
Adenocarcinomas | 35 (45.5) | 0.0119±0.0016 | 0.0707 |
|
Squamous cell carcinomas | 29 (37.7) | 0.0073±0.0010 |
|
|
Others | 13 (16.8) | 0.0075±0.0011 |
|
| Smokers |
|
Yes | 44 (57.1) | 0.0098±0.0012 | 0.6264 |
| No | 33 (42.9) | 0.0090±0.0016 |
|
| Clinical stage |
| I | 21 (27.3) | 0.0077±0.0012 | 0.2214 |
| II | 19 (24.7) | 0.0112±0.0017 |
|
|
III | 26 (33.8) | 0.0078±0.0012 |
|
| IV | 11 (14.2) | 0.0134±0.0036 |
|
| Lymph node |
|
Yes | 36 (46.8) | 0.0095±0.0012 | 0.9264 |
| No | 41 (53.2) | 0.0093±0.0012 |
|
| Distant
metastases |
|
Yes | 11 (14.3) | 0.0134±0.0036 | 0.0562 |
| No | 66 (85.7) | 0.0088±0.0008 |
|
hOGG1 mRNA expression is downregulated
in NSCLC cell lines and associated with DNA methylation
As shown in Fig. 2A,
hOGG1 mRNA levels were significantly lower in A549, H1650,
H460, 95C, 95D, H226 and SK-MES-1 cells compared with control HBE
cells, except for SPC-A-1. Our previous study supported the idea
that DNA methylation could be epigenetically responsible for
inactivation of tumor suppressor genes in NSCLC, and the
methylation of the hOGG1 gene promoter region occurs
frequently in NSCLC (10,19). Therefore, to determine whether
methylation of hOGG1 gene promoter is an alternative
mechanism underlying inactivation of hOGG1 mRNA expression,
we detected the mRNA expression after using demethylating agent
5-Aza on NSCLC cell lines. As illustrated in Fig. 2B, after 5-Aza treatment,
hOGG1 mRNA expression was increased in NSCLC cell lines
(A549, H460, SPC-A1, 95D and SK-MES-1); therefore, we suggest that
hOGG1 expression is silenced by DNA methylation.
Methylation levels of the +322–327 CpG
site is higher in NSCLC than adjacent non-cancerous lung tissues
and inversely correlated with hOGG1 mRNA expression
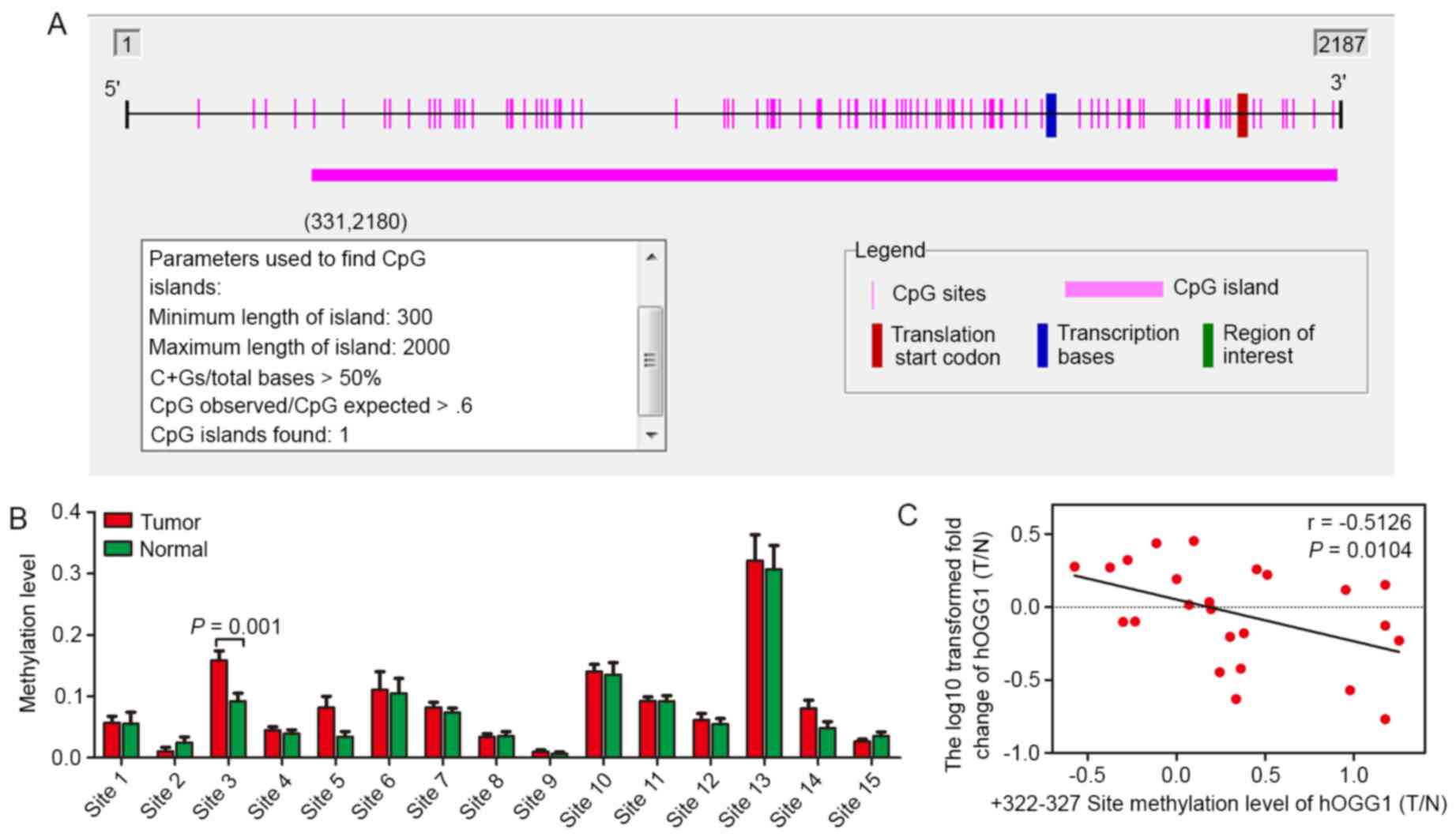
It is known that CpG islands are located −200 to
−1,000 bp from the transcription start site of a gene. Based on
this knowledge, we used Methyl Primer Express® software
to identify potential CpG sites in the hOGG1 promoter and
observed a GC-rich region (Fig.
3A). Furthermore, by using the MassARRAY EpiTYPER application,
methylation levels of CpG sites in the hOGG1 gene were
observed in 10 paired NSCLC tissues and adjacent non-cancerous lung
tissues (Tables II–IV). In the present study, we detected
three separate regions (position −1000 - −643, −463 - +34 and +35 -
+412), including 68 CpG sites, in the hOGG1 promoter.
Several CpG sites were detected between positions −1000 - −643 and
−463 - +34, and differences in methylation levels were detect
(P<0.05). However, we found that these regions have no
transcriptional binding sites or have lower frequency of
methylation. Consequently, we expanded the sample size to 25-paired
tissues to detect potential CpG sites within the third area
(position +35 - +412). We observed significantly higher methylation
of CpG site-3 in NSCLC patients compared with the control group
(Fig. 3B; Table V). Notably, the methylation level of
+322 - 327 site (T/N) was inversely correlated with hOGG1
mRNA level (T/N) in 25 paired tissues (P=0.0104; Fig. 4C).
 | Table II.CpG methylation of −1000 - −643 of
the hOGG1 promoter in NSCLC and paired normal tissues. |
Table II.
CpG methylation of −1000 - −643 of
the hOGG1 promoter in NSCLC and paired normal tissues.
|
|
| Tumor | Paried-normal |
|
|---|
|
|
|
|
|
|
|---|
| No. | CpG site | n | ∑X/n±s | n | ∑X/n±s | P-value |
|---|
| 1 | CpG_1 | 10 | 0.417±0.175 | 10 | 0.261±0.061 | 0.016 |
| 2 | CpG_3 | 10 | 1 | 10 | 0.983±0.018 | 0.010 |
| 3 | CpG_4.5 | 10 | 0.933±0.018 | 10 | 0.929±0.014 | 0.594 |
| 4 | CpG_6 | 10 | 0.900±0.024 | 10 | 0.889±0.026 | 0.339 |
| 5 | CpG_7.8 | 10 | 0.951±0.015 | 10 | 0.960±0.009 | 0.129 |
| 6 | CpG_10 | 10 | 0.835±0.064 | 10 | 0.828±0.082 | 0.835 |
| 7 | CpG_11.12 | 10 | 0.938±0.013 | 10 | 0.937±0.012 | 0.863 |
| 8 | CpG_13 | 10 | 1 | 10 | 1 |
|
 | Table IV.CpG methylation of +35 - +412 of the
hOGG1 promoter in NSCLC and paired normal tissues. |
Table IV.
CpG methylation of +35 - +412 of the
hOGG1 promoter in NSCLC and paired normal tissues.
|
|
| Tumor | Paried-normal |
|
|---|
|
|
|
|
|
|
|---|
| No. | CpG site | n | ∑X/n±s | n | ∑X/n±s | P-value |
|---|
| 1 | CpG_2 | 9 | 0.088±0.114 | 9 | 0.101±0.137 | 0.840 |
| 2 | CpG_3.4.5 | 9 | 0.031±0.056 | 9 | 0.048±0.068 | 0.554 |
| 3 | CpG_6.7 | 9 | 0.356±0.201 | 9 | 0.291±0.221 | 0.520 |
| 4 | CpG_8 | 9 | 0.035±0.040 | 9 | 0.021±0.035 | 0.431 |
| 5 | CpG_9.10 | 9 | 0.035±0.044 | 9 | 0.023±0.035 | 0.529 |
| 6 | CpG_11 | 9 | 0.195±0.196 | 9 | 0.17±0.166 | 0.769 |
| 7 | CpG_12 | 9 | 0.071±0.055 | 9 | 0.063±0.046 | 0.750 |
| 8 | CpG_14 | 9 | 0.022±0.017 | 9 | 0.021±0.018 | 0.898 |
| 9 | CpG_15 | 9 | 0.012±0.016 | 9 | 0.017±0.019 | 0.526 |
| 10 | CpG_16.17 | 9 | 0.101±0.075 | 9 | 0.073±0.062 | 0.405 |
| 11 | CpG_18.19 | 9 | 0.058±0.032 | 9 | 0.054±0.034 | 0.780 |
| 12 | CpG_20 | 9 | 0.025±0.026 | 9 | 0.032±0.025 | 0.599 |
| 13 | CpG_21.22 | 9 | 0.133±0.073 | 9 | 0.078±0.065 | 0.117 |
| 14 | CpG_23 | 9 | 0.051±0.039 | 9 | 0.035±0.031 | 0.370 |
| 15 | CpG_24.25 | 9 | 0.034±0.030 | 9 | 0.025±0.019 | 0.471 |
 | Table V.CpG methylation of +35 - +412 of the
hOGG1 promoter in NSCLC and paired normal tissues. |
Table V.
CpG methylation of +35 - +412 of the
hOGG1 promoter in NSCLC and paired normal tissues.
|
|
| Tumor | Paried-normal |
|
|---|
|
|
|
|
|
|
|---|
| No. | CpG site | n | ∑X/n±s | n | ∑X/n±s | P-value |
|---|
| 1 | CpG_2 | 25 | 0.057±0.050 | 25 | 0.055±0.092 | 0.945 |
| 2 | CpG_3.4.5 | 25 | 0.010±0.033 | 25 | 0.024±0.045 | 0.210 |
| 3 | CpG_6.7 | 24 | 0.158±0.079 | 24 | 0.092±0.066 | 0.001 |
| 4 | CpG_8 | 25 | 0.044±0.028 | 25 | 0.039±0.030 | 0.535 |
| 5 | CpG_9.10 | 25 | 0.082±0.091 | 25 | 0.034±0.042 | 0.226 |
| 6 | CpG_11 | 25 | 0.111±0.145 | 25 | 0.105±0.120 | 0.874 |
| 7 | CpG_12 | 25 | 0.082±0.043 | 25 | 0.074±0.035 | 0.475 |
| 8 | CpG_14 | 25 | 0.033±0.024 | 25 | 0.035±0.035 | 0.815 |
| 9 | CpG_15 | 24 | 0.011±0.015 | 24 | 0.006±0.013 | 0.276 |
| 10 | CpG_16.17 | 24 | 0.140±0.058 | 24 | 0.135±0.095 | 0.827 |
| 11 | CpG_18.19 | 24 | 0.092±0.032 | 24 | 0.092±0.043 | 0.955 |
| 12 | CpG_20 | 25 | 0.061±0.052 | 25 | 0.054±0.044 | 0.625 |
| 13 | CpG_21.22 | 25 | 0.321±0.209 | 25 | 0.306±0.195 | 0.802 |
| 14 | CpG_23 | 25 | 0.080±0.064 | 25 | 0.048±0.046 | 0.073 |
| 15 | CpG_24.25 | 25 | 0.026±0.017 | 25 | 0.035±0.032 | 0.245 |
Methylation of the +322–327 CpG site
inhibits hOGG1 mRNA expression by inhibiting Sp1 binding to the
hOGG1 promoter region
Because methylation of individual CpG dinucleotides
may contribute to cancer development (20,30),
we postulated that site-specific CpG methylation could alter the
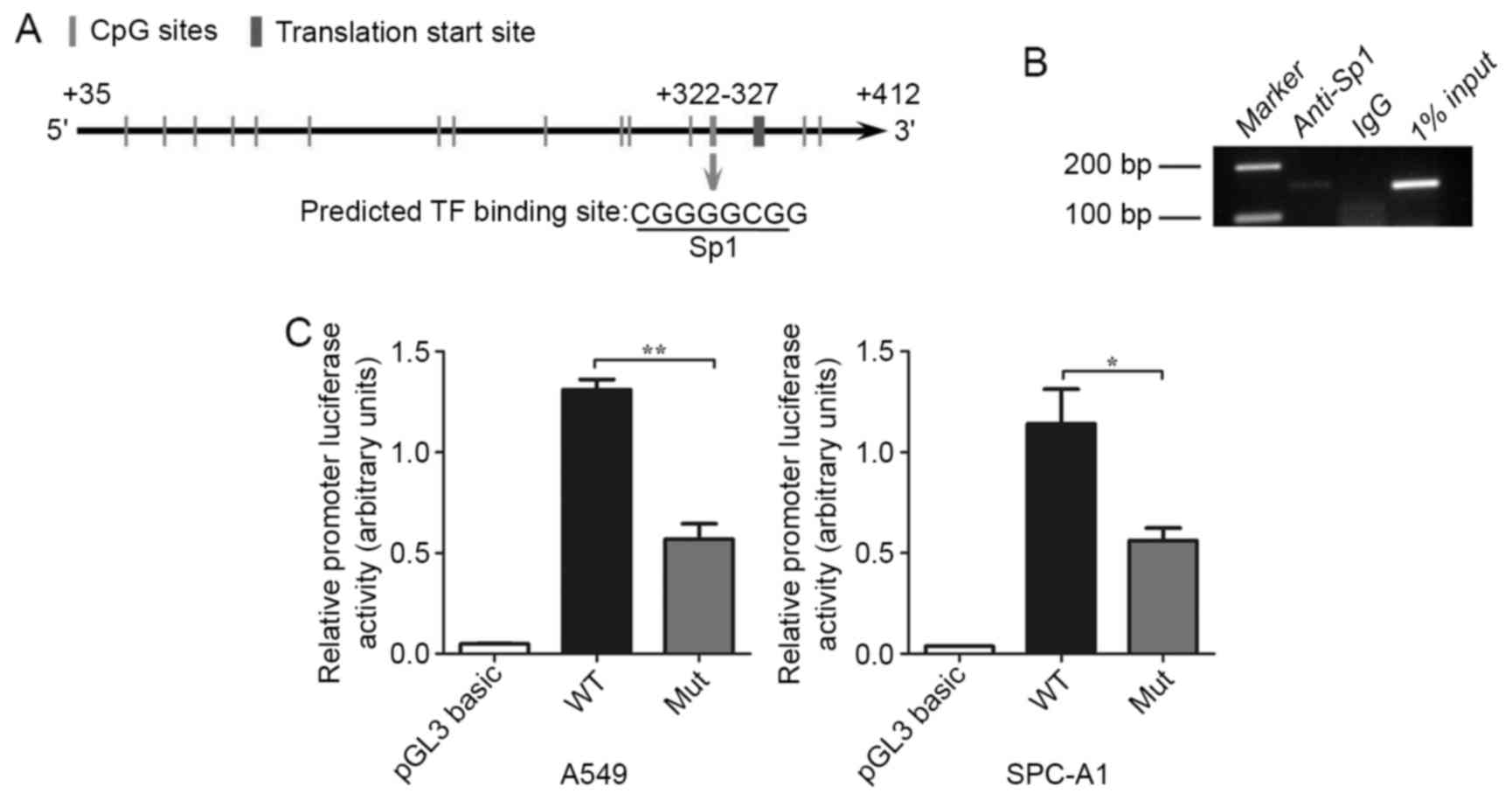
expression of hOGG1 in NSCLC. We found one putative
functional CpG site at position +322–327 in the proximal promoter
region of hOGG1 that was located within a transcription
factor Sp1-binding sequence (5-CGGGGCGG-3) using TRANSFAC, TFSEARCH
and Methyl Primer Express® software (Fig. 4A). We utilized ChIP analysis to
examine whether Sp1 binds to the hOGG1 proximal promoter
region at the +322–327 CpG site, and found that Sp1 was recruited
to the +322–327 CpG site in A549 cells (Fig. 4B). Collectively, the results
demonstrated that Sp1 may be target hOGG1 5-promoter
containing the +322–327 CpG site and thereby upregulate
hOGG1 expression in NSCLC cells. Subsequently, we
synthesized segments of the hOGG1 promoter that contained a
wild-type and a mutant Sp1 binding site for use in luciferase
reporter constructs; these constructs were transfected into A549
and SPC-A1 cells. The mutant Sp1 binding site construct displayed a
significant decrease in luciferase activity compared with the
wild-type construct (Fig. 4C). In
summary, our results suggest that the methylation of the +322–327
site in the hOGG1 promoter represses Sp1 binding and
regulates the expression of hOGG1 in NSCLC cells.
Discussion
Lung cancer is a leading cause of death throughout
the world. The morbidity and mortality of lung cancer has
significantly increased in the past decade in China (2). Studies have shown that NSCLC may
result from the accumulation of multiple genetic and/or epigenetic
aberrations. DNA methylation could be responsible for the
inactivation of the tumor suppressor genes found in NSCLC.
Endogenous and exogenous sources cause oxidatively
induced DNA damage in living organisms by a variety of mechanisms.
The resulting DNA lesions are mutagenic and, unless repaired, lead
to a variety of mutations and consequently to genetic instability,
which is a hallmark of cancer. The BER is known to preserve genome
integrity by removing damaged bases. It is the main pathway for the
repair of oxidized modifications in both nuclear and mitochondrial
DNA. Compelling evidence has shown that hOGG1 plays an important
role in tumorigenesis (11–13). Lower hOGG1 activity has been
reported in patients with NSCLC and downregulation of hOGG1 mRNA
and protein levels are compromised in their ability to remove
8-oxoG from their DNA (31,32). Despite such functional importance,
it remains largely unknown how hOGG1 is regulated at the
transcriptional level in human NSCLC, particularly through
epigenetic mechanisms, such as DNA methylation. Genetic
polymorphisms in individuals have recently been implicated to
account for some of the observed differences in lung cancer
susceptibility. The Ser326Cys hOGG1 polymorphism may be the most
frequently reported; it is associated with increased risk for lung
cancers, but its function is still controversial (33). Our previous studies showed that DNA
methylation could underlie epigenetic inactivation of the CpG
island methylator phenotype (CIMP) involving TSGs on 3p, suggesting
that this is a frequent epigenetic event that may confer an
increased risk of NSCLC (19,20). A
previous study reported that the CpG methylation of an adjacent
cytosine could moderately decrease the oxoGua excision rate,
whereas methylation opposite oxoGua could lower the rate of product
release (34).
We showed that +322–327 CpG methylation in the
hOGG1 5′-UTR decreased hOGG1 mRNA expression in NSCLC
tissues and cells using MassARRAY Epi-TYPER applications. Our
findings revealed that +322–327 CpG methylation may reduce the
recruitment of the transcriptional activator Sp1 to the
hOGG1 5′-UTR. Sp1 is a well-characterized sequence-specific
transcriptional factor that regulates a large number of
housekeeping and tissue-specific genes by binding to GC-rich DNA
sequences in the promoter region of many human genes (35,36).
Our findings support the idea that site-specific CpG
methylation may play an important role in cancer progression
(14,18,20,31).
To date, the mechanistic roles of individual CpG site methylation
are rarely reported in cancer (20,31).
This encourages us to investigate how the +322–327 CpG site
epigenetically affects the regulation of hOGG1 expression.
In the present study, we identified that the +322–327 CpG in the
hOGG1 proximal promoter is within a putative transcription
factor Sp1-binding sequence. Furthermore, cell-based and
biochemical analyses revealed that +322–327 CpG methylation can
inhibit hOGG1 transcriptional expression by interfering with
the recruitment of Sp1 to the hOGG1 promoter. However, we
cannot exclude the possibility of the roles of other functional CpG
sites in the hOGG1 promoter region.
Acknowledgements
We are grateful to all the patients who participated
in the present study. This study was supported by grants from the
National Natural Science Foundation of China (no. 31270940 to
J.-A.H., no. 81201575 to Z.-Y.L.), the Jiangsu Province Colleges
and Universities Natural Science Research Foundation (No.1 4KJB0017
to Z.L.), the Science and Technology Plan Projects of Suzhou (no.
SYS201612 to Z.-Y.L.) the Foundation of Health Care Rejuvenation by
Science and education (KJXW2016003 to Y.-Y.Z.), Huaian City Science
and Technology Support Program (no. HAS2015013-4), the Clinical
Medicine Center of Suzhou (no. Szzx201502), the Suzhou Key
Laboratory for Respiratory Medicine (no. SZS201617), the Societal
and Developmental Project of Suzhou (no. SS201630) and the Clinical
Key Speciality Project of China.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mulshine JL and Sullivan DC: Clinical
practice. Lung cancer screening. N Engl J Med. 352:2714–2720. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fortini P, Pascucci B, Parlanti E, DErrico
M, Simonelli V and Dogliotti E: 8-Oxoguanine DNA damage: At the
crossroad of alternative repair pathways. Mutat Res. 531:127–139.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Evans MD, Dizdaroglu M and Cooke MS:
Oxidative DNA damage and disease: induction, repair and
significance. Mutat Res. 567:1–61. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fortini P and Dogliotti E: Base damage and
single-strand break repair: Mechanisms and functional significance
of short- and long-patch repair subpathways. DNA Repair (Amst).
6:398–409. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Klungland A, Rosewell I, Hollenbach S,
Larsen E, Daly G, Epe B, Seeberg E, Lindahl T and Barnes DE:
Accumulation of premutagenic DNA lesions in mice defective in
removal of oxidative base damage. Proc Natl Acad Sci USA.
96:13300–13305. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Radicella JP, Dherin C, Desmaze C, Fox MS
and Boiteux S: Cloning and characterization of hOGG1, a human
homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl
Acad Sci USA. 94:8010–8015. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rosenquist TA, Zharkov DO and Grollman AP:
Cloning and characterization of a mammalian 8-oxoguanine DNA
glycosylase. Proc Natl Acad Sci USA. 94:7429–7434. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Z, Li W, Lei Z, Zhao J, Chen XF, Liu
R, Peng X, Wu ZH, Chen J, Liu H, et al: CpG island methylator
phenotype involving chromosome 3p confers an increased risk of
non-small cell lung cancer. J Thorac Oncol. 5:790–797. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sampath H, Vartanian V, Rollins MR, Sakumi
K, Nakabeppu Y and Lloyd RS: 8-Oxoguanine DNA glycosylase (OGG1)
deficiency increases susceptibility to obesity and metabolic
dysfunction. PLoS One. 7:e516972012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sheng Z, Oka S, Tsuchimoto D, Abolhassani
N, Nomaru H, Sakumi K, Yamada H and Nakabeppu Y: 8-Oxoguanine
causes neurodegeneration during MUTYH-mediated DNA base excision
repair. J Clin Invest. 122:4344–4361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peng Y, Li Z, Zhang S, Xiong Y, Cun Y,
Qian C, Li M, Ren T, Xia L, Cheng Y, et al: Association of DNA base
excision repair genes (OGG1, APE1 and XRCC1) polymorphisms with
outcome to platinum-based chemotherapy in advanced nonsmall-cell
lung cancer patients. Int J Cancer. 135:2687–2696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo- and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Das PM and Singal R: DNA methylation and
cancer. J Clin Oncol. 22:4632–4642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sekido Y, Fong KM and Minna JD: Progress
in understanding the molecular pathogenesis of human lung cancer.
Biochim Biophys Acta. 1378:F21–F59. 1998.PubMed/NCBI
|
|
18
|
Doi A, Park IH, Wen B, Murakami P, Aryee
MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al:
Differential methylation of tissue- and cancer-specific CpG island
shores distinguishes human induced pluripotent stem cells,
embryonic stem cells and fibroblasts. Nat Genet. 41:1350–1353.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Z, Zhao J, Chen XF, Li W, Liu R, Lei
Z, Liu X, Peng X, Xu K, Chen J, et al: CpG island methylator
phenotype involving tumor suppressor genes located on chromosome 3p
in non-small cell lung cancer. Lung Cancer. 62:15–22. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qian Q, Shi X, Lei Z, Zhan L, Liu RY, Zhao
J, Yang B, Liu Z and Zhang HT: Methylated +58CpG site decreases DCN
mRNA expression and enhances TGF-β/Smad signaling in NSCLC cells
with high metastatic potential. Int J Oncol. 44:874–882.
2014.PubMed/NCBI
|
|
21
|
Loft S, Vistisen K, Ewertz M, Tjønneland
A, Overvad K and Poulsen HE: Oxidative DNA damage estimated by
8-hydroxydeoxyguanosine excretion in humans: Influence of smoking,
gender and body mass index. Carcinogenesis. 13:2241–2247. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Loft S, Poulsen HE, Vistisen K and Knudsen
LE: Increased urinary excretion of 8-oxo-2-deoxyguanosine, a
biomarker of oxidative DNA damage, in urban bus drivers. Mutat Res.
441:11–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bhat HK and Singh B: Induction of
NAD(P)H-quinone oxidoreductase 1 by antioxidants in female ACI rats
is associated with decrease in oxidative DNA damage and inhibition
of estrogen-induced breast cancer. Carcinogenesis. 3:156–163.
2012.
|
|
24
|
Tzortzaki EG, Dimakou K, Neofytou E,
Tsikritsaki K, Samara K, Avgousti M, Amargianitakis V, Gousiou A,
Menikou S and Siafakas NM: Oxidative DNA damage and somatic
mutations: A link to the molecular pathogenesis of chronic
inflammatory airway diseases. Chest. 141:1243–1250. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Burnham EL, McCord JM, Bose S, Brown LA,
House R, Moss M and Gaydos J: Protandim does not influence alveolar
epithelial permeability or intrapulmonary oxidative stress in human
subjects with alcohol use disorders. Am J Physiol Lung Cell Mol
Physiol. 302:L688–L699. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ehrich M, Nelson MR, Stanssens P, Zabeau
M, Liloglou T, Xinarianos G, Cantor CR, Field JK and van den Boom
D: Quantitative high-throughput analysis of DNA methylation
patterns by base-specific cleavage and mass spectrometry. Proc Natl
Acad Sci USA. 102:15785–15790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park J, Chen L, Tockman MS, Elahi A and
Lazarus P: The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA
repair enzyme and its association with lung cancer risk.
Pharmacogenetics. 14:103–109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iida T, Furuta A, Nishioka K, Nakabeppu Y
and Iwaki T: Expression of 8-oxoguanine DNA glycosylase is reduced
and associated with neurofibrillary tangles in Alzheimers disease
brain. Acta Neuropathol. 103:20–25. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mambo E, Chatterjee A, De Souza-Pinto NC,
Mayard S, Hogue BA, Hoque MO, Dizdaroglu M, Bohr VA and Sidransky
D: Oxidized guanine lesions and hOgg1 activity in lung cancer.
Oncogene. 24:4496–4508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen H, Yang T, Lei Z, Wang L, Yang H,
Tong X, Yang WT, Zhao J, Gu Y, Chen Y, et al: RNF111/Arkadia is
regulated by DNA methylation and affects TGF-β/Smad signaling
associated invasion in NSCLC cells. Lung Cancer. 90:32–40. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sevilya Z, Leitner-Dagan Y, Pinchev M,
Kremer R, Elinger D, Rennert HS, Schechtman E, Freedman LS, Rennert
G, Paz-Elizur T, et al: Low integrated DNA repair score and lung
cancer risk. Cancer Prev Res (Phila). 7:398–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leitner-Dagan Y, Sevilya Z, Pinchev M,
Kramer R, Elinger D, Roisman LC, Rennert HS, Schechtman E, Freedman
L, Rennert G, et al: N-methylpurine DNA glycosylase and OGG1 DNA
repair activities: Opposite associations with lung cancer risk. J
Natl Cancer Inst. 104:1765–1769. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li H, Hao X, Zhang W, Wei Q and Chen K:
The hOGG1 Ser326Cys polymorphism and lung cancer risk: A
meta-analysis. Cancer Epidemiol Biomarkers Prev. 17:1739–1745.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kasymov RD, Grin IR, Endutkin AV, Smirnov
SL, Ishchenko AA, Saparbaev MK and Zharkov DO: Excision of
8-oxoguanine from methylated CpG dinucleotides by human
8-oxoguanine DNA glycosylase. FEBS Lett. 587:3129–3134. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and Krüppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suske G: The Sp-family of transcription
factors. Gene. 238:291–300. 1999. View Article : Google Scholar : PubMed/NCBI
|