Introduction
Colorectal cancer (CRC) is a highly common
malignancy in European countries (1–3) and
worldwide (3). According to
GLOBOCAN data (3), 1.36 million new
cases affecting 17.2/100,000 individuals (746,000 men and 614,000
women) are diagnosed worldwide each year; of these patients,
693,000 (373,000 men and 320,000 women) die, accounting for a
yearly mortality rate of 8.4/100,000. Crucially, more than 95% of
CRC patients may benefit from surgical treatment when diagnosed
early (4). CRC is sporadic in 90%
of patients, whereas in <10% of cases it is inherited (5,6) or is
a complication of inflammatory bowel disease (IBD), either
ulcerative colitis (UC) or Crohn's disease (CD) (7–9). In
the majority of cases, CRC develops from adenoma, a preclinical
benign precursor; the progression from early adenoma to invasive
cancer takes years (10,11).
A growing body of evidence supports the notion that
inflammation and CRC are interrelated (12,13).
The confirmed risk factors for the association between IBD and CRC
are duration, severity, and extent of the inflammatory disease,
concurrent primary sclerosing cholangitis (PSC), and a family
history of CRC (14,15). Whereas the link between chronic
inflammation and cancer is well recognized, the molecular
mechanisms involved in the association between IBD and CRC are
largely unknown. The need for a greater knowledge of their
underlying molecular biology, immune pathobiology, and genetic
processes has been driving an intense research effort (16–19).
High-temperature requirement serine protease A
(HtrA) is a heat-induced gene, indispensable for bacterial
survival at elevated temperatures, that was first identified in
Escherichia coli (20).
Subsequent studies have indicated that it degrades misfolded
protein in the periplasm at high temperatures (21). HtrA1, also called PRSS11 or L56, is
one of the 4 members of the human HtrA protein family (HtrA 1–4)
that has been identified by Zumbrunn and Trueb (22) in human embryonic lung fibroblasts.
As a serine protease, HtrA1 is a conserved protein that is widely
expressed in normal tissue in several species (22).
HtrA1 plays a pivotal role in the fibroblast growth
factor pathway, downregulating tumour progression (24). Changes in its expression have been
reported in conditions such as osteoarthritis, age-related macular
degeneration and cancer (25–27).
It tends to be downregulated in the metastatic foci of various
tumours compared with the primary tumour in a number of
malignancies, such as melanoma, sarcoma, neuroblastoma and lung
cancer (23,24), while its overexpression has been
hypothesized to inhibit growth and proliferation processes by
acting on tumour cell apoptosis, invasiveness and migration
(28). There are currently no data
on the immunohistochemical localization of HtrA1 in CRC, colorectal
adenoma or IBD.
In the present study, HtrA1 concentrations were
determined in colon mucosa from patients with CRC, adenoma with
high-grade dysplasia (AHD), adenoma with low-grade dysplasia (ALD),
UC of >10 year duration (UCL), UC of <5 year duration (UCS)
and colonic diverticulitis (D), and compared with the expression
found in normal colon tissues (NCTs) collected 5 cm from the CRC
lesions and in colon tissues from healthy controls (HC), to test
its ability to serve as a biomarker of these diseases.
Materials and methods
A total of 250 colon tissue specimens collected from
July 1, 2014 to July 30, 2015, and fixed with haematoxylin and
eosin according to a standard protocol were retrieved from the
pathology archives of the Sections of Pathological Anatomy of the
University of L'Aquila (L'Aquila, Italy), the Hospital of Teramo
(Teramo, Italy) and the Università Politecnica Delle Marche
(Ancona, Italy). Specimens were reviewed by 3 pathologists (G.C.,
G.Q. and R.M.) to confirm the histological diagnosis and select the
more representative areas for immunohistochemical analysis. The
study protocol was approved by the Ethics Board of Teramo Health
Service (26 June 2013) and was in line with the ethical standards
laid down in the 1964 Declaration of Helsinki.
Specimen characteristics
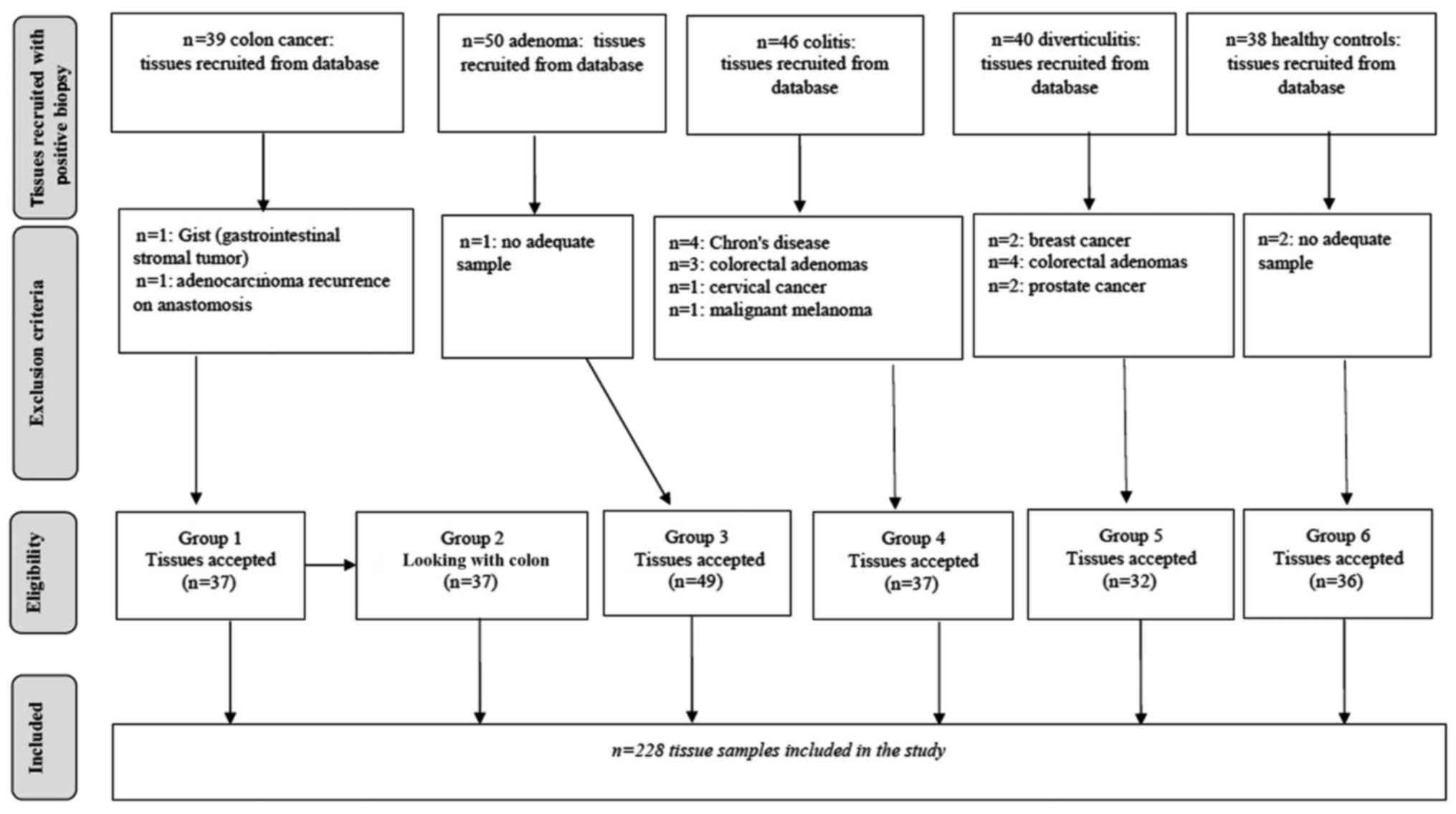
The 250 samples of colon mucosa examined are
described in Fig. 1. Of these, 22
could not be evaluated for the reasons reported in the figure,
leaving 228 CRC, AHD, ALD, UCL, UCS, D, NCT and HC specimens for
the analyses.
All specimens came from Italian Caucasian subjects
who suffered from a single disease. The 37 CRC tissue specimens, 17
(45.9%) from the right colon and 20 (54.1%) from the left colon,
were obtained from patients subjected to radical surgical resection
of the primary tumour. Two different tissue samples were obtained
from these specimens; one representative of CRC and another of NCT.
The latter samples were collected at a distance of 5 cm from the
neoplastic lesion and their normal features were established by
histology.
CRC specimens
Tumour staging was based on the TNM classification,
the Dukes system and histological grading. The latter included the
following classification: well differentiated (G1), moderately
differentiated (G2) and poorly differentiated (G3). Genetic and
epigenetic alterations were not assessed in these specimens.
Adenoma specimens
The 49 specimens of conventional adenoma (tubular,
villous, tubulovillous) were divided into 2 groups according to the
grade of dysplasia: 22 (44.9%) were adenomas with high-grade
dysplasia and focal low-grade dysplasia (AHD) and 27 (55.1%) were
adenomas with low-grade dysplasia (ALD). All adenomas were removed
by colonoscopy. Sessile serrated adenomas/polyps were not included
in this series.
Ulcerative colitis specimens
The 37 endoscopic biopsies from patients with UC
were divided according to disease duration. There were 18 (48.6%)
samples from patients with disease duration >10 years (UCL) and
19 (51.4%) from patients with disease duration <5 years (UCS).
All UC patients showed mild/moderate clinical activity and
mild/moderate mucosal lesions on colonoscopy.
Colonic diverticulitis specimens
The 32 samples from D patients had an average length
of 23 cm (sigmoid colon; range, 12–30 cm).
Healthy specimens
The HC mucosal biopsies were from 36 subjects
without a family history of CRC or IBD who underwent colonoscopy
due to clinical signs related to haemorrhoid disease or IBS. This
group did not include macroscopic lesions of large bowel
mucosa.
The same exclusion criteria were applied to all
tissue samples. Subjects with a personal or family history of
malignancy and those previously subjected to adjuvant therapy
(chemotherapy or radiotherapy) were excluded. Subjects with a
personal or family history of hereditary or familial CRC or
familial adenomatous polyposis were also excluded, since the
genetic and epigenetic mechanisms underpinning these forms are
different from those of sporadic and UC-associated CRC.
Immunohistochemistry
Paraffin sections were processed for HtrA1 analysis
as previously described (29).
Briefly, they were deparaffinized and rehydrated via xylene and a
graded series of ethyl alcohol, and treated with Tween 0.3% in
phosphate-buffered saline (PBS) for 25 min at room temperature.
Sections were incubated for 50 min with 3% hydrogen peroxide in
deionized water to inhibit endogenous peroxidase activity. To block
non-specific background, they were incubated for 1 h at room
temperature with normal serum. Sections were then incubated
overnight at 4°C with rabbit polyclonal HtrA1 antibody diluted 1:20
(ab38610; Abcam PLC, Cambridge, UK). After washing in PBS, they
were incubated with biotinylated secondary antibody (Vector
Laboratories, Burlingame, CA, USA). The peroxidase ABC method
(Vector Laboratories) was performed for 1 h at room temperature;
3′,3′-diaminobenzidine hydrochloride (Sigma, St. Louis, MO, USA)
was used as the chromogen. Sections were counterstained with
Mayer's haematoxylin, dehydrated and mounted with Eukitt solution
(Kindler GmbH and Co., Freiburg, Germany). Negative controls were
performed by omitting the primary or the secondary antibody.
Further negative controls were performed using an isotype control
antibody (rabbit IgG, cat. ab27478; Abcam). Placenta tissue was
used as a positive control.
Immunostaining evaluation
Immunohistochemical evaluations were independently
performed by 3 histologists (M.M., D.M. and C.L.). Immunostaining
was evaluated in mucosal glandular and surface epithelial and in
mucosal stromal cells and mucosal extracellular matrix. HtrA1
staining was scored as positive when brown precipitate was detected
in the epithelial or in the stromal compartment of the mucosa. The
expression level of HtrA1-stained cells by light microscopy was
evaluated in a semi-quantitative manner and ranked as follows: 0
(negative, 0% of positive cells), 1 (weak positive, 1–25% of
positive cells), 2 (moderate positive, 26–60% of positive cells),
and 3 (intense positive, 61–100% positive cells) at a magnification
of ×20, as previously described (30). A value 1 was used as the cut-off.
Accordingly, samples ranked 0 and 1 were classified as ‘low
expression’ and those ranked 2 and 3 were classified as ‘high
expression’.
Statistical analysis
Patient age is shown as the mean ± standard
deviation (SD). As in our previous study of HtrA1 (31), the Kolmogorov-Smirnov test was
performed in advance to check the normality of variables.
Differences in HtrA1 expression levels among groups were evaluated
using the non-parametric Kruskal-Wallis test. The inter-observer
agreement of the immunohistochemical evaluations (colon mucosa) was
calculated as follows: intensity, weak-high, of brown staining in
epithelial cells in regards to the epithelial compartment, and
intensity of the brown staining in cells and extracellular matrix
(ECM) in regards to the stromal compartment (lamina propria). The
inter-observer agreement was expressed using Cohen's κ statistic as
follows: κ, <0.20 poor, 0.21–0.40 fair, 0.41–0.60 moderate,
0.61–0.80 good and 0.81–1.00 very good agreement. The agreement was
required, since the HtrA1 antibody is not applied in routine
diagnosis and cannot therefore be determined with an automatic
analyzer. In addition, the observers were able to evaluate the
background in the sections. The inter-observer agreement was
calculated for the main groups (CRC, NCT, HC, UC, D and adenoma)
not for the subgroups, since the histological elements that were
evaluated were always the same.
Cohn's κ expresses the degree of agreement beyond
chance. Cell counts were performed by 3 observers with different
experience in light microscopy (LM) who were not aware of the
histopathological diagnosis. Their LM experience was expressed by a
score that took into account the number of years spent specializing
in LM (score: 1, <5 years; 2, >5 years); their daily LM work
(1, <3 h/day; 2, >3 h/day), and the number of
workshops/seminars attended (1, <5; 2, >5). These criteria
allowed classifying the observers into those with strong experience
(A and B) and limited experience (C). All evaluations were
performed in a blinded manner. Global agreement was estimated for
each group of samples.
The statistical significance of contingency tables
was verified by the χ2 or Fisher's exact test as
appropriate.
The present study had 91% power considering the
following parameters: p=0.70 (expected proportion in CRC group),
p=0.30 (expected proportion in HC group), sample size=37 (CRC
group), sample=36 (HC group), α=0.05.
All p-values <0.05 were considered statistically
significant. There were no missing data. All analyses were carried
out using SAS/STAT statistical software.
Results
A total number of 228 tissue samples were examined
and divided into 6 groups (Fig. 1):
group 1, 37 samples from CRC patients; group 2, 37 NCT samples from
group 1 patients; group 3, 49 adenoma samples subdivided into
low-degree dysplasia (ALD) and high-degree dysplasia (AHD); group
4, 37 UC samples subdivided into short (UCS) and long disease
duration (UCL); group 5, 32 samples from patients with D; group 6,
36 HC samples. The characteristics of each group are summarized in
Table I. Since HtrA1 is a secreted
protein, immunohistochemical HtrA1 staining was detected either in
the cytoplasm of colonic epithelial or in stromal cells and ECM of
the colon mucosa. HtrA1 staining was scored as low or high
expression both in the epithelial and in the stromal
compartment.
 | Table I.Characteristics and site of tissue
samples studied. |
Table I.
Characteristics and site of tissue
samples studied.
|
Characteristics | CRC | AHD | ALD | UCL | UCS | D | NCT | HC |
|---|
| Total no. of
cases | 37 | 22 | 27 | 18 | 19 | 32 | 37 | 36 |
| Age (years) (mean ±
SD) | 65.5±9.4 | 66.6±9.1 | 68.9±9.2 | 55.4±14.5 | 53.1±17.4 | 58.4±16.2 | 65.5±9.4 | 53.0±10.0 |
| Sex, n (%) |
|
Female | 15 (40.5) | 6 (27.3) | 8 (29.6) | 10 (55.6) | 12 (63.2) | 13 | 15 (40.5) | 23 (63.9) |
|
Male | 22 (59.5) | 16 (72.7) | 19 (70.4) | 8 (44.4) | 7 (36.8) | 19 | 22 (59.5) | 13 (36.1) |
| Anatomic site, n
(%) sampling |
| Right
colon | 17 (45.9) | 5 (22.7) | 7 (25.9) |
|
|
| 17 | 18 (50) |
| Left
colon | 20 (54.1) | 9 (40.9) | 14 (51.9) |
|
|
| 20 | 17 (47.2) |
|
Transversum |
| 2 (9.1) | 5 (18.5) |
|
|
|
| 1 (2.8) |
|
Rectum |
| 6 (27.3) | 1 (3.7) |
|
|
|
|
|
| Left
colitis |
|
|
| 11 (61.1) | 12 (31.6) |
|
|
|
|
Sub-total colitis |
|
|
| 1
(5.6) | 1 (5.3) |
|
|
|
| Total
colitis |
|
|
| 6
(33.3) | 6 (63.1) |
|
|
|
| Sigmoid
colon |
|
|
|
|
| 32 |
|
|
| Grading, n (%) |
| G1 | 12 (32.4) |
|
|
|
|
|
|
|
| G2 | 22 (59.5) |
|
|
|
|
|
|
|
| G3 | 3 (8.1) |
|
|
|
|
|
|
|
| TNM, n (%) |
| pT1
(N0) | 7 (19.0) |
|
|
|
|
|
|
|
| pT2 (11
N0 and 2 N1) | 13 (35.1) |
|
|
|
|
|
|
|
| pT3 (10
N0, 3 N1, 1 N1c, 1 N2a, 1 N2b) | 16 (43.2) |
|
|
|
|
|
|
|
| pT4
(N2a) | 1 (2.7) |
|
|
|
|
|
|
|
| Dukes stage |
| A | 18 (48.6) |
|
|
|
|
|
|
|
| B | 12 (32.4) |
|
|
|
|
|
|
|
| C | 7 (19.0) |
|
|
|
|
|
|
|
In HC and NCT specimens, HtrA1 expression was
homogeneous in all segments of the colon and rectum, suggesting
that its expression patterns in the different conditions are not
affected by lesion site. HtrA1 expression did not differ in
relation to age. HtrA1 was absent or weakly expressed in the
epithelium and in the stromal compartment of CRC sections (Fig. 2A), whereas NCT and HC samples showed
a strong positive reaction in both compartments (Fig. 2G and H). In the adenoma samples,
HtrA1 expression was weaker in the epithelium than in the stroma,
without clear differences between AHD (Fig. 2B) and ALD (Fig. 2C) specimens. The immunostaining
pattern of UCL tissue was very similar to that of CRC samples, with
very weak staining in the epithelium and stroma (Fig. 2D), whereas UCS and D tissues showed
moderate-high staining in the stroma and moderate-weak staining in
the epithelium (Fig. 2E and F).
 | Figure 2.HtrA1 immunostaining in (A) CRC, (B)
AHD, (C) ALD, (D) UCL, (E) UCS, (F) D, (G) NCT and (H) HC tissue.
HtrA1 was weakly expressed in the epithelial and stromal
compartment in (A) CRC compared with (G) NCT and (H) HC specimens.
HC (H) shows strong HtrA1 immunostaining. Staining is clearly
evident in the stroma compartment of (B) AHD and (C) ALD samples
and is moderate and inhomogeneous in the epithelium. HtrA1 is
weakly expressed in (D) UCL, whereas (E) UCS specimens show
homogenous staining in the stroma and moderate staining in the
epithelial compartment. (F) Moderate and homogeneous immunostaining
is observed in diverticulitis samples (magnification, ×20; green
scale bar, 65 µm). CRC, colorectal cancer; AHD, adenoma with
high-grade dysplasia; ALD, adenoma with low-grade dysplasia; UCL,
ulcerative colitis of >10 year duration; UCS, ulcerative colitis
of <5 year duration; D, colonic diverticulitis; NCT, normal
colon tissues collected 5 cm from the CRC lesion; HC, healthy colon
mucosa. |
The results of HtrA1 expression in the tissue groups
are summarized in Table II
according to staining intensity (low/high). Statistically
significant differences among the groups were found both in the
stromal (p<0.0001) and the epithelial (p<0.0002) compartment.
In the stromal compartment, low HtrA1 expression was most frequent
in samples from CRC and UCL patients (83.8 and 83.3%) and least
frequent in HC tissue (2.8%); the difference was significant
(p<0.0001). In the epithelial compartment, low HtrA1 expression
was most frequent in UC patients with disease duration >10 years
(94.4%) and least common in HC samples (8.3%).
 | Table II.HtrA1 expression according to the
pathologies considered. |
Table II.
HtrA1 expression according to the
pathologies considered.
|
| Low expression | High
expression |
|
|---|
|
|
|
|
|
|---|
| Groups | No. | (%) | No. | (%) |
P-valuea |
|---|
| Stromal
compartment |
|
CRC | 31 | 83.8 | 6 | 16.2 | <0.0001 |
|
AHD | 1 |
4.4 | 22 | 95.6 |
|
|
ALD | 1 |
3.9 | 25 | 96.1 |
|
|
UCL | 15 | 83.3 | 3 | 16.7 |
|
|
UCS | 5 | 26.3 | 14 | 73.7 |
|
| D | 18 | 56.2 | 14 | 43.8 |
|
|
NCT | 10 | 27.0 | 27 | 73.0 |
|
| HC | 1 |
2.8 | 35 | 97.2 |
|
| Epithelial
compartment |
|
CRC | 24 | 64.9 | 13 | 35.1 | <0.0001 |
|
AHD | 10 | 43.5 | 13 | 56.5 |
|
|
ALD | 6 | 23.1 | 20 | 76.9 |
|
|
UCL | 17 | 94.4 | 1 |
5.6 |
|
|
UCS | 10 | 52.6 | 9 | 47.4 |
|
| D | 17 | 53.1 | 15 | 46.9 |
|
|
NCT | 9 | 24.3 | 28 | 75.7 |
|
| HC | 3 |
8.3 | 33 | 91.7 |
|
Testing for differences in HtrA1 expression between
diseases highlighted a number of statistically significant
differences. Stromal compartment: CRC vs. AHD (p=0.0001); CRC vs.
ALD (p=0.0001); CRC vs. UCL (p=1.0); CRC vs. UCS (p=0.0001); CRC
vs. D (p=0.03); CRC vs. NCT (p=0.0001); CRC vs. HC (p=0.0001); UCL
vs. UCS (p=0.0008); UCL vs. HC (p=0.0001); UCL vs. AHD (p=0.0001);
and UCL vs. ALD (p=0.0001) (Table
III). Epithelial compartment: CRC vs. ALD (p=0.007); CRC vs.
UCL (p=0.04); CRC vs. NCT (p=0.001); CRC vs. HC (p=0.0001); and AHD
vs. HC (p=0.02); UCL vs. UCS (p=0.01); UCL vs. HC (p=0.0001); UCL
vs. AHD (p=0.001); UCL vs. ALD (p=0.0001) (Table III).
 | Table III.HtrA1 expression in stromal and
epithelial compartments: comparison between the different groups
investigated. |
Table III.
HtrA1 expression in stromal and
epithelial compartments: comparison between the different groups
investigated.
| Disease | Stromal
P-value | Epithelium
P-value |
|---|
| CRC vs. AHD | 0.0001 | 0.09 |
| CRC vs. ALD | 0.0001 | 0.007 |
| CRC vs. UCL | 1.0 | 0.04 |
| CRC vs. UCS | 0.0001 | 0.57 |
| CRC vs. D | 0.03 | 0.32 |
| CRC vs. NCT | 0.0001 | 0.001 |
| CRC vs. HC | 0.0001 | 0.0001 |
| ALD vs. AHD | 0.52 | 0.54 |
| AHD vs. HC | 1.0 | 0.02 |
| ALD vs. HC | 1.0 | 0.30 |
| UCL vs. UCS | 0.0008 | 0.01 |
| UCL vs. HC | 0.0001 | 0.0001 |
| UCL vs. AHD | 0.0001 | 0.001 |
| UCL vs. ALD | 0.0001 | 0.0001 |
| HC vs. NCT | 0.14 | 0.56 |
The data regarding overall and pairwise
inter-observer agreement in the stromal and epithelial compartment
are reported in Table IV. In UC
patients, full agreement (k=100%) was found for immunostaining
evaluation of the stromal compartment and very good agreement for
the epithelial compartment (k=94.6%). In the stromal compartment,
pairwise agreement (A vs. B) was very good for CRC (k=89%), adenoma
(k=93%), HC (k=84%), and NCT (k=83%), and good for D (k=75%),
whereas in the epithelial compartment it was very good for D
(k=81%) and moderate for the other diseases (Table IV). The differences in the level of
agreement between the evaluation of epithelial and stromal
immunostaining are essentially due to the histological
characteristics of the colon epithelium, which is particularly rich
in goblet cells. These cells contain mucin drops that occupy nearly
the whole cell cytoplasm, making the assessment of cytoplasmic
immunostaining more difficult in the epithelial compartment than in
the stroma, where cells do not contain mucin drops.
 | Table IV.Agreement on immunostaining
assessment. |
Table IV.
Agreement on immunostaining
assessment.
|
|
| Agreement among
observers A, B and C Pairwise comparisons (Cohen's κ
statistic) |
|---|
|
|
|
|
|---|
| Disease | Overall agreement
(%) | A vs. B (95%
CI) | A vs. C (95%
CI) | B vs. C (95%
CI) |
|---|
| Stromal
compartment |
| CRC |
92.8 | 0.89
(0.69–1.0) | 0.65
(0.35–0.95) | 0.75
(0.49–1.0) |
| A |
95.2 | 93.7
(93.1–94.3) | 93.7
(93.1–94.3) | 93.7
(93.1–94.3) |
| UC | 100.0 | 1.0 | 1.0 | 1.0 |
| D |
74.0 | 0.75
(0.52–0.98) | 0.57
(0.30–0.84) | 0.69
(0.46–0.93) |
| NCT |
79.3 | 0.83
(0.65–1.0) | 0.77
(0.57–0.98) | 0.82
(0.62–1.0) |
| HC |
92.8 | 0.84
(0.67–1.0) | 0.53
(0.28–0.78) | 0.5
(0.25–0.75) |
| Epithelial
compartment |
| CRC |
83.8 | 0.63
(0.40–0.86) | 0.63
(0.40–0.86) | 0.77
(0.54–0.98) |
| A |
89.1 | 75.6
(0.56–0.96) | 82.6
(0.66–0.98) | 68.7
(0.48–0.89) |
| UC |
94.6 | 0.66
(0.35–0.96) | 0.51
(0.13–0.88) | 0.73
(0.46–1.0) |
| D |
87.4 | 0.81
(0.61–1.0) | 0.69
(0.45–0.93) | 0.62
(0.35–0.88) |
| NCT |
87.4 | 0.65
(0.35–0.95) | 0.65
(0.35–0.95) | 0.56
(0.26–0.87) |
| HC |
85.6 | 0.77
(0.47–1.0) | 0.54
(0.14–0.93) | 0.60
(0.25–0.95) |
Discussion
To the best of our knowledge, the present study is
the first to examine HtrA1 expression in CRC, AHD, ALD, UCL, UCS,
NCT and HC samples by immunohistochemistry. HtrA1 immunostaining
was detected in the epithelium and stroma (lamina propria) of the
colon mucosa in all the tissue samples analysed. Its expression
patterns in the different HC and NCT colon segments examined showed
no differences, suggesting that the various conditions depend on
disease type rather than colon segment. Age was also unrelated to
HtrA1 expression characteristics. The main finding of the present
study was the significantly reduced HtrA1 expression found in CRC
and UCL tissues compared with that in the NCT and HC samples and
with the tissue specimens from the other patients. In particular,
HtrA1 expression was similar in the stromal compartment of UCL and
CRC sections whereas in the epithelial compartment it was weaker in
UCL than CRC tissue. HtrA1 expression was also significantly lower
in UCL than UCS samples, suggesting that it declines with disease
duration, particularly in the stromal compartment.
HtrA1 has a functional role both in epithelium and
stroma compartments. HtrA1 is a serine protease involved in
important physiological processes, including maintenance of
mitochondrial homeostasis, apoptosis and cell signalling (32,33).
HtrA1 has also the capacity to degrade numerous extracellular
matrix (ECM) proteins, produced by the stromal cells, as well as to
decrease biological function of vascular endothelial cells, and
thus, able to act even in the stromal compartment (34,35).
The effects of HtrA1 on proliferation and apoptosis of epithelial
cells and the effects on the ECM remodelling can intervene at
different phases of colorectal carcinogenesis. In UCL, both the
epithelial proliferation and apoptosis and ECM components in the
mucosa and submucosa are altered (18,36,37),
findings that may link the risk of CRC of UCL to the altered
expression of HtrA1 observed in this subset of UC patients.
UCL is a well-known risk factor for CRC development,
whereas UCS and D patients are not at great risk (16,18,38,39).
The similar HtrA1 expression patterns found in UCS and D samples in
the present study provide support for this concept. It may thus be
hypothesized that different pathological inflammatory
microenvironments induce a reduction in HtrA1 expression in CRC and
UCL, and that such low HtrA1 levels underpin the risk of
progression to CRC. Moreover, different inflammatory soluble
factors are found in acute and chronic inflammation. In
long-standing chronic inflammation some molecules foster epithelial
hyperproliferation and the genetic and epigenetic alterations that
promote CRC (17–19,38–40).
It is clearly useful to establish which of the specific chronic
inflammatory mediators of UCL correlate with HtrA1 expression, but
this was outside the scope of the present study. However, the
present data do suggest that HtrA1 may provide an important link
between UCL and CRC, and that epithelium and stroma are
differentially affected (41).
Notably, it has been suggested that HtrA1 responds to different
environments in different ways, probably due to its secretory
characteristic (42), and that high
HtrA1 levels are associated with chronic inflammatory conditions
such as rheumatoid arthritis, osteoarthritis and macular
degeneration (26,27,43–45).
The latter data appear to contrast with the decreased HtrA1
expression found in UCL, where patterns were similar to those
observed in CRC. However, it should be noted that different
cellular and molecular mechanisms act in rheumatoid arthritis,
osteoarthritis and macular degeneration, which are not associated
with an increased risk of cancer (45). Furthermore, unlike the case of UC
and CRC (17), the intestinal
microbiota do not appear to play an important pathogenic role in
these inflammatory diseases.
Concerning AHD and ALD tissues, HtrA1 levels were
not significantly different compared with those measured in HC
samples in the two mucosal compartments, except for a slight
reduction found in the epithelial compartment. Since adenoma is
considered as the main risk factor for sporadic CRC, the present
data appear to exclude a direct involvement of HtrA1 in the
progression from adenoma to CRC.
HtrA1 does not appear to be involved in the first
stage of adenoma formation (which is mainly related to APC
gene mutations) or in the first stage of development of dysplasia
in adenoma, but rather in a later stage of carcinogenesis. This
notion is partly supported by the distribution of percentage, found
in the present study, of HtrA1 expression, histological grade and
CRC stage. We did not apply a statistical test due to the small
numbers (data not shown). It is therefore likely that different
molecular mechanisms are responsible for the development of
dysplasia in adenoma and chronic UC.
There are important clinical and biological
differences between the adenoma-carcinoma sequence and the
UCL-carcinoma sequence (37,46).
The molecular basis of the adenoma-carcinoma sequence involves
first APC gene mutations and secondarily p53 mutations
(46), whereas in the UCL-carcinoma
sequence the p53 mutations come first (37). The different sequence of events
correlated with the different behaviour of HtrA1 in UCL and adenoma
(AHD and ALD). In addition, the fact that adenomatous tissue does
not contain inflammatory components could explain the normal HtrA1
expression found in adenomas and its downregulation in UCL.
The present findings suggest that HtrA1 may be a
promising marker to identify those UCL patients who are at high
risk of developing CRC. Studies of larger patient samples are also
required to demonstrate the ability of HtrA1 to screen patients at
high risk of developing CRC, who may therefore require intensive
endoscopic and histological monitoring.
References
|
1
|
Altobelli E, Lattanzi A, Paduano R,
Varassi G and di Orio F: Colorectal cancer prevention in Europe:
Burden of disease and status of screening programs. Prev Med.
62:132–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Altobelli E, D'Aloisio F and Angeletti PM:
Colorectal cancer screening in countries of European Council
outside of the EU-28. World J Gastroenterol. 22:4946–4957. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pawa N, Arulampalam T and Norton JD:
Screening for colorectal cancer: Established and emerging
modalities. Nat Rev Gastroenterol Hepatol. 8:711–722. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rubenstein JH, Enns R, Heidelbaugh J,
Barkun A, Adams MA, Dorn SD, Dudley-Brown SL, Flamm SL, Gellad ZF,
Gruss CB, et al: Clinical Guidelines Committee: American
Gastroenterological Association Institute Guideline on the
Diagnosis and Management of Lynch Syndrome. Gastroenterology.
149:777–782, quiz e16-e17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Samadder NJ, Jasperson K and Burt RW:
Hereditary and common familial colorectal cancer: Evidence for
colorectal screening. Dig Dis Sci. 60:734–747. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sebastian S, Hernández V, Myrelid P, Kariv
R, Tsianos E, Toruner M, Marti-Gallostra M, Spinelli A, van der
Meulen-de Jong AE, Yuksel ES, et al: Colorectal cancer in
inflammatory bowel disease: Results of the 3rd ECCO pathogenesis
scientific workshop (I). J Crohns Colitis. 8:5–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parian A and Lazarev M: Who and how to
screen for cancer in at-risk inflammatory bowel disease patients.
Expert Rev Gastroenterol Hepatol. 9:731–746. 2015.PubMed/NCBI
|
|
9
|
Breynaert C, Vermeire S, Rutgeerts P and
Van Assche G: Dysplasia and colorectal cancer in inflammatory bowel
disease: A result of inflammation or an intrinsic risk? Acta
Gastroenterol Belg. 71:367–372. 2008.PubMed/NCBI
|
|
10
|
Brenner H, Hoffmeister M, Stegmaier C,
Brenner G, Altenhofen L and Haug U: Risk of progression of advanced
adenomas to colorectal cancer by age and sex: Estimates based on
840,149 screening colonoscopies. Gut. 56:1585–1589. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuntz KM, Lansdorp-Vogelaar I, Rutter CM,
Knudsen AB, van Ballegooijen M, Savarino JE, Feuer EJ and Zauber
AG: A systematic comparison of microsimulation models of colorectal
cancer: The role of assumptions about adenoma progression. Med
Decis Making. 31:530–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Terzić J, Grivennikov S, Karin E and Karin
M: Inflammation and colon cancer. Gastroenterology.
138:2101–2114.e5. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sakellakis M, Makatsoris T, Gkermpesi M,
Peroukidis S and Kalofonos H: Ulcerative colitis six years after
colon cancer: Only a coincidence? Int Med Case Rep J. 7:85–88.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dyson JK and Rutter MD: Colorectal cancer
in inflammatory bowel disease: What is the real magnitude of the
risk? World J Gastroenterol. 18:3839–3848. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herszényi L, Barabás L, Miheller P and
Tulassay Z: Colorectal cancer in patients with inflammatory bowel
disease: The true impact of the risk. Dig Dis. 33:52–57. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
O'Connor PM, Lapointe TK, Beck PL and
Buret AG: Mechanisms by which inflammation may increase intestinal
cancer risk in inflammatory bowel disease. Inflamm Bowel Dis.
16:1411–1420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Azer SA: Overview of molecular pathways in
inflammatory bowel disease associated with colorectal cancer
development. Eur J Gastroenterol Hepatol. 25:271–281. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Foersch S and Neurath MF:
Colitis-associated neoplasia: Molecular basis and clinical
translation. Cell Mol Life Sci. 71:3523–3535. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hartnett L and Egan LJ: Inflammation, DNA
methylation and colitis-associated cancer. Carcinogenesis.
33:723–731. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lipinska B, Zylicz M and Georgopoulos C:
The HtrA (DegP) protein, essential for Escherichia coli survival at
high temperatures, is an endopeptidase. J Bacteriol. 172:1791–1797.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Spiess C, Beil A and Ehrmann M: A
temperature-dependent switch from chaperone to protease in a widely
conserved heat shock protein. Cell. 97:339–347. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zumbrunn J and Trueb B: Primary structure
of a putative serine protease specific for IGF-binding proteins.
FEBS Lett. 398:187–192. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao Z, Li H, Wang C, Xu W, Sun J and Zhao
W: Serine protease HtrA1 as an inhibitor on proliferation invasion
and migration of gastric cancer. Med Oncol. 32:1122015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baldi A, De Luca A, Morini M, Battista T,
Felsani A, Baldi F, Catricalà C, Amantea A, Noonan DM, Albini A, et
al: The HtrA1 serine protease is down-regulated during human
melanoma progression and represses growth of metastatic melanoma
cells. Oncogene. 21:6684–6688. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grau S, Richards PJ, Kerr B, Hughes C,
Caterson B, Williams AS, Junker U, Jones SA, Clausen T and Ehrmann
M: The role of human HtrA1 in arthritic disease. J Biol Chem.
281:6124–6129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Altobelli E, Marzioni D, Lattanzi A and
Angeletti PM: HtrA1: Its future potential as a novel biomarker for
cancer. Oncol Rep. 34:555–566. 2015.PubMed/NCBI
|
|
27
|
Weger M, Renner W, Steinbrugger I, Köfer
K, Wedrich A, Groselj-Strele A, El-Shabrawi Y, Schmut O and Haas A:
Association of the HTRA1 −625G>A promoter gene polymorphism with
exudative age-related macular degeneration in a Central European
population. Mol Vis. 13:1274–1279. 2007.PubMed/NCBI
|
|
28
|
Xia J, Wang F, Wang L and Fan Q: Elevated
serine protease HtrA1 inhibits cell proliferation, reduces
invasion, and induces apoptosis in esophageal squamous cell
carcinoma by blocking the nuclear factor-κB signaling pathway.
Tumour Biol. 34:317–328. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lorenzi T, Lorenzi M, Altobelli E,
Marzioni D, Mensà E, Quaranta A, Paolinelli F, Morroni M,
Mazzucchelli R, De Luca A, et al: HtrA1 in human urothelial bladder
cancer: A secreted protein and a potential novel biomarker. Int J
Cancer. 133:2650–2661. 2013.PubMed/NCBI
|
|
30
|
De Luca A, De Falco M, Severino A,
Campioni M, Santini D, Baldi F, Paggi MG and Baldi A: Distribution
of the serine protease HtrA1 in normal human tissues. J Histochem
Cytochem. 51:1279–1284. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marzioni D, Lorenzi T, Altobelli E,
Giannubilo SR, Paolinelli F, Tersigni C, Crescimanno C, Monsurrò V,
Tranquilli AL, Di Simone N, et al: Alterations of maternal plasma
HTRA1 level in preeclampsia complicated by IUGR. Placenta.
33:1036–1038. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zurawa-Janicka D, Skorko-Glonek J and
Lipinska B: HtrA proteins as targets in therapy of cancer and other
diseases. Expert Opin Ther Targets. 14:665–679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schmidt N, Irle I, Ripkens K, Lux V,
Nelles J, Johannes C, Parry L, Greenow K, Amir S, Campioni M, et
al: Epigenetic silencing of serine protease HTRA1 drives
polyploidy. BMC Cancer. 16:3992016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tiaden AN and Richards PJ: The emerging
roles of HTRA1 in musculoskeletal disease. Am J Pathol.
182:1482–1488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang J, Huang L, Yu W, Wu X, Zhou P and
Li X: Overexpression of HTRA1 leads to down-regulation of
fibronectin and functional changes in RF/6A cells and HUVECs. PLoS
One. 7:e461152012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Triantafillidis JK, Nasioulas G and
Kosmidis PA: Colorectal cancer and inflammatory bowel disease:
Epidemiology, risk factors, mechanisms of carcinogenesis and
prevention strategies. Anticancer Res. 29:2727–2737.
2009.PubMed/NCBI
|
|
37
|
Ullman TA and Itzkowitz SH: Intestinal
inflammation and cancer. Gastroenterology. 140:1807–1816. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Antoniou E, Margonis GA, Angelou A,
Zografos GC and Pikoulis E: Cytokine networks in animal models of
colitis-associated cancer. Anticancer Res. 35:19–24.
2015.PubMed/NCBI
|
|
39
|
Waldner MJ, Neurath MF and Markus M:
Cytokines in colitis associated cancer: Potential drug targets?
Inflamm Allergy Drug Targets. 7:187–194. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goel GA, Kandiel A, Achkar JP and Lashner
B: Molecular pathways underlying IBD-associated colorectal
neoplasia: Therapeutic implications. Am J Gastroenterol.
106:719–730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ishiguro K, Yoshida T, Yagishita H, Numata
Y and Okayasu T: Epithelial and stromal genetic instability
contributes to genesis of colorectal adenomas. Gut. 55:695–702.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Clausen T, Southan C and Ehrmann M: The
HtrA family of proteases: Implications for protein composition and
cell fate. Mol Cell. 10:443–455. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hou Y, Lin H, Zhu L, Liu Z, Hu F, Shi J,
Yang T, Shi X, Guo H, Tan X, et al: The inhibitory effect of IFN-γ
on protease HTRA1 expression in rheumatoid arthritis. J Immunol.
193:130–138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu J, Liu W, Bemis A, Wang E, Qiu Y,
Morris EA, Flannery CR and Yang Z: Comparative proteomic
characterization of articular cartilage tissue from normal donors
and patients with osteoarthritis. Arthritis Rheum. 56:3675–3684.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tomasello G, Tralongo P, Damiani P,
Sinagra E, Di Trapani B, Zeenny MN, Hussein IH, Jurjus A and Leone
A: Dismicrobism in inflammatory bowel disease and colorectal
cancer: Changes in response of colocytes. World J Gastroenterol.
20:18121–18130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hardy RG, Meltzer SJ and Jankowski JA: ABC
of colorectal cancer. Molecular basis for risk factors. BMJ.
321:886–889. 2000. View Article : Google Scholar : PubMed/NCBI
|