Introduction
Hepatocellular carcinoma (HCC) is a common malignant
tumor of the digestive tract (1).
The incidence of HCC has increased significantly in recent years,
and the overall survival of patients with HCC remains
unsatisfactory. Similar to other malignancies, the development of
HCC is a long-term, multistep process characterized by the
alteration of genes. However, the molecular mechanisms involved in
the initiation and progression of HCC are still poorly understood.
Recently, the role of epigenetic regulation, particularly DNA
methylation and the aberrant expression of miRNAs, in the
occurrence and development of cancers is gradually being recognized
(2,3).
MicroRNAs (miRNAs) are single stranded, small
non-coding RNAs of 18–25 nucleotides in length. They can negatively
regulate gene expression through base-pairing to the 3′
untranslated region (3′UTR) of target mRNAs, resulting in
translation inhibition or mRNA degradation (4–6). It is
currently estimated that ~30% of coding genes in humans are
regulated by miRNAs (7). More and
more studies suggest that beyond involvement in various biological
processes, including cell growth, differentiation and apoptosis
(8–10), dysregulation or dysfunction of
miRNAs contributes to the genesis and progression of cancer.
Several studies have revealed that miRNAs participate in the
initiation and progression of HCC. miRNA-375 inhibited the invasion
and differentiation of HCC cells (11), and miRNA-199a-3p induced G1 phase
arrest and promoted the efficacy of doxorubicin in HCC cells
(12). It is expected that by
exploring the role of miRNAs in tumorigenesis and progression a new
approach for the early diagnosis and treatment of HCC may be
provided.
miR-1247-5p, located at the distal end of human
chromosome 14 (13) and
conservatively expressed in mammalian species, was found to be
differentially expressed in cartilage (14) as well as in breast (15), colorectal (16) pancreatic (17), and prostatic cancer cells (18), and had different effects on
proliferation and invasion in various cancer cells. Currently, one
study reported that the miR-1247-5p gene was hypermethylated in
clinical samples of patients with HCC (19). However, the expression level and
functional effects of miR-1247-5p in HCC are largely unknown.
In the present study, we demonstrated a significant
downregulation of miR-1247-5p in the clinical samples of HCC
patients and HCC cell lines. Ectopic overexpression of miR-1247-5p
inhibited the proliferation and invasion of HepG2 cells, induced
cell apoptosis in vitro, and suppressed the growth of
transplanted tumors in vivo. Notably, we demonstrated that
miR-1247-5p directly targeted the 3′UTR of the Wnt3 gene, and that
the expression of miR-1247-5p could be regulated by DNA
methylation. Our findings provide valuable evidence elucidating the
function and regulatory mechanisms of miR-1247-5p in human HCC, and
indicate that miR-1247-5p can be used as a potential therapeutic
target as well as a diagnostic marker of HCC.
Materials and methods
Patients and tissue specimens
Paraffin-embedded samples, including 16 HCC tumor
tissues and 10 non-tumor tissues, as well as serum samples from 41
HCC patients and 41 healthy volunteers were obtained at the General
Hospital of Ningxia Medical University (Yinchuan, Ningxia, China).
All of the tumor samples were confirmed by pathologists and none of
the patients had undergone any therapy before recruitment to this
study. The use of clinical material for all experiments was
approved by the Ethics Committee of the General Hospital of Ningxia
Medical University.
Cell lines and cell culture
The human HCC cell lines, HepG2, HCCLM3 and
SMMC-7721, and normal liver cell line LO2 were purchased from
Shanghai Fu Meng Biotechnology Inc. (Shanghai, China). HEK-293T
cells were stored at our laboratory. HepG2, HCCLM3 and SMMC-7721
cells were cultured in Dulbecco's modified Eagle's medium (DMEM),
while LO2 cells were cultured with RPMI-1640 medium, containing 10%
heat-inactivated fetal bovine serum (FBS) (both from Gibco/Life
Technologies, Grand Island, NY, USA), 100 U/ml penicillin, 100
µg/ml streptomycin, 15 mmol/l HEPES and 200 mmol/l L-glutamine. All
of the cells were incubated at 37°C in a humidified incubator
containing 5% CO2.
Quantitative reverse
transcriptase-polymerase chain reaction (qRT-PCR)
Total RNA was extracted using an RNA extraction kit
(Omega Bio-Tek, Inc., Norcross, GA, USA) from paraffin-embedded
tissues. An miRNA purification kit (Kangweishiji Biotech Co., Ltd.,
Beijing, China) was used for serum samples and TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) was used for cells according to the
manufacturer's protocol. Then, 1 µg of total RNA from each sample
was reverse transcribed to single-stranded cDNA with EasyScript
First-Strand cDNA Synthesis SuperMix (TransGen Biotech Co., Ltd.,
Beijing, China) and the miR-1247-5p stem-loop RT primer sequence
was: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCGGGGAC-3′. The mRNA
expression of miR-1247-5p was quantified with the TransStart Top
Green qPCR SuperMix (TransGen Biotech Co., Ltd.) and the
StepOnePlus qPCR system (ABI, Carlsbad, CA, USA) under the
following conditions: 95°C for 5 min followed by 40 cycles
consisting of 95°C for 15 sec, 60°C for 15 sec and 72°C for 15 sec.
The relative gene expression levels of miR-1247-5p were calculated
using the 2−ΔΔCt method, and U6 snRNA was used for
normalization. The oligonucleotide sequences of the primers are
shown in Table I.
 | Table I.Oligonucleotide sequences. |
Table I.
Oligonucleotide sequences.
| Name | Sequence (5′ to
3′) |
|---|
| miR-1247-5p-F |
ACACTCCAGCTGGGACCC |
| U6-F |
CTCGCTTCGGCAGCACA |
| U6-R |
AACGCTTCACGAATTTGCGT |
| Wnt3-utr-F and
Wnt3-mut-F |
CCCAAGCTTGGGGGATTCAGCGAAGTCTCA |
| Wnt3-utr-R |
GACTAGTCAAGCCTCAGGTCTGTTCC |
| Wnt3-mut-R |
GACTAGTCTATTGTCACAGGCGAGTTGGGTCTGG |
|
Methylation-specific primer-F |
AGGGAGTTGTTTCGTATTTTTAAAC |
|
Methylation-specific primer-R |
GAACGTTACTCTCTACCCCGAA |
| Unmethylation
primer-F |
GGAGTTGTTTTGTATTTTTAAATGT |
| Unmethylation
primer-R |
CAAACATTACTCTCTACCCCAAA |
Plasmid construction and
transfection
Oligonucleotide sequences of miR-1247-5p precursor
(MIMAT0005899, miRBase) were designed using the Ambion Company
online software: forward sequence,
5′-GTT^AACACCCGTCCCGCTTGTCCCCGGATTCAAGAGATCCGGGGACGAACGGGACGGGTTTTTTTC^TCGAG-3′
and reverse sequence,
5′-CAA^TTGTGGGCAGGGCAAGCAGGGGCCTAAGTTCTTCTAGGCCCCTGCTTGCCCTGCCCAAAAAAAGAGCT^C-3′,
and synthesized by Sangon Biotech Inc. (Shanghai, China). After
annealing, miR-1247-5p precursor was subcloned between
HpaI-XhoI restriction sites in lentiviral vector
pSicoR to generate pSicoR-miR-1247-5p (LV-miR-1247-5P) and pSicoR
empty vector was used as a negative control (LV-NC). The
miR-1247-5p inhibitor (Inh-miR-1247-5p) and its negative control
(Inh-NC) were purchased from GenePharma Inc. (Shanghai, China). The
3′UTR and the mutant 3′UTR of the Wnt3 gene were amplified (primer
sequences are shown in Table I) and
subcloned into the pMIR-Luc reporter plasmid (Promega, Madison, WI,
USA) between SpeI-HindIII to generate pMIR-Luc-Wnt3
and pMIR-Luc-mut-Wnt3 recombinant plasmid. All of the constructs
were verified by sequencing.
The recombinant lentiviral vectors
pSicoR-miR-1247-5p were co-transfected with pCMV–VSV-G and pCMV-dR
8.91 into HEK-293T cells using Lipofectamine 3000 (Invitrogen)
according to the manufacturer's instructions. After 48 h of
incubation, the supernatant of the cultures was collected and
concentrated and then used to infect HepG2 cells. The pSicoR empty
vector was used as a negative control using the same protocol. The
miR-1247-5p inhibitor and its negative control, pMIR-Luc-Wnt3 and
pMIR-Luc-mut-Wnt3 recombinant plasmid were transfected into HepG2
cells using Lipofectamine 3000 for 48 h.
Cell proliferation assay
Cell proliferation activity was detected using the
TransDetect Cell Counting Kit-8 (CCK-8) (TransGen Biotech Co.,
Ltd.). In brief, HepG2 cells were seeded at 5×103
cells/well in 96-well plates. At various time-points, 10 µl of
CCK-8 solution was added to each well and incubated for another 2 h
at 37°C. The absorbance was assessed at a wavelength of 450 nm
using the MK3 ELISA reader (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Cell invasion assay
Transwell assay was performed to analyze the role of
miR-1247-5p in the invasive activity of HepG2 cells. Briefly, cells
were trypsinized and added to the upper Transwell chambers coated
with Matrigel (BD, Franklin Lakes, NJ, USA). The lower chambers
were filled with fresh medium containing 15% FBS. After a 48-h
incubation, the cells on the upper surface were removed and the
cells on the lower surface were fixed in 4% formaldehyde
(Sigma-Aldrich, St. Louis, MI, USA). Then, the fixed cells were
stained with 0.1% crystal violet (Sigma-Aldrich). Subsequently, the
cells in each group were photographed in five randomly selected
fields using an Olympus IX71 microscope (Olympus, Shinjuku-ku,
Tokyo, Japan). The number of invading cells was calculated and
analyzed with GraphPad Prism 6 (Graph Pad Software, La Jolla, CA,
USA).
Cell cycle assay
The cell cycle was analyzed using flow cytometry. In
brief, the cells were seeded at 1×104
cells/cm2 in 6-well plates and incubated for 72 h. Then,
the cells were harvested by trypsinization, washed in ice-cold
phosphate-buffered saline (PBS), and fixed in 75% ethanol at 4°C
overnight. Subsequently, the cells were rinsed twice in chilled
PBS, incubated with 2 µg/ml of RNAase at 37°C for 30 min and then
rinsed twice and incubated with 20 µg/ml of propidium iodide
(Beyotime Biotech, Beijing, China) at room temperature for 1 h.
Finally, the cells in each group were analyzed by flow cytometry
(FACSCalibur; BD Biosciences, San Jose, CA, USA).
Hoechst staining assay
HepG2 cells were plated with 1×105
cells/ml into 6-well plates. Then, the cells were infected with
lentiviral-mediated miR-1247-5p or its negative control for 48 h,
or transfected with inhibitors of miR-1247-5p or its negative
control using Lipofectamine 3000 for 48 h. Subsequently, the cells
in each group were stained with a Hoechst staining kit (Beyotime,
Shanghai, China). The nuclear morphology was observed under an
Olympus IX71 fluorescence microscope.
Tumorigenicity assay
Ten BALB/c nude mice, purchased from the
Experimental Animal Center of Ningxia Medical University, were
randomly divided into two groups. All mice were housed and
maintained under specific pathogen-free conditions, and the use of
mice for this study was approved by the Ethics Committee of the
General Hospital of Nigxia Medical University. In the experimental
group, the mice were subcutaneously injected with 1×107
HepG2 cells infected with LV-miR-1247-5p at the lateral axillary.
From the first day of injection, the tumor size was assessed once
every two days for four weeks. Then, the mice were sacrificed and
the tumors were photographed. The mice in the control group were
injected with HepG2 cells infected with lentiviral empty vectors
using the same protocol.
Luciferase assay
The specificity of miR-1247-5p targeting the 3′UTR
of the Wnt3 gene was ascertained by co-transfection of plasmid DNA
of pMIR-Luc-Wnt3 or pMIR-Luc-mut-Wnt3 recombinant plasmid with
pSicoR-miR-1247-5P (LV-miR-1247-5P) or pSicoR empty plasmid
(LV-NC), inhibitor of miR-1247-5p (Inh-miR-1247-5p) or negative
control of inhibitor (Inh-NC) into HepG2 cells, and determined by
the relative firefly luciferase activity expressed in relative
light units (RLU) at 48 h post-transfection using a Dual-Luciferase
Reporter Assay kit (Promega). A Renilla luciferase
expressing plasmid pRL-TK (Promega) was always included in the
transfection to normalize the efficiency of each transfection.
Western blot analysis
Total proteins were extracted from cells using RIPA
lysis buffer containing 1X protease inhibitor cocktail (Beyotime),
and the protein concentration was calculated using the BCA protein
assay kit (KeyGen Biotech Inc., Nanjing, China). Western blot
analysis was carried out according to the standard protocol.
Briefly, 40 µg of proteins were separated by 10% SDS-PAGE gel
electrophoresis and transferred to polyvinylidine fluoride (PVDF)
membranes (Millipore, Bedford, MA, USA) using 95 mA at 4°C for 2 h.
After blocking in 5% non-fat dry milk in Tris-buffered saline
(TBS), the membranes were incubated with a rabbit polyclonal
antibody against β-catenin (51067-2-AP), a rabbit polyclonal
against GAPDH (10494-1-AP) (both from Proteintech, Rosemont, IL,
USA), a goat polyclonal against Wnt3 (sc5213; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) and a rabbit polyclonal
against β-actin (ab8227; Abcam, Cambridge, MA, USA) at 1:500
dilution in TBS overnight at 4°C. The membranes were then washed
three times with TBS-Tween-20, and subsequently incubated with
secondary antibodies [anti-rabbit IgG-horseradish
peroxidase-conjugated and anti-goat IgG-horseradish
peroxidase-conjugated (ZSGB-Bio Inc., Beijing, China)], conjugated
with horseradish peroxidase at a 1:1,000 dilution in TBS for 2 h at
room temperature. The membranes were washed again in TBS-Tween-20
six times at room temperature. The protein bands were visualized
using the Luminol reagent (Thermo Fisher Scientific, Inc.) and
detected using an enhanced chemiluminescence detection system.
DNA methylation analysis
The sequence of the promoter region of the
miR-1247-5p gene was analyzed using UCSC Genome Database
(http://genome.ucsc.edu/). Genomic DNA of HepG2
cells, and HepG2 cells treated with 5 or 10 µmol/l of 5-azacytidine
for 48 h were extracted using a DNA extraction kit (Promega) and
sulfated using the EpiTect Bisulfite kit (Qiagen,
Schnackenburgallee, Hamburg, Germany) according to the
manufacturer's instructions. Then, the bisulfite-modified DNA was
used as a template for the amplification of DNA
methylation-specific PCR (MSP). Normal liver cell line LO2 was used
as a normal control. Methylation-specific primers and unmethylation
primers were designed using MethPrimer online software and
synthesized by GenePharma Inc. (primer sequences are shown in
Table I). The reactions were
incubated at 95°C for 5 min, followed by 40 cycles of 95°C for 30
sec, 58°C for 30 sec (methylation-specific primers) or 56°C for 30
sec (unmethylation primers), 72°C for 30 sec, and then 72°C for 10
min. PCR products were detected by 2.5% agarose gel
electrophoresis, and photographed using a gel imaging system
(Bio-Rad, Hercules, CA, USA).
Statistical analysis
All data collected in the present study, were
obtained from at least three independent experiments for each
condition. SPSS 19.0 analysis software (SPSS, Inc., Chicago, IL,
USA) and GraphPad Prism 6 (Graph Pad Software, La Jolla, CA, USA)
were used for the statistical analysis. Statistical evaluation of
the data was performed by one-way ANOVA when more than two groups
were compared with a single control, and t-test for comparison of
differences between the two groups. Significant differences were
assigned to p-values <0.05, <0.01, <0.0005 and <0.0001.
Data are presented as the mean ± standard deviation (SD).
Results
miR-1247-5p is downregulated in
HCC
The mRNA expression of miR-1247-5p in clinical
samples of HCC patients and HCC cell lines was analyzed by qRT-PCR.
The results revealed that miR-1247-5p was significantly
downregulated in tumor samples (n=16) and serum samples (n=41) of
HCC patients compared to non-tumor samples (n=10) and serum samples
(n=41) of healthy volunteers (Fig. 1A
and B). Subsequently, the mRNA expression level of miR-1247-5p
was assessed in HCC cell lines, HepG2, HCCLM3 and SMMC-7721, and
normal liver cell line LO2. The results revealed that the mRNA
expression of miR-1247-5p was significantly decreased in the HCC
cell lines, HepG2 (0.721±0.090%; p<0.0005), HCCLM3
(0.890±0.097%; p<0.0005) and SMMC-7721 (8.185±0.355%;
p<0.0005) normalized to the normal liver cell line LO2 (Fig. 1C). The HepG2 cells were chosen for
the subsequent experiments due to the lowest expression of
miR-1247-5p.
miR-1247-5p inhibits cell
proliferation and invasion in vitro
The role of miR-1247-5p in the progression of HCC
was investigated using CCK-8 and Transwell assays. The mRNA
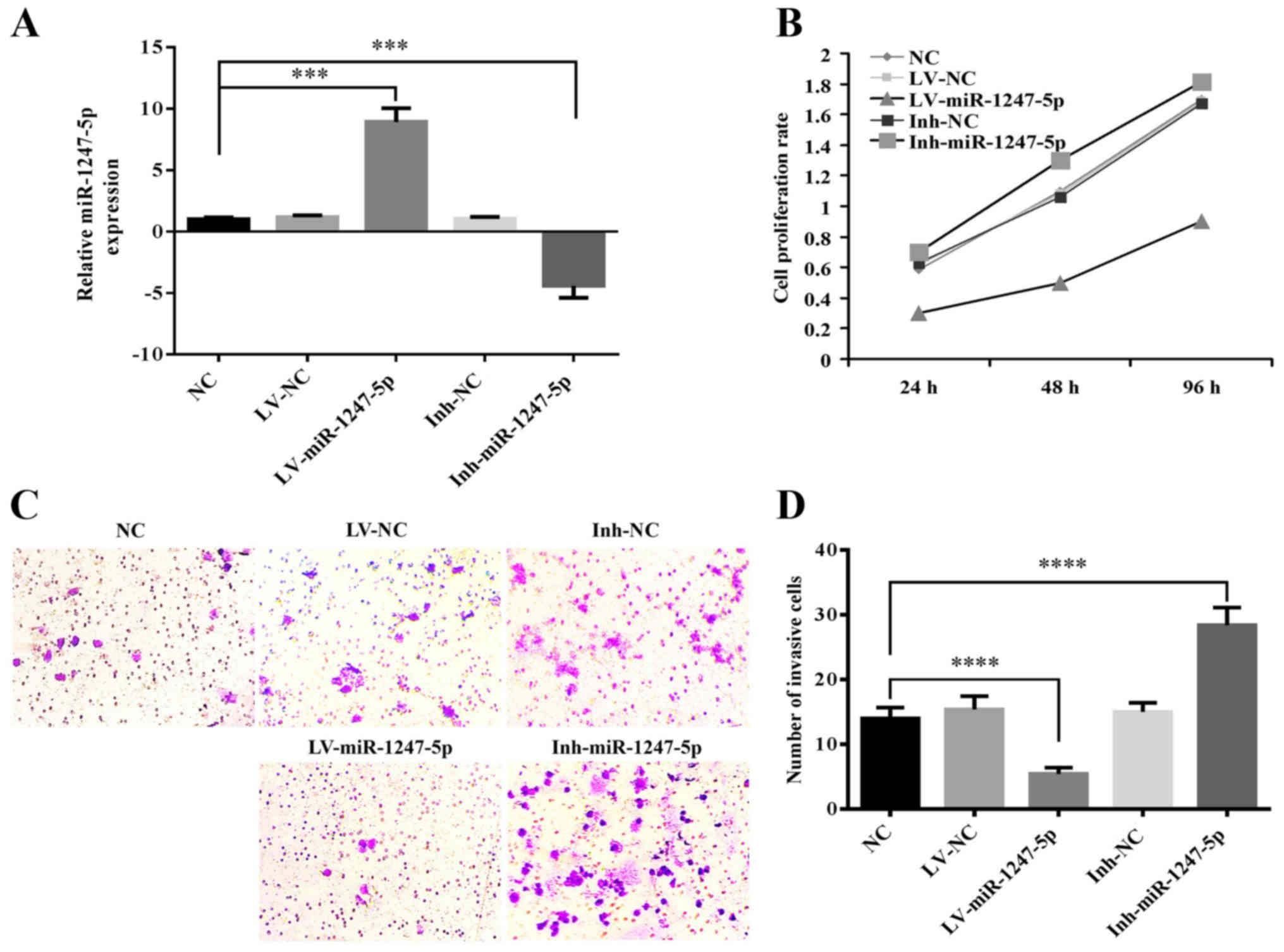
expression of miR-1247-5p in HepG2 cells infected with
lentiviral-mediated miR-1247-5p (LV-miR-1247-5p) or its negative
control (LV-NC), and HepG2 cells transfected with the inhibitor of
miR-1247-5p (Inh-miR-1247-5p) or its negative control (Ihn-NC) were
detected by qRT-PCR. Normal HepG2 cells were used as the normal
control (NC). The results revealed that the mRNA expression of
miR-1247-5p was 8.9 times higher in the LV-miR-1247-5p-infected
group and 5.4 times lower in the Inh-miR1247-5p-transfected group
compared to that of the NC group (p<0.0005) (Fig. 2A). These results indicated that the
cell models of miR-1247-5p were successfully established. Then, the
CCK-8 assay was performed to explore the role of miR-1247-5p in
cell proliferation and the results revealed that the ectopic
overexpression of miR-1247-5p significantly decreased the
proliferative rate of HepG2 cells, whereas the decreased expression
of miR-1247-5p induced the enhancement of cell proliferation
(Fig. 2B). Then, the role of
miR-1247-5p in cell invasion was assessed using the Transwell
assay. The results revealed that the invasive activity in the
LV-miR-1247-5p-infected group was significantly decreased
(p<0.0001), whereas that in the Inh-miR-1247-5p-transfected goup
was significantly enhanced (p<0.0001) compared to that of the
control groups (Fig. 2C and D).
These results demonstrated that the ectopic overexpression of
miR-1247-5p inhibited the proliferation and invasion of HepG2 cells
in vitro.
miR-1247-5p induces cell apoptosis in
vitro
The role of miR-1247-5p in the regulation of the
cell cycle was analyzed using flow cytometry. The results revealed
that the number of apoptotic cells or cell debris increased
significantly in the LV-miR-1247-5p-infected group (p<0.05). In
contrast, that in the Inh-miR-1247-5p transfected group was
slightly decreased compared to that in the control group, and there
was no difference in the proportion of HepG2 cells in the G0/G1,
G2/M and S phases between the different groups (Fig. 3A and B). To further demonstrate
this, Hoechst staining assay was performed. The results revealed
that an increased abundance of cells with characteristics of
chromosome condensation was found in the cells infected with the
LV-miR-1247-5p (Fig. 3C). These
results demonstrated that the ectopic overexpression of miR-1247-5p
induced the apoptosis of HepG2 cells in vitro.
miR-1247-5p inhibits the growth of
tumors in vivo
To demonstrate that miR-1247-5p can inhibit the
growth of tumors in vivo, a nude mouse tumorigenicity assay
was performed. HepG2 cells infected with lentiviral-mediated
miR-1247-5p (LV-miR-1247-5p) or negative control vectors (LV-NC)
were subcutaneously injected at the lateral axillary of BALB/c nude
mice. The tumor size of each mouse was assessed once every two days
for four weeks. Subsequently, the experimental mice were sacrificed
and the tumors were photographed. The results revealed that the
tumor size of the experimental mice was significantly smaller than
that of the control group of mice (Fig.
4A and B). These results demonstrated that the ectopic
overexpression of miR-1247-5p inhibited the growth of HCC tumors
in vivo. Incidentally, during this experiment, one mouse in
the experimental group died for unknown reasons, and one mouse in
the control group did not form tumors.
Wnt3 is a target of miR-1247-5p in
HepG2 cells
It is generally understood that miRNAs execute
post-transcriptional regulation by binding to the 3′UTR of their
downstream genes. To explore the molecular mechanisms involved in
the miR-1247-5p-mediated tumor suppression, target genes of
miR-1247-5p were analyzed using the online target prediction
software DIANA-MICROT (http://diana.imis.athena-innovation.gr) and miRanda
(www.micorrna.org), and the correlative signaling
pathway regulated by miR-1247-5p was analyzed by CytoScape 3.0
software. The results revealed that Wnt/β-catenin and MAPK were the
two most related pathways, and wingless-type MMTV integration site
family, member 3 (Wnt3), a potential target gene of miR-1247-5p,
was chosen as the focus of our research in the present study based
on the complementarity of its sequence to miR-1247-5p and the
important role of the Wnt/β-catenin pathway in HCC. In contrast to
the sequence of miR-1247-5p, there were seven consecutive binding
sites in the 3′UTR of wild-type Wnt3 (Wnt3 3′UTR) and four
consecutive binding sites in mutant Wnt3 3′UTR (mut-Wnt3 3′UTR)
(Fig. 5A). To further confirm that
there was a direct interaction between miR-1247-5p and Wnt3, a
luciferase reporter assay was performed in HepG2 cells
co-transfected with the luciferase reporter vector expressing the
3′UTR of Wnt3 and LV-miR-1247-5p, Inh-miR-1247-5p or negative
control vectors. Vectors expressing the mutant 3′UTR of the Wnt3
gene were used as the control. A significant decrease in the
luciferase signal was observed in cells co-transfected with the
Wnt3 3′UTR vector and the LV-miR-1247-5p. In contrast, the
inhibition was fully rescued when target sites were mutated
(Fig. 5B). These results revealed
that miR-1247-5p could inhibit the transcription activity of the
Wnt3 gene by targeting its 3′UTR. To further demonstrate the
inhibitory effect of miR-1247-5p on the expression of Wnt3, the
expression of Wnt3 and its downstream protein β-catenin in each
group was analyzed by western blotting. The results revealed that
Wnt3 and β-catenin were significantly decreased at the protein
level after ectopic overexpression of miR-1247-5p. In contrast, the
protein expression of Wnt3 and β-catenin was slightly increased in
the Inh-miR-1247-5p transfected group (Fig. 5C and D).
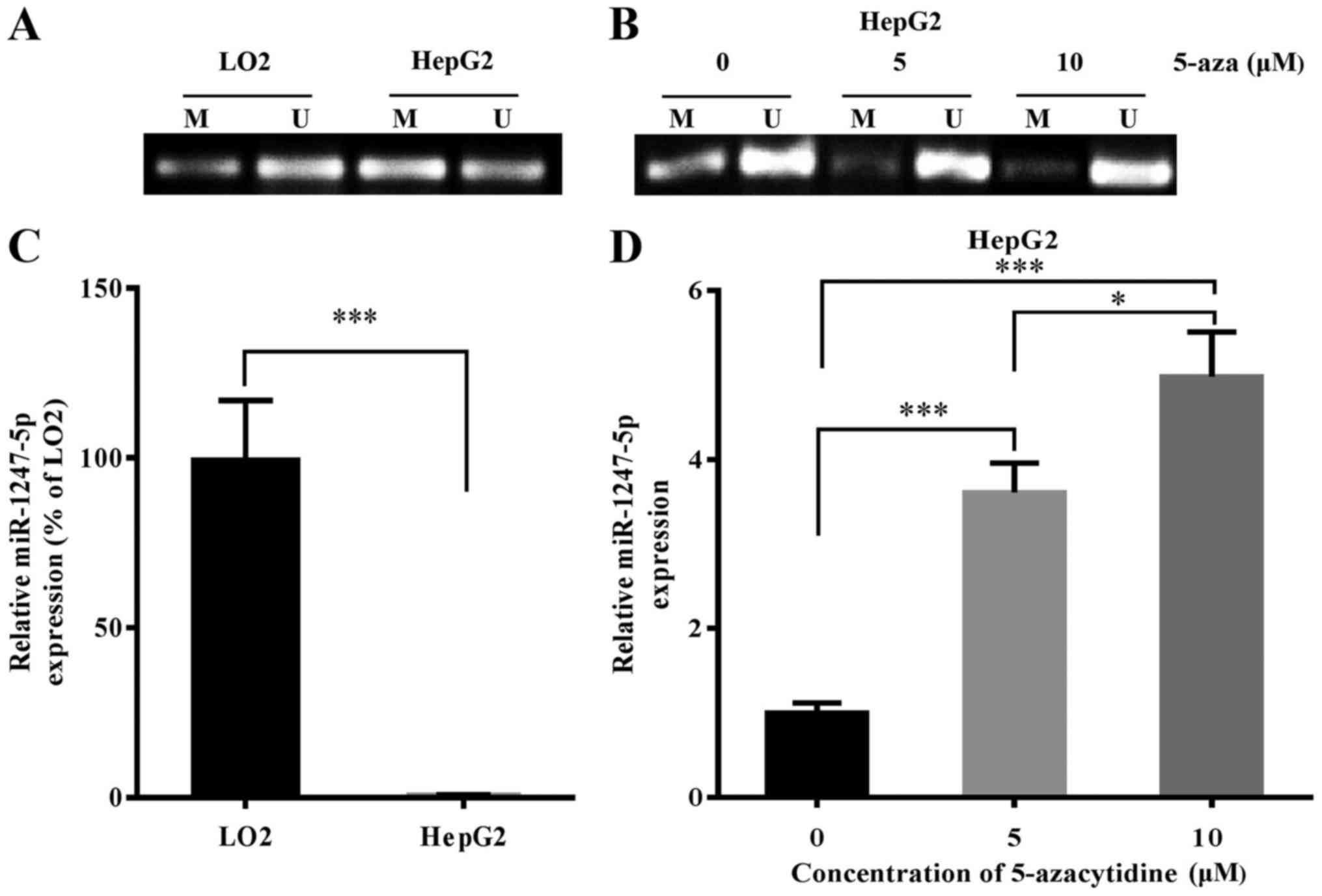
DNA methylation regulates the
expression of miR-1247-5p
Based on the study of Anwar et al that
revealed that the miR-1247-5p gene was hypermethylated in HCC, we
surmised that DNA methylation may play an important role in the
regulation of the expression of miR-1247-5p. To further explore the
role of DNA methylation in the regulation of miR-1247-5p expression
in HCC, the sequence of the miR-1247-5p gene was analyzed using
UCSC Genome Database (http://genome.ucsc.edu/). The results revealed that
the promoter region of the miR-1247-5p gene was 5,578 bp in length,
located on chromosome 14 between 101,559,653 up to 101,565,230 bp.
In addition, the CG content of this region accounted for 62.4%, and
the possibility of a CpG island existing in this region was 0.84
(data not shown). These results revealed that a CpG island most
probably existed in the promoter region of the miR-1247-5p gene. To
confirm that the expression of miR-1247-5p was regulated by DNA
methylation, the methylation level of the miR-1247-5p gene in HepG2
cells (an HCC cell line) and LO2 cells (a normal liver cell line)
was detected using MSP assay and the expression of miR-1247-5p in
HepG2 cells was assessed by qRT-PCR after treatment with
demethylating drug, 5-azacytidine. The results revealed that the
methylation level of miR-1247-5p increased significantly and the
expression of miR-1247-5p decreased significantly in the HepG2
cells compared to that of LO2 cells (Fig. 6A and C). Following treatment with
5-azacytidine, the methylation level of miR-1247-5p in HepG2 cells
decreased and the expression of miR-1247-5p in these cells
significantly increased (Fig. 6B and
D). Based on these results, we demonstrated that the expression
of miR-1247-5p could be regulated by DNA methylation.
Discussion
Dysregulation of miRNAs has been demonstrated to be
involved in tumorigenesis and progression in various types of
tumors, and the role of miRNAs in the development of cancers has
gradually been recognized (20–22).
However, the elucidation of the role of miRNAs in HCC is still in
the early developmental stage. miR-1247-5p, transcripted by
DLKI-DIO3 domain, is located at the distal end of human chromosome
14 and conservatively expressed at a high level in mammalian
species. Previous studies have reported that miR-1247-5p is
differentially expressed in various types of cancer, and had
positive or negative effects on the proliferation, invasion and
apoptosis of different cancer cells (15–18).
However, the role of miR-1247-5p in HCC is largely unknown.
In the present study, it was demonstrated that
miR-1247-5p was significantly downregulated in serum and tumor
samples of patients with HCC and in HCC cell lines. miRNAs existing
in the sera are usually called circulating miRNAs. Circulating
miRNAs have tumor-marker characteristics, and tumors at different
stages of development have different miRNA expression profiles
(23). A recent study revealed that
circulating miRNAs may be derived from active secretion of tissue
cells (24). It has been reported
that circulating miRNAs could be used in the early diagnosis of
many cancers, such as gastric (25), prostatic (26), esophageal (27), bladder (28), ovarian (29) and nasopharyngeal cancer (30). Due to the existence of miR-1247-5p
in sera, it is suggested that the detection of the expression of
miR-1247-5p in sera can be used as a non-invasive approach for the
early diagnosis of HCC. In a recent study, researchers found that
miR-1247-5p was downregulated in pancreatic cancer, and could
inhibit cancer cell proliferation by targeting neuropilins
(17). Thus, it was determined that
miR-1247-5p is a tumor-suppressor miRNA in pancreatic cancer.
However, another research group found a significant upregulation of
miR-1247-5p in castration-resistant prostate cancer (CRPC) samples
and prostate cancer (PC) cell lines, therefore miR-1247-5p
functioned as a onco-miRNA (18).
In the present study, miR-1247-5p was significantly downregulated
in clinical samples of patients with HCC and HCC cell lines.
Ectopic overexpression of miR-1247-5p significantly inhibited the
proliferation and invasion of HepG2 cells, induced cell apoptosis,
and suppressed tumor growth in vivo via the regulation of
the expression of Wnt3. These results indicated that miR-1247-5p
functioned as a tumor suppressor in HCC. We suggest that the
different effects of miR-1247-5p on cell proliferation and invasion
of cancer cells depended on the type of cancer or the genes it was
targeting.
To explore the regulatory mechanisms involved in
miR-1247-5p-mediated tumor suppression, putative target genes of
miR-1247-5p were analyzed by prediction software. Wingless-type
MMTV integration site family, member 3 (Wnt3), a secreted
glycoprotein, located on human chromosome 17 and an upstream
protein of the canonical Wnt/β-catenin pathway (31), was predicted as a potential target
gene of miR-1247-5p and this was ascertained by luciferase reporter
assay and western blotting. Previous studies demonstrated that the
canonical Wnt/β-catenin pathway participates in the genesis and
progression of several cancers by activating the transcription of
its downstream genes (32–34). In brief, when stimulated by signals,
Wnt proteins bind with transmembrane receptors to activate the
expression of dishevelled proteins (DVL), which inhibit the
activity of GSK3β in the degradation complex, preventing the
phosphorylation of β-catenin by GSK3β, thus avoiding its
identification and degradation from ubiquitin or proteasomes. Then,
β-catenin accumulates in the cytoplasm and translocates to the
nucleus, where it binds with nuclear transcription factors and
activates the expression of downstream target genes, which leads to
the abnormal proliferation of cells and the occurrence of tumors.
Our results indicated that the ectopic overexpression of
miR-1247-5p inhibited the expression of the Wnt3 protein in HepG2
cells by targeting the 3′UTR of the Wnt3 gene, and β-catenin, a
downstream protein of the Wnt/β-catenin pathway could also be
inhibited. These results demonstrated the role of miR-1247-5p in
the inhibition of the proliferation and invasion of HCC cells
partially achieved via the regulation of the activity of the
Wnt/β-catenin pathway.
DNA methylation in miRNA genes could serve as a new
biomarker for the early detection, diagnosis and prognosis of
malignant tumors. Generally, miRNAs, which function as tumor
suppressors, are silenced by DNA hypermethylation, which results in
the overexpression of downstream oncogenes and the occurrence of
tumors (35,36). However, knowledge concerning
microRNA gene methylation in human HCC is still limited. To explore
the role of DNA methylation in the regulation of miR-1247-5p
expression in HCC, the UCSC genome database (http://genome.ucsc.edu/) was used to analyze the
promoter region of the hsa-miR-1247-5p gene, and the methylation
level of the miR-1247-5p gene in HepG2 cells and normal liver cells
which was detected by MSP assay. The results revealed that a CpG
island most probably exists in the promoter region of the
miR-1247-5p gene. Τhe methylation level of the miR-1247-5p gene was
significantly increased in HepG2 cells compared to that of normal
liver cells. Following treatment with demethylation drug
5-azacytidine, the methylation level of the miR-1247-5p gene was
decreased and the expression of miR-1247-5p was significantly
increased. These results reveal that the expression of miR-1247-5p
could be regulated by DNA methylation.
In summary, these results demonstrated that
miR-1247-5p expression is downregulated in HCC tumors, and
influence the proliferation and invasion of HepG2 cells via the
regulation of the Wnt/β-catenin pathway, and can be regulated by
DNA methylation. These data reveal that miR-1247-5p functions as a
tumor suppressor in HCC, and can be used as a new biomarker and
potential target for the diagnosis and treatment of HCC. Further
studies should focus on the regulatory mechanism of miR-1247-5p in
the genesis and progression of HCC.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of China (no. 81460368), the Ningxia
High Education Science and Technology Important Project [(2014) no.
2014-70], and the Science and Technology Program of Ningxia
(2013).
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ and
Kang GH: Aberrant CpG island hypermethylation along multistep
hepatocarcinogenesis. Am J Pathol. 163:1371–1378. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Søkilde R, Vincent M, Møller AK, Hansen A,
Høiby PE, Blondal T, Nielsen BS, Daugaard G, Møller S and Litman T:
Efficient identification of miRNAs for classification of tumor
origin. J Mol Diagn. 16:106–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Osman A: MicroRNAs in health and disease -
basic science and clinical applications. Clin Lab. 58:393–402.
2012.PubMed/NCBI
|
|
5
|
Zhao G, Cai C, Yang T, Qiu X, Liao B, Li
W, Ji Z, Zhao J, Zhao H, Guo M, et al: MicroRNA-221 induces cell
survival and cisplatin resistance through PI3K/Akt pathway in human
osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Rooij E and Kauppinen S: Development
of microRNA therapeutics is coming of age. EMBO Mol Med. 6:851–864.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nugent M: MicroRNA function and
dysregulation in bone tumors: The evidence to date. Cancer Manag
Res. 6:15–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu AM, Poon RT and Luk JM: MicroRNA-375
targets Hippo-signaling effector YAP in liver cancer and inhibits
tumor properties. Biochem Biophys Res Commun. 394:623–627. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fornari F, Milazzo M, Chieco P, Negrini M,
Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L and Gramantieri
L: MiR-199a-3p regulates mTOR and c-Met to influence the
doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res.
70:5184–5193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morin RD, O'Connor MD, Griffith M,
Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T,
Hirst M, et al: Application of massively parallel sequencing to
microRNA profiling and discovery in human embryonic stem cells.
Genome Res. 18:610–621. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martinez-Sanchez A and Murphy CL: miR-1247
functions by targeting cartilage transcription factor SOX9. J Biol
Chem. 288:30802–30814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu Y, Ginther C, Kim J, Mosher N, Chung S,
Slamon D and Vadgama JV: Expression of Wnt3 activates Wnt/β-catenin
pathway and promotes EMT-like phenotype in trastuzumab-resistant
HER2-overexpressing breast cancer cells. Mol Cancer Res.
10:1597–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan H, Choi AJ, Lee BH and Ting AH:
Identification and functional analysis of epigenetically silenced
microRNAs in colorectal cancer cells. PLoS One. 6:e206282011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi S, Lu Y, Qin Y, Li W, Cheng H, Xu Y,
Xu J, Long J, Liu L, Liu C, et al: miR-1247 is correlated with
prognosis of pancreatic cancer and inhibits cell proliferation by
targeting neuropilins. Curr Mol Med. 14:316–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scaravilli M, Porkka KP, Brofeldt A,
Annala M, Tammela TL, Jenster GW, Nykter M and Visakorpi T:
MiR-1247-5p is overexpressed in castration resistant prostate
cancer and targets MYCBP2. Prostate. 75:798–805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Anwar SL, Albat C, Krech T, Hasemeier B,
Schipper E, Schweitzer N, Vogel A, Kreipe H and Lehmann U:
Concordant hypermethylation of intergenic microRNA genes in human
hepatocellular carcinoma as new diagnostic and prognostic marker.
Int J Cancer. 133:660–670. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han G, Wang Y, Bi W, Jia J and Wang W:
MicroRNA-124 functions as a tumor suppressor and indicates
prognosis in human osteosarcoma. Exp Ther Med. 9:679–684.
2015.PubMed/NCBI
|
|
21
|
Mardin WA and Mees ST: MicroRNAs: Novel
diagnostic and therapeutic tools for pancreatic ductal
adenocarcinoma? Ann Surg Oncol. 16:3183–3189. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rachagani S, Kumar S and Batra SK:
MicroRNA in pancreatic cancer: Pathological, diagnostic and
therapeutic implications. Cancer Lett. 292:8–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bianchi F, Nicassio F, Marzi M, Belloni E,
Dall'olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G and Di
Fiore PP: A serum circulating miRNA diagnostic test to identify
asymptomatic high-risk individuals with early stage lung cancer.
EMBO Mol Med. 3:495–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang
C, Huang D, Chen X, Zhang H, Zhuang R, et al: A five-microRNA
signature identified from genome-wide serum microRNA expression
profiling serves as a fingerprint for gastric cancer diagnosis. Eur
J Cancer. 47:784–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen ZH, Zhang GL, Li HR, Luo JD, Li ZX,
Chen GM and Yang J: A panel of five circulating microRNAs as
potential biomarkers for prostate cancer. Prostate. 72:1443–1452.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang C, Wang C, Chen X, Yang C, Li K,
Wang J, Dai J, Hu Z, Zhou X, Chen L, et al: Expression profile of
microRNAs in serum: A fingerprint for esophageal squamous cell
carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Long JD, Sullivan TB, Humphrey J,
Logvinenko T, Summerhayes KA, Kozinn S, Harty N, Summerhayes IC,
Libertino JA, Holway AH, et al: A non-invasive miRNA based assay to
detect bladder cancer in cell-free urine. Am J Transl Res.
7:2500–2509. 2015.PubMed/NCBI
|
|
29
|
Suryawanshi S, Vlad AM, Lin HM,
Mantia-Smaldone G, Laskey R, Lee M, Lin Y, Donnellan N, Klein-Patel
M, Lee T, et al: Plasma microRNAs as novel biomarkers for
endometriosis and endometriosis-associated ovarian cancer. Clin
Cancer Res. 19:1213–1224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zeng X, Xiang J, Wu M, Xiong W, Tang H,
Deng M, Li X, Liao Q, Su B, Luo Z, et al: Circulating miR-17,
miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers
in nasopharyngeal carcinoma. PLoS One. 7:e463672012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakashima N, Liu D, Huang CL, Ueno M,
Zhang X and Yokomise H: Wnt3 gene expression promotes tumor
progression in non-small cell lung cancer. Lung Cancer. 76:228–234.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
White BD, Chien AJ and Dawson DW:
Dysregulation of Wnt/β-catenin signaling in gastrointestinal
cancers. Gastroenterology. 142:219–232. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lachenmayer A, Alsinet C, Savic R,
Cabellos L, Toffanin S, Hoshida Y, Villanueva A, Minguez B, Newell
P, Tsai HW, et al: Wnt-pathway activation in two molecular classes
of hepatocellular carcinoma and experimental modulation by
sorafenib. Clin Cancer Res. 18:4997–5007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Voloshanenko O, Erdmann G, Dubash TD,
Augustin I, Metzig M, Moffa G, Hundsrucker C, Kerr G, Sandmann T,
Anchang B, et al: Wnt secretion is required to maintain high levels
of Wnt activity in colon cancer cells. Nat Commun. 4:26102013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kunej T, Godnic I, Ferdin J, Horvat S,
Dovc P and Calin GA: Epigenetic regulation of microRNAs in cancer:
An integrated review of literature. Mutat Res. 717:77–84. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kozaki K and Inazawa J: Tumor-suppressive
microRNA silenced by tumor-specific DNA hypermethylation in cancer
cells. Cancer Sci. 103:837–845. 2012. View Article : Google Scholar : PubMed/NCBI
|