Introduction
Liver cancer is one of the most common cancers in
the world with an estimated 782,500 new liver cancer cases and
745,500 deaths occurring worldwide during 2012. China accounted for
approximately 50% of the total number of cases and deaths in the
world (1). Most (70–90%) primary
live cancers occurring worldwide are hepatocellular carcinoma (HCC)
(2). Currently, liver cancer rates
are decreasing in some historically high-risk areas, including
China and Japan, because of the decreasing HBV infection in China
and HCV infection in Japan by elevating hygiene and sanitation
(3). Even then, 54% of HCC cases
worldwide are related to HBV (4).
Therefore, research on the mechanism of HBV inducing HCC is still a
crucial field to reduce the morbidity and find new therapies.
HBV contains four overlapping open reading frames
(ORFs), coding the viral envelope (pre-S1/pre-S2/S), core proteins
(pre-C/C), viral polymerase and hepatitis B virus × protein (HBx)
(5). HBx, a ~17 kDa protein with
154 amino acids, is an essential factor for HBV replication through
accelerating mitochondrial calcium uptake and significantly
contributes to HCC tumorigenesis (6). The properties of HBx in
hepatocarcinogenesis is diversiform, some studies show that HBx can
influence proliferation, apoptosis, anti-apoptosis, invasion,
motility, mitochondria DNA damage, oxidative stress and epigenetic
changes (7). HBx protein is an
enigmatic and promiscuous transactivator that can activate a
variety of viral and cellular promoters and enhancers (8). Except for protein-protein interaction
mediating transcriptional activity, HBx, targeting microRNAs
directly or through other bridge protein indirectly, plays its role
in HBV-associated HCC development.
MicroRNAs, that have been discovered in humans total
more than 2000 species playing important roles in all biological
processes by post-transcription regulation of protein-coding genes
(9). As regulator, the mature miRNA
is incorporated into the RNA-induced silencing complex and promotes
target mRNA degradation or repression of translation. Via these
mechanism, a number of studies associate miRNAs with numerous
functions in tumorigenesis including cell proliferation,
differentiation, apoptosis, invasion, metastasis, autophagy
(10), epithelial-mesenchymal
transition (EMT) (11), lipogenesis
(12) and epigenetics (13). Similarly to other cancers,
involvement of miRNA in HCC has been demonstrated, aberrant miRNA
expression can affect many crucial cancer-associated pathways,
including p53, RAS/MAPK, PI3K/AKT/mTOR, WNT/β-catenin and
transforming growth factor β (TGF-β) (14). Therefore, in view of the importance
of HBV infection and dysregulation miRNA in HCC generation, the
interaction effects between HBV and miRNA is particularly
significant.
Based upon recent studies, the present study aims to
review the mechanism and result of the interplay between HBx and
miRNA in HBV-related HCC, which provides help to further clarify
the regulation role of HBx protein for miRNA in tumor
development.
The mechanism and result of HBx acting on
miRNA
The mechanism of HBx acting on
miRNA
Here, we summarize three mainly mechanism for HBx
acting upon miRNAs. First of all, HBx alters the expressions of
miRNAs by playing a role in the miRNA production process. miRNAs
are transcribed by RNA polymerase II to produce a long primary
miRNAs transcripts (pri-miRNAs), and then is cleaved into ~70-nt
precursor miRNAs (pre-miRNAs) by Drosha which is a member of the
RNase III family, the key to it is, Drosha has a cofactor DGCR8,
which is decreased by HBx through influencing its promoter activity
(15). Some studies have indicated
that HBV downregulates the expression of DGCR8 by modulating its
promoter activity directly, besides, data indicate that HBV induces
the expression of YY1, which directly results in inhibition of
DGCR8 promoter activity (16).
Notably, downregulation of DGCR8 induced by HBV can result in
partial repression of miRNA expression, because of the mirtrons
locating in the introns of coding genes and using splicing to
bypass Drosha-DGCR8 cleavage, are alternative precursors for miRNAs
biogenesis and which essential for the generation of miRNAs
(16–18).
Except for the afore-mentioned, studies have shown
that the HBx RNA appears to be sufficient to downregulate its
targeting microRNAs such as the deregulation of miR-15a/miR-16
(19), the data strongly indicate
that the HBx RNA, with or without the coding potential, is able to
bind and trigger the decay of the corresponding microRNAs, which is
reminiscent of induced downregulation of miR-122 by HBV RNA
(20) and enhanced microRNA
instability by the herpesvirus non-coding RNA (21).
HBx does not bind DNA directly, its multiregulative
functions are mediated by its interactions with host factors, such
as activator protein-1 and −2, NF-κB and ATF2 (22). For instance, NF-κB transcriptional
activity was significantly enhanced through interacting with HBx,
resulting in the increasing of miR-143 (23) and miR-146a (24). In previous studies, HBx was shown to
repress the expression of miR-148a transcription through reduced
recruitment of p53 to the miR-148a promoter (25). HBx was found to inhibit the normal
function of p53 and p53 inhibited the expression of hepatocyte
nuclear factor 4a (HNF4a), a key regulator of miR-122 (26) and miR-548p (27) expression in the liver. Besides, via
inhibiting the activity of p53 directly, HBx decreases the
expression of miR-216b (28) and
miR-122. Notably, one miRNA could be regulated by various host
proteins, which are the targets of HBx simultaneously, such as
miR-122, which could be repressed by p53 and germline development 2
(Gld2, also called PAPD4) (26).
HBx alters host gene expression by constitutively activating
cytoplasmic signal transduction pathways, and previous studies have
shown that HBx could induce the expression of c-Myc which is the
crucial protein in tumorigenesis, hence, c-Myc mediated the
HBx-induced repression of miR-15a/16 (29). In addition, it has been indicated
that there is a HBx-c-Myc-Lin28B axis existing in let-7 regulation
pathways, in turn, let-7 inhibited the expression of c-Myc and
Lin28B. This antagonism, maintains a balancing equilibrium between
these factors in tumorigenesis (30). Beyond that, HBx has previously been
shown to activate activator protein-1 (AP-1), and the upregulation
of miR-21 by HBx is through AP-1. Moreover, AP-1 activity is
reported to be negatively regulated by PDCD4, leading to the
formation of positive feedback loop between AP-1-miR-21-PDCD4
(31). In fact, HBx alone is
considered a poor transformer of human and rodent hepatic cells,
usually, the function of HBx regulating the expression of miRNA is
achieved by a protein complex. Some researchers concluded that HBx
downregulates miR-520b through interacting with Sp1 with survivin
(32). Also, regulating the level
of DNA methylation on the promoter of miRNA is another essential
method for HBx dysregulating the expression of miRNA. In reality,
HBx could induce the expression of DNA methyltransferase (DNMTs)
which would increase the level of DNA methylation on the promoter
of miR-132 resulting in the decreasing of miR-132 (33). Finally, HBx could upregulate the
URG11 which stimulates hepatocellular growth by transcriptionally
activating the β-catenin promoter, resulting in the increasing of
miR-148a (34). In general, HBx
activates some signal pathways or key protein which could be acting
on miRNA specifically to dysregulate the expression of miRNA, and
induces hepatocarcinogenesis conclusively.
The results of HBx acting on
miRNA
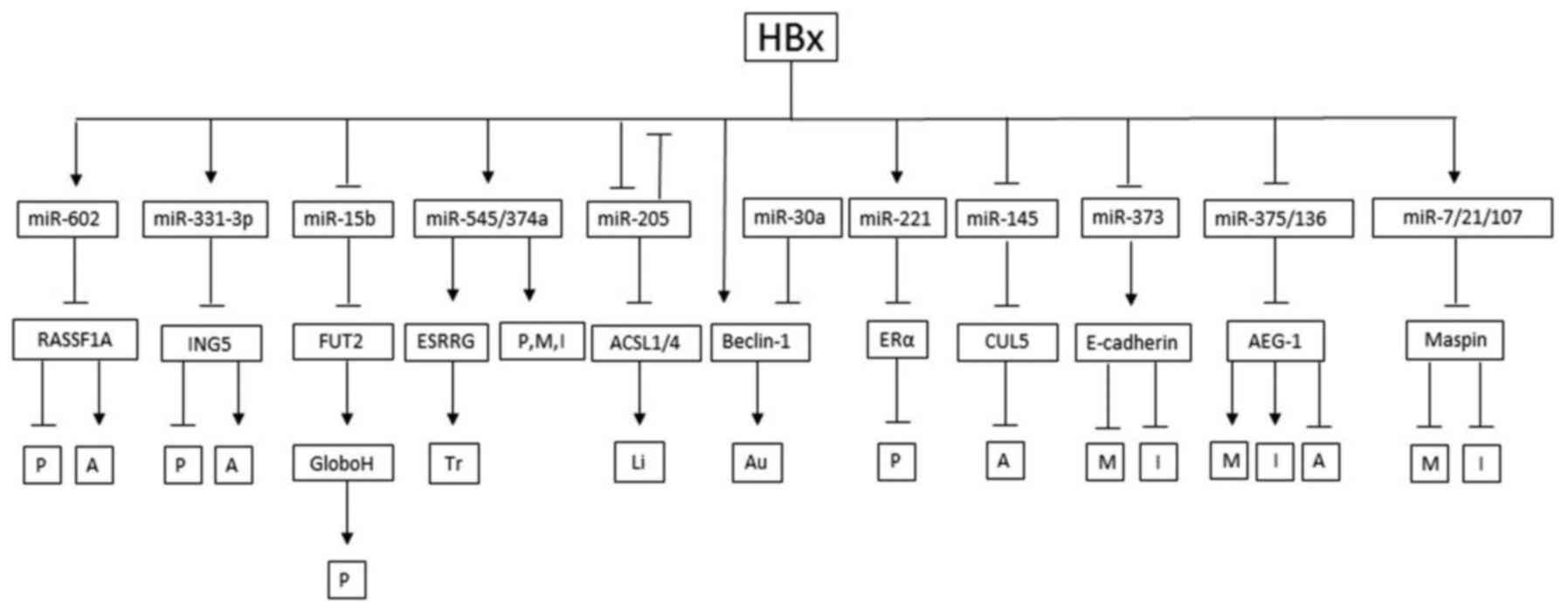
The effect of HBx acting on miRNA has two aspects,
upregulation and downregulation. The detailed results are shown
below (Table I).
 | Table I.The results of HBx acting on
miRNA. |
Table I.
The results of HBx acting on
miRNA.
| Name | Biological
function | Refs. |
|---|
| Upregulated miRNA
by HBx |
|
miR-7 | Proliferation↓ | (35,36) |
|
|
Migration↑/Invasion↑ |
|
|
| Anoikis
resistance↑ |
|
|
|
Chemoresistance↑ |
|
|
miR-21 | Proliferation↑ | (31,36,37,38) |
|
| Apoptosis↓ |
|
|
|
Migration↑/Invasion↑ |
|
|
| Malignant
transformation↑ |
|
|
miR-29a | Migration↑ | (41) |
|
| Invasion↑ |
|
|
miR-107 |
Migration↑/Invasion↑ | (36) |
|
| Anoikis
resistance↑ |
|
|
|
Chemoresistance↑ |
|
|
miR-125a | HBV
replication↓ | (42) |
|
miR-143 |
Migration↑/Invasion↑ | (23) |
|
miR-146a | Inflammation↑ | (24) |
|
miR-148a | Proliferation↑ | (25,34) |
|
|
Migration↑/Invasion↑ |
|
|
| EMT↑ |
|
|
miR-181 | Malignant
transformation↑ | (47) |
|
miR-215 | Proliferation↑ | (49) |
|
miR-221 | Proliferation↑ | (51) |
|
miR-224 |
Migration↑/Invasion↑ | (55) |
|
miR-331-3p | Proliferation↑ | (56) |
|
| Apoptosis↓ |
|
|
miR-374a | Proliferation↑ | (58) |
|
|
Migration↑/Invasion↑ |
|
|
miR-545 | Proliferation↑ | (58) |
|
|
Migration↑/Invasion↑ |
|
|
miR-602 | Proliferation↑ | (61) |
|
| Apoptosis↓ |
|
|
Downregulated miRNA by
HBx |
|
|
Let-7 | Proliferation↑ | (30) |
|
| Apoptosis↓ |
|
|
| Cell cycle
arrest↓ |
|
|
miR-15a | HBV
replication↓ | (19) |
|
miR-15b | Proliferation↑ | (39,40) |
|
| HBV
replication↓ |
|
|
miR-16 | Proliferation↑ | (29) |
|
miR-16-1 | HBV replication
↓ | (19) |
|
miR-101 | DNA
methylation↑ | (43) |
|
miR-122 | Proliferation↑ | (26,44) |
|
| Adipogenesis↑ |
|
|
| HBV
replication↓ |
|
|
| EMT↑ |
|
|
miR-132 | Proliferation↑ | (33) |
|
miR-136 |
Migration↑/Invasion↑ | (45) |
|
miR-145 | Apoptosis↓ | (46) |
|
Downregulated miRNA by
HBx |
|
miR-152 | DNA
Methylation↑ | (48) |
|
miR-193b | Proliferation↑ | (50) |
|
| Apoptosis↓ |
|
|
miR-205 | Adipogenesis↑ | (52–54) |
|
miR-216b | Triglyceride
accumulation↑ | (28) |
|
| Proliferation↑ |
|
|
|
Migration↑/Invasion↑ |
|
|
miR-373 |
Migration↑/Invasion↑ | (57) |
|
miR-375 |
Migration↑/Invasion↑ | (45) |
|
miR-429 | Proliferation↑ | (59) |
|
| Adipogenesis↑ |
|
|
miR-520b | Proliferation↑ | (32) |
|
| Apoptosis↓ |
|
|
miR-548p | Proliferation↑ | (27) |
|
| Apoptosis↓ |
|
|
miR-661 | Inflammation↑ | (60) |
The effect of the particular HBx form on
miRNA
Full length HBx protein (wild-type HBx, wtHBx) and
its C-terminal truncated variants (trHBx proteins or Ct-HBx) are
both contributing to the development of HCC. trHBx proteins are
detected in HCC and differ significantly with wtHBx in their
biological activities (62).
The C-terminal region of wtHBx, which is
indispensable for wtHBx stability and contributed to the
wtHBx-mediated stimulation of HBV replication, starts from residues
58 and ends up with 140 (63).
Under normal circumstances, wtHBx could stimulate the replication
of HBV (63), induce apoptosis
(64), inhibit cell growth
(64), inhibit cell transformation
(64) and exert transactivation
function (66), as a contrast,
trHBx could not play a part in the above-mentioned processes.
Nonetheless, wtHBx and trHBx are expressed in hepatocellular
carcinoma cells and enhance the invasiveness and metastasis
formation simultaneously (67,68).
Researchers previously identified a natural mutant
of the HBx gene with C-terminal-deletion from 382 to 401 bp (termed
HBxΔ127), which strongly enhanced cell proliferation and migration
in HCC relative to wild-type HBx (69). The specific mechanism is that
HBxΔ127 strongly upregulates miR-215 in hepatoma cells, and the
latter directly targets PTPRT (protein tyrosine phosphatase,
receptor type T) mRNA which normally acts as a tumor suppressor
(49). Another important
C-terminal-deletion formation is HBxΔ35. It showed that the
upregulation of miR-21 which induces hepatocarcinogenesis (70) was mediated by HBxΔ35-induced
interleukin-6 pathway followed by activation of STAT3
transcriptional factor (37).
Except for the C-terminal-deletion formation of HBx, recent studies
suggest that viral-human hybrid RNA transcripts, which play a
critical role in promoting HCC progression, may be the molecules
that link HBV infection to HCC development. High-throughput
next-generation sequencing studies suggest that HBV DNA integration
does not occur at random locations but rather often occurs within
or near-repetitive, noncoding sequences, such as long interspersed
nuclear elements (LINEs) and short interspersed nuclear elements
(SINEs) (71). As a result, there
is a possibility that HBV DNA may integrate into host chromosomes
at locations that yield particular host/viral fusion products.
Researches recently indicated a viral-human hybrid RNA transcript,
HBx-LINE1-674 (denoted as HBx-LINE1), in HBV-positive
HCC (72). HBx-LINE1 contains six
miR-122-binding sites, and overexpression of HBx-LINE1 effectively
depleted cellular miR-122, promoting hepatic cell
epithelial-mesenchymal transition (EMT), proliferation, hepatic
injury, β-catenin signaling activation, E-cadherin reduction and
cell migration (44). Besides,
HBx-LINE1 acts as a sponge to sequester cellular miR-122 and thus
affects the expression and function of miR-122 target genes.
miR-122 is a tumor suppressor and a crucial inhibitor factor for
HBV infection (73–75). HBx-LINE1 rapidly decreases cellular
mature miR-122 level which is an important step for HBV to
successfully infect hepatocytes. Interestingly, the same phenomenon
was reported in HCV infection about miR-122 sequestration, in other
words, HCV RNA resulted in a miR-122 sequestration directly and
induced a global de-repression of target genes by miR-122,
ultimately, creating an environment fertile for the long-term
oncogenic potential of HCV (76).
The influence of HBx through regulating
miRNA for hepatocarcinogenesis
HBx regulating the expression of miRNA to alter the
characteristic of hepatocyte biological behavior induces a
complicated program of hepatocarcinogenesis. In this regulatory
network, the concrete mechanism is described in detail below
(Figs. 1–3).
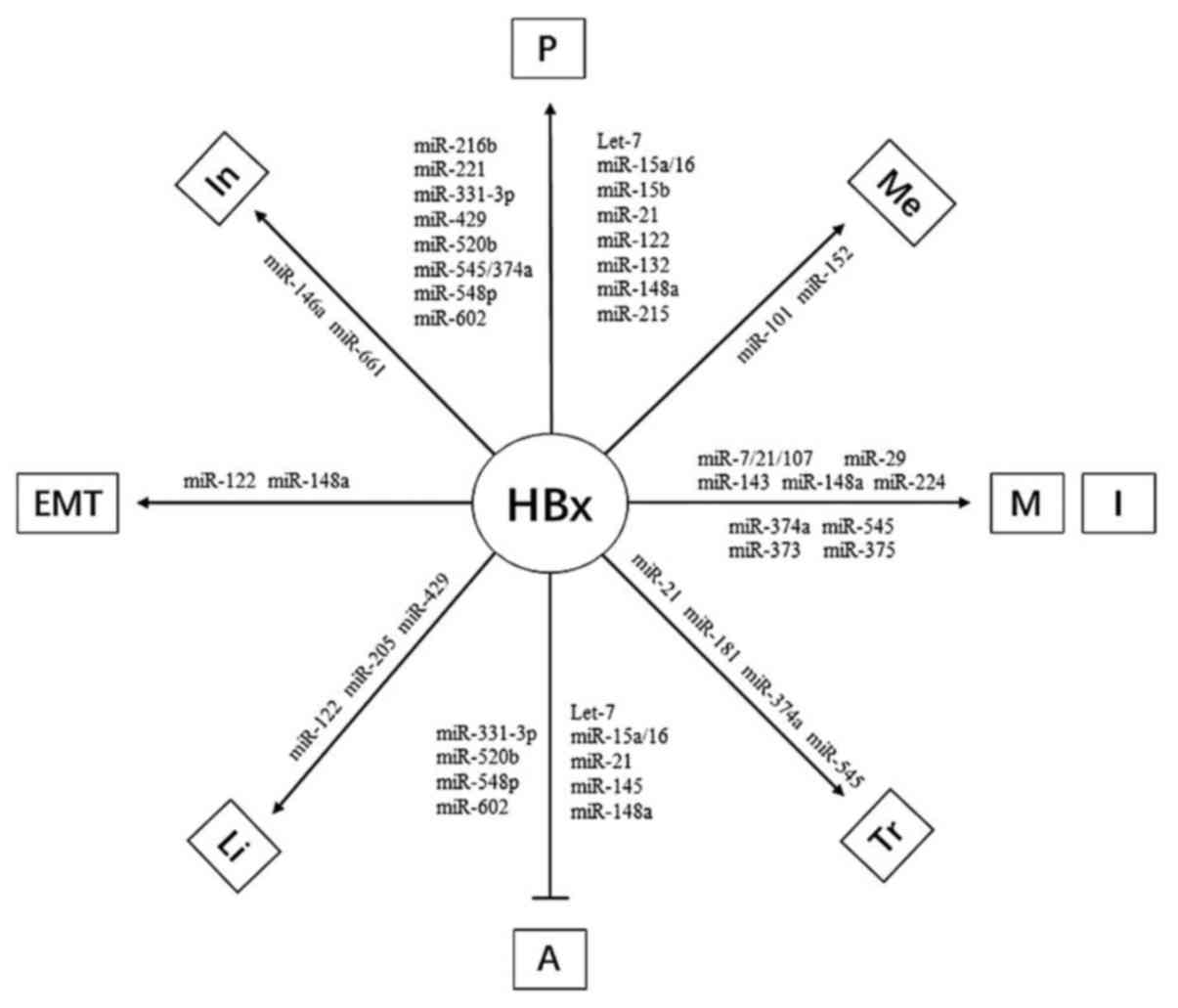 | Figure 3.The final result of HBx influencing
hepatocarcinogenesis through miRNAs. →, promote; ˧, inhibit; A,
apoptosis; P, proliferation; M, migration; I, invasion; Tr,
transformation; In, inflammation; Li, lipogenesis; Me, methylation;
EMT, epithelial mesenchymal transition. |
Proliferation
The results in previous study suggest that
microRNA-602 plays an important regulatory role in HBx-mediated
hepatocarcinogenesis by inhibiting the tumor suppressive function
of RASSF1A, from very early stage of chronic HBV hepatitis and
HBV-positive cirrhosis through HCC (61). A similar mechanism also exists in
miR-331-3p which is upregulated by HBx via enhancing its promoter
activity, and miR-331-3p promotes proliferation of HCC cells
through targeting ING5 (56). In
patients with HCC, miR-216b, which could be reduced by HBx,
functions as a tumor suppressor by targeting IGF2BP2 and
subsequently suppressing the downstream IGF2, AKT/mTOR and MAPK/ERK
signaling pathways, finally, inducing the proliferation (28). Several long non-coding RNAs
(lncRNAs) also play various roles in HCC progression. The Ftx
transcript, which is a conserved functional lncRNA, encodes 4
microRNAs in its introns (77), and
intron 12 encodes 1 cluster of 2 microRNAs (miR-374b and miR-421)
while intron b encodes 1 related cluster of 2 microRNAs (miR-374a
and miR-545). miR-545/374a is positively regulated by HBx, and
promotes HCC cell proliferation via downregulating ESRRG
(estrogen-related receptor gamma) (58).
miR-16 family (miR-15a, miR-15b and miR-16) members
are frequently suppressed by HBx expression. CCND1 promotes cell
cycle progression, and is downregulated by miR-16 directly
(29). Moreover, FUT2, which is the
primary synthetase of Globo H, is the target of miR-15b (40). Globo H is a member of the family of
antigenic carbohydrates that are highly expressed in various types
of cancers as cancer-associated carbohydrate antigens (78) and inducing the proliferation.
miR-148a, inhibited by HBx through decreasing the recruitment of
p53 in its promoter region, decreases the expression of PTEN which
can repress the activity of PI3K and HPIP (hematopoietic pre-B cell
leukemia transcription factor-interacting protein) which regulates
cancer cell growth through activation of AKT and ERK (25,34),
in addition, miR-132, another target of HBx and downregulated
through methylation of its promoter region, represses the
expression of Akt directly (33).
In HCC, HBx activates Rab18 through downregulating
miR-429 directly, synchronously, HBx activates Rab18 promoter
through COX-2 and 5-LOX involving AP-1 and CREB, in brief, there
are two pathways such as HBx/COX-2/5-LOX/AP-1/CREB/Rab18 and
HBx/miR-429/Rab18 involved in hepatocarcinogenesis (59). Similar regulatory mechanism presents
in regulation of HBx for miR-520b. Actually, HBx alone is
considered a poor transformer in human hepatic cells, some
oncogenes, such as H-ras or c-Myc, is necessary for accelerating
hepatocarcinogenesis (79).
Experimental investigation shows that HBx downregulates miR-520b
through interacting with Sp1 with survivin, and mammalian hepatitis
B X-interacting protein (HBXIP) which was originally identified as
a binding protein of HBx is the target protein of miR-520b and
induces the proliferation, in a word, HBX-Sp1-survivin complex
increases the expression of HBXIP by repressing the miR-520b
(32). Notably, HBx also directly
upregulate HBXIP in hepatoma cells through inducing demethylation
of CpG islands of HBXIP promoter with its partner survivin.
Adversely, miR-520b targets mitogen activated protein kinase kinase
kinase 2 (MEKK2) and cyclin D1 inhibits the proliferation of liver
cancer cells (80), indicating the
multiple function of HBx in hepatocarcinogenesis. With regard to
HBXIP, it is a new oncoprotein which was originally identified by
its interaction with the C-terminus of HBx. The function of HBXIP
protein is to negatively regulate HBx activity and thus to alter
the replication life cycle of the virus (81). Besides, miR-548p, which is reduced
by hepatocyte nuclear factor-4a (HNF4A) mediated by HBx
downregulates HBXIP by directly targeting 3UTR of HBXIP mRNA and
induces the tumorigenesis finally. Interestingly, except for
HBx/HNF4A/miR-548p/HBXIP regulation axes, HBx suppresses the
expression of miR-548p directly, furthermore, HNF4A directly
downregulates the transcription of HBXIP by binding to the first
exon, which is not dependent on post-transcriptional regulation
pathway by miR-548p (27).
Moreover, the identification of potent mutant of HBx
in hepatocarcinogenesis is significant, such as HBxΔ127 and HBxΔ35.
For regulating miR-21, HBx and HBxΔ35 play a part concurrently.
Discriminatively, HBx protein increases the expression of miR-21
directly, on the contrary, HBxΔ35 depends on IL-6 and STAT3 which
are the targets of HBxΔ127 at the same time, and HBxΔ127 regulates
the STAT3 through inducing the miR-215 and repressing protein
tyrosine phosphatase receptor type T (PTPRT) which is a tumor
suppressor (37,48,49).
Previous data indicate that estrogen receptor-α
(ERα) has protective effects on HCC (82). As a result, miR-221 negatively
regulates ERα expression by interaction with the 3′-untranslated
region (3′UTR) of ERα in breast cancer (83). To date, it has been found that
upregulation of miR-221 was unregulated by HBx protein in HCC, and
the overexpression of miR-221 was found to induce cell
proliferation by suppressing ERα (51). Estrogens are well accepted as
cancer-promoting agents in both the breast and the uterus (84). However, they play a part in
contradictory influence to protect individuals against HCC possibly
dependent on the estrogens through tissue-specific selective ER
modulators (85).
Interestingly, HBx is not only inducing
proliferation. In a previous study, data surprisingly revealed that
HBx decreases cell proliferation of HCC cells by suppressing EGFR
protein expression, mechanically, targeting EGFR mRNA 3UTR by
upregulated microRNA-7 (miR-7) in response to HBx accounts for the
suppression of EGFR protein level in HBx-expressing HCC cells
(35). Furthermore, EGFR has also
been reported to induce miR-7 transcription relying on its tyrosine
kinase activity (86), suggesting
miR-7 as a negative feedback regulator of EGFR expression. Thus,
the roles of HBx in proliferation through regulating miRNA remain
controversial, there is a delicate balance between inducement and
inhibition.
Apoptosis
In hepatocarcinogenesis, HBx regulates apoptosis by
increasing or decreasing miRNAs. For example, as mentioned above,
HBx regulates miR-21, miR-331-3p, miR-520b, miR-548p and miR-602
via corresponding mechanism to induce the proliferation of
hepatocytes, these regulatory networks could repress apoptosis of
hepatocyte coinstantaneous. Main reason of this phenomenon is that
the target proteins of this miRNA, such as RASSF1A and HBXIP, could
not only influence proliferation but also apoptosis. Furthermore,
as a member of the Cullin-RING E3 ubiquitin family, Cullin-5 (CUL5)
has been reported to be functionally involved in numerous cellular
activities including the cell cycle and apoptosis (87). CUL5 is the targets of miR-145 which
is downregulated by HBx, finally, the overexpression of CUL5
reduces apoptosis.
Anoikis, from the Greek word ‘homelessness’, is a
particular programmed apoptotic death due to loss or inappropriate
cell adhesion (88). In spite of
its unique definition, anoikis is essentially an apoptotic process
(89). Mammary serine protease
inhibitor (Maspin, also named serpin B5) which is a member
belonging to the serine protease inhibitor (serpin) superfamily
(90) has been shown to reduce
tumor growth, metastasis and angiogenesis (91). For hepatocarcinogenesis, taken
together, recent study demonstrated that HBx enhanced the levels of
microRNA-7, miR-107 and miR-21 to promote HCC tumor progression
involving anoikis resistance by directly targeting and suppressing
maspin expression (36). As a
result, the increase of anoikis resistance of HCC cell would lead
to acquirement of the ability to survive in circulation, and
finally presents distant metastasis. In a word, proliferation,
apoptosis, invasion and metastasis supplement each other, and that
they could not play a part isolated in tumorigenesis without the
others.
For regulating apoptosis of HCC cells, feedback loop
exists in HBx, miRNA targeting proteins. Programmed cell death 4
(PDCD4), a novel tumor suppressor gene (TSG), which is expressed
ubiquitously in normal tissues with highest levels in liver, was
recently reported to be downregulated in HCC (92). A recent study reported for the first
time that HBx downregulates PDCD4 and upregulates miR-21
expression, and the low-expression of PDCD4 induced by miR-21 could
suppress apoptosis (31).
Interestingly, HBx has previously been shown to activate tumor
promoting signals including protein kinase C (PKC) and activator
protein-1 (AP-1) (93) and miR-21
is positively regulated by AP-1 (94). The upregulation of miR-21 by HBx may
be through AP-1. Moreover, AP-1 activity is reported to be
negatively regulated by PDCD4 (95), leading to the formation of positive
feedback loop between AP-1/miR-21/PDCD4. Besides, Lin28, which is
oncofetal gene, is thought to promote tumorigenesis upon
re-expression in somatic cells. Lin28B, as a homologue of Lin28,
was first cloned from and shown to be overexpressed in human
hepatocellular carcinoma cells and clinical samples (96). Due to the pleiotropic functions of
HBx, it demonstrated that HBx induced the upregulation of c-Myc in
HepG2 cells which is capable of trans-activating Lin28B (29). In brief, HBx/c-Myc/Lin28B axis
mediated the repression of let-7, and both c-Myc and Lin28B
themselves are the targets of let-7, that is to say, c-Myc/Lin28B
and let-7 antagonize each other, maintaining a balancing
equilibrium between these factors in apoptosis (30).
Actually, the relationship between HBx and apoptosis
is controversial. On the one hand, it has been reported that HBx
could inhibit hepatocyte apoptosis by miRNAs and target proteins,
and on the other hand, there is a p53-independent pro-apoptotic
effect of HBx in vivo and in vitro (97). In short, the regulation of apoptosis
for hepatocarcinogenesis by HBx targeting miRNA is complicated and
multidirectional.
Migration and invasion
HBx does not bind DNA directly, its multi-regulative
functions are mediated by its interactions with host factors, such
as NF-κB and p53 (22,98). NF-κB transcriptional activity was
significantly enhanced through interacting with HBx in HBV-HCC
(99). Research has shown that
upregulation of miR-143 expression transcribed by NF-κB in HBV-HCC
promotes cancer cell invasion/migration and tumor metastasis by
repression of FNDC3B expression (23). Similarly, HBx inhibits miR-148a
transcription through reduced recruitment of p53 to the miR-148a
promoter, and the latter suppresses the mTOR pathway through
inhibition of HPIP/AKT and HPIP/ERK pathways, furthermore, mTOR is
a serine/threonine protein kinase that regulates cell
proliferation, migration and invasion. Taken together, HBx
suppresses p53-mediated activation of miR-148a and miR-148a
suppresses liver cancer cell migration and invasion in vitro
through inhibition of HPIP expression, which targets the mTOR
pathway (25). Importantly,
miR-148a could repress PTEN directly which is a phosphoinositide
phosphatase that originally was identified as a tumor suppressor
frequently promoting tumorigenesis. PTEN was indicated to be able
to inhibit migration and invasion through regulation of PI3K/Akt
pathway or SRC family kinases (100). Recent studies have indicated that
PTEN was also shown to be a direct target of miR-148a and miR-29a,
both of them could be upregulated by HBx. Thus, HBx induces the
invasion and migration by promoting miR-29a and miR-148a
expression, and both of them suppress the expression of PTEN
(34,41).
The transmembrane glycoprotein E-cadherin provides a
physical link between adjacent cells and is crucial for cell
polarity and the structural integrity of tissue (101), loss of E-cadherin is associated
with acquisition of metastatic capacity. HBx represses the
expression of miR-373 which is a positive regulator of E-cadherin
expression, finally, decreased E-cadherin mediated by HBx induces
the invasion and migration in HCC (57). Maspin increases cellular adherence
to fibronectin via inducing integrin expression, leading to a
reduction of invasion (102). HBx
induces microRNA-7/103/107 and miR-21 to suppress maspin
expression, finally, decreased Maspin promote tumor progression by
downregulated cell adhesion and increased cell motility (36). Besides, HBx increases AEG-1
expression by downregulating miR-375 and miR-136, and increased
AEG-1 could induce the migration and invasion in HCC (45). In parallel, expression of
miR-545/374a which promotes HCC cell migration and invasion, is
positively regulated by HBV infection and can be induced by HBx
expression (58).
Methylation
Growing evidence additionally supports a role of
miRNAs as both targets and effectors in aberrant mechanisms of DNA
hypermethylation (103). In
mammalian cells, the DNA methyltransferase (DNMT) family contains
DNMT1, DNMT3A, and DNMT3B which catalyze the addition of a methyl
group to the 5-CpG dinucleotide of the cytosine ring. Our present
study indicated that miR-101 is frequently downregulated in an
HBx-expressing HCC cell line, and DNMT3A is a direct target of
miR-101, finally, HBx is sufficient to upregulate DNMT3A expression
(43). In addition, a study showed
that HBx can repress miR-152 which can inhibit DNMT1, miR-152
induces a decrease in global DNA hypermethylation and an increase
in the methylation level of two tumor suppressor genes, GSTP1 and
E-cadherin (48). In a recent study
it was found that HBx was able to suppress miR-205 expression in
hepatoma and liver cells through inducing hypermethylation of
miR-205 promoter, resulting in the proliferation of hepatoma.
Interestingly, miR-205 can directly target HBx mRNA, and repress
the expression of HBx. Thereby, HBx and miR-205 presents a negative
feedback control in stimulating the tumor growth process (52).
Taken together, HBx, by dysregulating the DNMTs
targets miRNAs, and finally changing the methylation level of tumor
suppressor genes and influencing hepatocarcinogenesis.
Epithelial-mesenchymal transition
(EMT)
In recent studies, it has been suggested that
epithelial cancer cells may convert to motile mesenchymal ones by
undergoing an epithelial-mesenchymal transition (EMT) in
tumorigenesis, which is characterized by loss of cell adhesion,
repression of E-cadherin expression, acquisition of mesenchymal
markers (including N-cadherin, vimentin and fibronectin) and
increased cell motility and invasiveness (11).
As mentioned above, HBx-LINE1 promotes
hepatocellular carcinoma progression via depleting cellular miR-122
and miR-122 serves as a tumor suppressor and negatively regulates
cancer cell proliferation, invasion and metastasis. Reduced miR-122
can activate β-catenin signaling and induce EMT (44). Besides, RhoA, a member of Ras
homolog gene family, is a potential functional target of miR-122,
suggesting that miR-122 may block EMT through the direct inhibition
of cellular RhoA simultaneously (104).
Except for inducing cell growth, invasion and
migration, HBx/miR-148a/HPIP/mTOR axis also promotes EMT. Moreover,
miR-148a increases expression of the epithelial marker E-cadherin
and decreases the E-cadherin repressor Snail as well as N-cadherin
and vimentin, which are mesenchymal markers, accompanied by the
inhibition of mTOR signaling. In conclusion, HBx suppresses
p53-mediated activation of miR-148a and promotes EMT mediated HPIP
(25).
Autophagy
Autophagy is a natural catabolic cellular process
for damaged organelles, misfolded proteins and pathogen clearing
(105). A previous study has shown
that HBV survival and its replication needs autophagy (106), and the HBx protein has been
demonstrated to be the major molecule involved in inducing
autophagy during HBV infection (107).
Autophagosome formation requires beclin-1 to
activate PI3 kinase class III into PI3K-phosphate which in turn
recruits effectors such as double FYV6 to initiate this process
(108). Previously, it has been
shown that HBx induces autophagosome formation via upregulation of
beclin-1 expression (107).
Besides, HBx-induced autophagosome formation is inhibited by
miRNA-30a overexpression which inhibits both mRNA and protein
expression of beclin-1 directly (109). Simultaneously, the study shows
that miR-30a has no effect on the expression of HBx. Briefly,
miRNA-30a could successfully inhibit HBx-induced autophagosome
formation and induce apoptosis in hepatic cells when HBx was
co-expressed along with miRNA-30a which could successfully negate
the procarcinogenic effects of HBx and protect the cells against
HBV infection.
Inflammation
In the past decade, abundant research showed that
chronic hepatitis B virus (HBV) or hepatitis C virus infection
accounts for >75% hepatocellular carcinoma (HCC) cases (110). Furthermore, HBx has also been
implicated in inflammatory response in chronic HBV infection during
HCC development (111). In this
process, HBx activates NF-κB signaling (112) and enhances the expression of its
downstream inflammatory targets, such as inducible nitric-oxide
synthase (iNOS) (113) which is
induced in a calcium/calmodulin-independent manner and generates NO
in a sustained manner (114).
Recent studies suggest that HBx requires MTA1 (metastatic tumor
antigen 1) which plays significant roles in both tumor biology and
inflammation to stimulate the iNOS. Besides, MTA1 can be suppressed
directly by miRNA-661 which is downregulated by HBx. Interestingly,
although the levels of miR-661 transcript were markedly reduced by
HBx, there was no effect of HBx upon the miR-661 promoter
luciferase activity, probably, HBx affects the levels of miR-661 by
modulating the stability of miR-661. In conclusion, in chronic
inflammatory response, HBx upregulates the expression of MTA1 by
repressing the level of miR-661 to induce iNOS, resulting in the
increasing of NO which has been implicated in the pathogenesis of
inflammatory disorders, including hepatitis B virus-associated
hepatocellular carcinoma (60).
The infection of HBV for hepatocytes do not account
for the cytopathic, instead, repeating immune responses of the host
results in continuous cycles of low-level liver cell destruction
and regeneration (115). Because
of this mechanism, 15–40% chronic hepatitis B (CHB) carriers
develop liver cirrhosis and hepatocellular carcinoma (HCC)
(116). Complement factor H (CFH),
an important negative regulator of the alternative pathway of
complement activation, were differentially expressed in
HBV-expressing and HBV-free hepatocytes. In hepatocytes and hepatic
tissue, HBx downregulates CFH expression, and lower CFH levels in
hepatocytes to be a direct cause of the liver inflammation
(24). The mechanism of HBx
regulating CFH is dependent on miR-146a which is an innate
immunity-related miRNA and induced by HBx through enhancing its
promoter activity. Luciferase reporter assays demonstrated that
miR-146a downregulated CFH mRNA expression in hepatocytes via
3-untranslated-region (UTR) pairing (24). In brief, aforementioned studies
demonstrate that the HBx/miR-146a/CFH/complement activation
regulation pathway axis might play an important role in the
immunopathogenesis of chronic HBV infection.
Malignant transformation
Accumulating evidence has shown that cancer stem
cells (CSCs), such as hepatic stem cells (HSC), have a
tumor-initiating capacity and play crucial roles in tumor
metastasis (117). Epithelial cell
adhesion molecule (EpCAM), containing intracellular domain, EpICD,
is a marker of HSCs and CSCs (118) and acts as a mitogenic signal
transducer via proteolysis and nuclear translocation, which binds
to DNA by a complex with the scaffolding protein FHL2, β-catenin
and lymphoid enhancer factor (LEF)-1 and regulates cell
proliferation (119).
Recent studies show that EpCAM can be regulated by
miR-181 (120). Furthermore,
experimental results suggest that miR-181 is an epigenetic target
of HBx (47). In general, HBx
induced expression of EpCAM by upregulating miR-181 to promote
stemness. Chronic HBV infection often extends for decades, but the
afore-mentioned stemness readily appear in liver just prior to the
appearance of HCC, actually, the HBx levels of HBV carriers
increase with the length of time in the liver, that is to say, the
highest levels of HBx expression is always seen in the cirrhotic
liver. In a word, the mechanism of HCC arising most often from
cirrhotic livers can be accounted for HBx promoting ‘stemness’ by
EpCAM through miR-181 in pathologic setting which is central to
early-stage tumor development.
Except for miR-181, HBx-miR-21 pathway was prevalent
in HCC cells, it was reported that upregulation of miR-21 was
mediated by HBx-induced interleukin-6 pathway followed by
activation of STAT3 transcriptional factor, finally, increased
miR-21 is able to induce cell transformation (37,72).
Lipid metabolism
A hallmark of cancer cells is a higher rate of
metabolism to sustain cell proliferation and metastasis (121). Cancer cells present excessive
caloric intake to keep a positive balance of energy and
biosynthetic requirements. Therefore, cancer cells get through
shifting lipid acquisition from lipid uptake toward de novo
lipogenesis to change membrane properties and protects cells from
both endogenous and exogenous damage, which is able to maintain
tumorigenesis (122). In brief,
cancer cells are always endowed with increasing lipogenesis.
Rab guanosine triphosphatases, which are regulators
of vesicular transport in both exocytic and endocytic pathways in
eukaryotic cells, are members of the Ras oncogene superfamily
(123). Rab18, a member of Ras
family, has been reported to be involved in lipogenesis of 3T3-L1
adipocytes (124). Recent studies
indicated that miR-429 was able to directly target the
3′-untranslated region of Rab18, showing that Rab18 is one of the
target genes of miR-429, furthermore, HBx was able to downregulate
miR-429 in hepatoma cells, in conclusion, HBx induces
hepatocarcinogenesis through leading to the dysregulation of
lipogenesis of hepatoma cells, depending on HBx/miR-429/Rab18
regulatory axis (59).
The development of liver cancer might be associated
with the accumulation of cholesterol in the tissues as well
(125). Acyl-CoA synthetase
long-chain family member (ACSL) catalyzes the ATP-dependent
acylation of fatty acids into long-chain acyl CoAs (LCA-CoAs),
which is the first step in lipid metabolism after fatty acid entry
into the cell (126). ACSL1 is one
of five isoforms, which is important in activating fatty acid
destined for triacylglycerol synthesis (127). Simultaneously, ACSL4 is an
essential enzyme in the steroid synthesis (128). Previous research has shown that
miR-205 is capable of downregulating ACSL1 and ACSL4 via targeting
its 3UTR in hepatoma cells, furthermore, HBx can repress the
expression of miR-205, so as a consequence, HBx induces the
accumulation of cholesterol and acceleration of lipogenesis via
increasing ACSL1 and ACSL4 as well (53,54).
Except for aforementioned miRNA, miR-122 is the
other crucial regulation molecule for cholesterol and fatty-acid
metabolism (129). Downregulated
miR-122 increases the genes involved in cholesterol biosynthesis
such as HMG-CoA reductase and lipid synthesis gene such as Agpat1,
Mogat1, Agpat3, Agpat9, Ppap2a and Ppap2c, besides, low-expressed
miR-122 enhances TG accumulation through increasing Cidec, also
known as FSP27, which is a protein that localizes to lipid
droplets, negatively regulating lipolysis (129). In fact, HBx could reduce the
miR-122 transcription level via binding and sequestering endogenous
miR-122 other than affecting its promoter activity, in addition,
HBx reduced the Gld2 promoter activity which could increase the
specific miRNA stabilization by monoadenylation (26). Recently, a study demonstrated that
miR-122 could be stabilized by Gld2 (130), as a result, HBx is a critical
protein for lipid metabolism, which regulates miR-122 via
downregulating Gld2 (26).
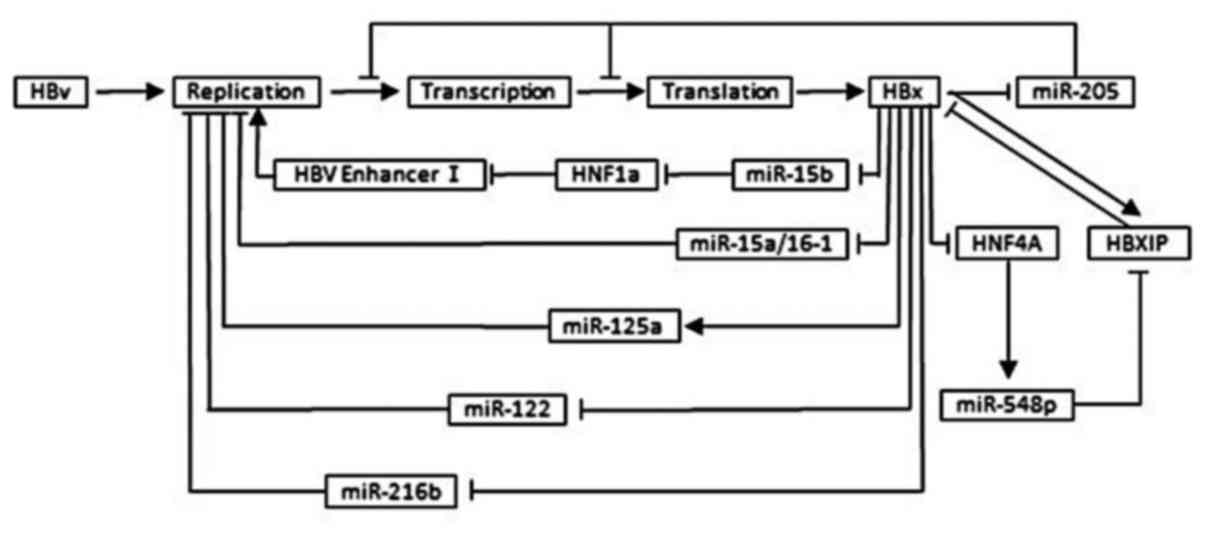
The effect of miRNA on HBV replication and
HBx expression
Depending on targeting viral transcripts or cellular
factors, specific microRNAs can inhibit viral replication, on the
other hand, it enhances viral replication by compromising the host
defense system or negative regulators of viral replication as well.
For HBV, plenty of miRNAs was suggested to target specific HBV
transcripts to inhibit HBV replication being the reason of chronic
viral infection, which in turn, alters the expression of specific
microRNAs (131). In a word, this
regulatory circuit helps HBV to fine-tune its replication, keeping
it low to escape the immune system and establish a chronic
infection. This mechanism balances the relationship between virus
and host to sustain long-term survival of the virus to damage
hepatocytes and spread in humans (42) (Fig.
4).
A large amount of evidence has documented that
specific microRNAs can target viral transcripts, which was
initially thought to serve as an innate immune system against viral
infection. It was shown that miR-15a, functions as part of the RISC
complex, suppress HBV replication in hepatocytes, simultaneously,
miR-15a could efficiently repress HBx expression through its
targeting site within the HBx coding region, interestingly, both
the protein and RNA versions of HBx were able to downregulate
miR-15a. In conclusion, miR-15a represses the expression of HBx by
reducing the replication of HBV and targeting the HBx coding
region, in turn, HBx clearly induced the repression of miR-15a
(19). Importantly, this feedback
loop and self-suppression maintain the appropriate level of HBV
quantity to induce chronic infection. Except for miR-15a, miR-15b,
miR-125a, miR-205 and miR-548p present the same interaction
inhibition and self-suppression with HBV replication or HBx
expression. miR-15b, another members of miR-15/16 family, directly
binds to hepatocyte nuclear factor 1α (HNF1α) mRNA, a negative
regulator of HBV enhancer I, to attenuate HNF1α expression,
resulting in transactivation of HBV Enhancer I, causing the
enhancement of HBV replication and expression of HBV antigens,
including HBx protein (39).
Interestingly, this study indicated that HBx downregulates the
level of intracellular miR-15b at well, also, it has been reported
that HBx protein can stimulate the DNA binding activity of HNF1α
(132). As a consequence, miR-15b
and HBx forms a complex interaction network to sustain, to a
certain extent, expression of HBx and virus replication. As for
miR-125a, researchers found a self-inhibitory feedback loop in
which HBV, through HBx, increases the expression of miR-125a, that
in turn interferes with expression of HBV surface antigen, thus
repressing viral replication. Besides, the function of miR-205
seems simpler and more efficient in regulating HBV replication and
HBx expression, because recent study demonstrated that HBx was able
to suppress miR-205 expression in hepatoma and liver cells, in
turn, miR-205 can directly target HBx mRNA and downregulated HBx.
More complicated, HBx can repress the expression of miR-548p
(133,134), depending on inducing HF4A which is
the important regulatory molecule for hepatocyte transformation and
hepatocarcinogenesis (27).
Furthermore, miR-548p downregulates HBXIP by directly targeting
3UTR of HBXIP mRNA, and decreased miR-548p through the above
mechanism inducing the upregulation of HBXIP which is a new
oncoprotein. Interestingly, HBXIP can be upregulated by HBx
directly, furthermore, HBx can repress the expression of miR-548p
directly other than getting through HNF4a exclusively, the latter,
HNF4a can repress the HBXIP, and HBXIP, downstream element of this
regulatory network, can suppress the expression of HBx (27). To sum up, HBx/HNF4A/miR-548p/HBXIP
constitute a complex regulatory circuit to induce tumorigenesis and
sustain an appropriate level of HBx and chronic infection
state.
Except for self-suppression and interaction
suppression between HBx and miRNA to maintain the appropriate level
HBV replication and HBx expression, miRNA mediated by HBx also can
induce the replication of HBV, meanwhile, increasing the expression
of HBx. miR-122 and miR-216b can be repressed by HBx directly and
indirectly, further, this miRNA will repress the replication of HBV
(26,28). Hence, downregulation of miR-122 and
miR-216b can induce the replication of HBV, and as a result,
increasing the expression of HBx, to constitute a positive feedback
and promote tumorigenesis. Whether suppression or facilitation of
HBV replication and HBx expression, will eventually promote chronic
infection and hepatocarcinogenesis, or are merely methods to adapt
for HBV survival in human liver cells, to spread in human and
giving rise to more serious liver impairment, which leads
ultimately to liver cancer, need further clarification.
Conclusion
As mentioned above, in the present study we reviewed
the recent findings on the interaction between HBx and miRNA and
their significance in hepatocarcinogenesis. An increasing number of
research shows that the interaction of HBx and miRNA regulates
numerous biological processes such as proliferation, apoptosis,
migration, invasion, methylation, EMT, autophagy, inflammation,
transformation and lipogenesis. Moreover, at the same time, miRNA
can also regulate the expression of HBx, they constitute a feedback
in mediating HBV replication and HBx expression. Therefore, making
it clear that the interaction of HBx and miRNA will contribute to
an overall comprehension of HCC formation. Besides, by means of
acting on targets which are referred to in the present review, some
novel therapeutic strategies for HBV-associated HCC patients are
likely to be discovered.
Acknowledgements
Dr Dan Song (Department of Hepatobiliary Surgery,
The First Affiliated Hospital of Chongqing Medical University,
Chongqing Medical University) is gratefully acknowledged for his
invaluable cooperation in preparing this review.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGlynn KA, Petrick JL and London WT:
Global epidemiology of hepatocellular carcinoma: An emphasis on
demographic and regional variability. Clin Liver Dis. 19:223–238.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Center MM and Jemal A: International
trends in liver cancer incidence rates. Cancer Epidemiol Biomarkers
Prev. 20:2362–2368. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seeger C and Mason WS: Hepatitis B virus
biology. Microbiol Mol Biol Rev. 64:51–68. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang H, Oishi N, Kaneko S and Murakami S:
Molecular functions and biological roles of hepatitis B virus ×
protein. Cancer Sci. 97:977–983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu C, Zhou W, Wang Y and Qiao L: Hepatitis
B virus-induced hepatocellular carcinoma. Cancer Lett. 345:216–222.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kremsdorf D, Soussan P, Paterlini-Brechot
P and Brechot C: Hepatitis B virus-related hepatocellular
carcinoma: Paradigms for viral-related human carcinogenesis.
Oncogene. 25:3823–3833. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu SY, Lan SH and Liu HS: Autophagy and
microRNA in hepatitis B virus-related hepatocellular carcinoma.
World J Gastroenterol. 22:176–187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J and Ma L: MicroRNA control of
epithelial-mesenchymal transition and metastasis. Cancer Metastasis
Rev. 31:653–662. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Gómez Cedrón M and de Ramírez Molina A:
Microtargeting cancer metabolism: Opening new therapeutic windows
based on lipid metabolism. J Lipid Res. 57:193–206. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peschansky VJ and Wahlestedt C: Non-coding
RNAs as direct and indirect modulators of epigenetic regulation.
Epigenetics. 9:3–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Callegari E, Elamin BK, Sabbioni S,
Gramantieri L and Negrini M: Role of microRNAs in hepatocellular
carcinoma: A clinical perspective. Onco Targets Ther. 6:1167–1178.
2013.PubMed/NCBI
|
|
15
|
Shan X, Ren M, Chen K, Huang A and Tang H:
Regulation of the microRNA processor DGCR8 by hepatitis B virus
proteins via the transcription factor YY1. Arch Virol. 160:795–803.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:(10A). 1902–1910. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ruby JG, Jan CH and Bartel DP: Intronic
microRNA precursors that bypass Drosha processing. Nature.
448:83–86. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Okamura K, Hagen JW, Duan H, Tyler DM and
Lai EC: The mirtron pathway generates microRNA-class regulatory
RNAs in Drosophila. Cell. 130:89–100. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Jiang L, Ji X, Yang B, Zhang Y and
Fu XD: Hepatitis B viral RNA directly mediates down-regulation of
the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J
Biol Chem. 288:18484–18493. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li C, Wang Y, Wang S, Wu B, Hao J, Fan H,
Ju Y, Ding Y, Chen L, Chu X, et al: Hepatitis B virus mRNA-mediated
miR-122 inhibition upregulates PTTG1-binding protein, which
promotes hepatocellular carcinoma tumor growth and cell invasion. J
Virol. 87:2193–2205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cazalla D, Yario T, Steitz JA and Steitz
J: Down-regulation of a host microRNA by a Herpesvirus saimiri
noncoding RNA. Science. 328:1563–1566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feitelson MA and Lee J: Hepatitis B virus
integration, fragile sites, and hepatocarcinogenesis. Cancer Lett.
252:157–170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated microRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li JF, Dai XP, Zhang W, Sun SH, Zeng Y,
Zhao GY, Kou ZH, Guo Y, Yu H, Du LY, et al: Upregulation of
microRNA-146a by hepatitis B virus X protein contributes to
hepatitis development by downregulating complement factor H. MBio.
6:e02459–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng
X, Zhu Z, Jiao H, Lin J, Jiang K, et al: Hepatitis B virus X
protein represses miRNA-148a to enhance tumorigenesis. J Clin
Invest. 123:630–645. 2013.PubMed/NCBI
|
|
26
|
Peng F, Xiao X, Jiang Y, Luo K, Tian Y,
Peng M, Zhang M, Xu Y and Gong G: HBx down-regulated Gld2 plays a
critical role in HBV-related dysregulation of miR-122. PLoS One.
9:e929982014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu XM, Yan XH, Hu YW, Huang JL, Cao SW,
Ren TY, Tang YT, Lin L, Zheng L and Wang Q: MicroRNA-548p
suppresses hepatitis B virus X protein associated hepatocellular
carcinoma by downregulating oncoprotein HBXIP. Hepatol Res.
46:804–815. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang
EL, Wu ZB, Huang ZY and Chen XP: MiR-216b is involved in
pathogenesis and progression of hepatocellular carcinoma through
HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis.
6:e16702015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu G, Yu F, Xiao Z, Xu K, Xu J, Tang W,
Wang J and Song E: Hepatitis B virus X protein downregulates
expression of the miR-16 family in malignant hepatocytes in vitro.
Br J Cancer. 105:146–153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu G, Huang P, Ju X, Li Z and Wang Y:
Lin28B over-expression mediates the repression of let-7 by
hepatitis B virus X protein in hepatoma cells. Int J Clin Exp Med.
8:15108–15116. 2015.PubMed/NCBI
|
|
31
|
Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao
Y, Li Y, Zeng T, Hu J, Zhang L, et al: HBx-mediated miR-21
upregulation represses tumor-suppressor function of PDCD4 in
hepatocellular carcinoma. Oncogene. 32:3296–3305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang
Q, Cai N, Wang H, Liu F, Ye L, et al: Hepatitis B virus X protein
accelerates hepatocarcinogenesis with partner survivin through
modulating miR-520b and HBXIP. Mol Cancer. 13:1282014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu
Z, Liu R and Wu Z: Epigenetic repression of miR-132 expression by
the hepatitis B virus × protein in hepatitis B virus-related
hepatocellular carcinoma. Cell Signal. 25:1037–1043. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yuan K, Lian Z, Sun B, Clayton MM, Ng IO
and Feitelson MA: Role of miR-148a in hepatitis B associated
hepatocellular carcinoma. PLoS One. 7:e353312012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen YJ, Chien PH, Chen WS, Chien YF, Hsu
YY, Wang LY, Chen JY, Lin CW, Huang TC, Yu YL, et al: Hepatitis B
Virus-encoded X Protein downregulates EGFR expression via inducing
MicroRNA-7 in hepatocellular carcinoma cells. Evid Based Complement
Alternat Med. 2013:6823802013.PubMed/NCBI
|
|
36
|
Chen WS, Yen CJ, Chen YJ, Chen JY, Wang
LY, Chiu SJ, Shih WL, Ho CY, Wei TT, Pan HL, et al: miRNA-7/21/107
contribute to HBx-induced hepatocellular carcinoma progression
through suppression of maspin. Oncotarget. 6:25962–25974. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li CH, Xu F, Chow S, Feng L, Yin D, Ng TB
and Chen Y: Hepatitis B virus X protein promotes hepatocellular
carcinoma transformation through interleukin-6 activation of
microRNA-21 expression. Eur J Cancer. 50:2560–2569. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Damania P, Sen B, Dar SB, Kumar S, Kumari
A, Gupta E, Sarin SK and Venugopal SK: Hepatitis B virus induces
cell proliferation via HBx-induced microRNA-21 in hepatocellular
carcinoma by targeting programmed cell death protein4 (PDCD4) and
phosphatase and tensin homologue (PTEN). PLoS One. 9:e917452014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dai X, Zhang W, Zhang H, Sun S, Yu H, Guo
Y, Kou Z, Zhao G, Du L, Jiang S, et al: Modulation of HBV
replication by microRNA-15b through targeting hepatocyte nuclear
factor 1α. Nucleic Acids Res. 42:6578–6590. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu CS, Yen CJ, Chou RH, Chen JN, Huang WC,
Wu CY and Yu YL: Downregulation of microRNA-15b by hepatitis B
virus X enhances hepatocellular carcinoma proliferation via
fucosyltransferase 2-induced Globo H expression. Int J Cancer.
134:1638–1647. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kong G, Zhang J, Zhang S, Shan C, Ye L and
Zhang X: Upregulated microRNA-29a by hepatitis B virus X protein
enhances hepatoma cell migration by targeting PTEN in cell culture
model. PLoS One. 6:e195182011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mosca N, Castiello F, Coppola N, Trotta
MC, Sagnelli C, Pisaturo M, Sagnelli E, Russo A and Potenza N:
Functional interplay between hepatitis B virus X protein and human
miR-125a in HBV infection. Biochem Biophys Res Commun. 449:141–145.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X
and Wu Z: miR-101 is down-regulated by the hepatitis B virus ×
protein and induces aberrant DNA methylation by targeting DNA
methyltransferase 3A. Cell Signal. 25:439–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liang HW, Wang N, Wang Y, Wang F, Fu Z,
Yan X, Zhu H, Diao W, Ding Y, Chen X, et al: Hepatitis B
virus-human chimeric transcript HBx-LINE1 promotes hepatic injury
via sequestering cellular microRNA-122. J Hepatol. 64:278–291.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao J, Wang W, Huang Y, Wu J, Chen M, Cui
P, Zhang W and Zhang Y: HBx elevates oncoprotein AEG-1 expression
to promote cell migration by downregulating miR-375 and miR-136 in
malignant hepatocytes. DNA Cell Biol. 33:715–722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao F, Sun X, Wang L, Tang S and Yan C:
Downregulation of MicroRNA-145 caused by hepatitis B virus X
protein promotes expression of CUL5 and contributes to pathogenesis
of hepatitis B Virus-associated hepatocellular carcinoma. Cell
Physiol Biochem. 37:1547–1559. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Arzumanyan A, Friedman T, Ng IO, Clayton
MM, Lian Z and Feitelson MA: Does the hepatitis B antigen HBx
promote the appearance of liver cancer stem cells? Cancer Res.
71:3701–3708. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang J, Wang Y, Guo Y and Sun S:
Down-regulated microRNA-152 induces aberrant DNA methylation in
hepatitis B virus-related hepatocellular carcinoma by targeting DNA
methyltransferase 1. Hepatology. 52:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu F, You X, Chi X, Wang T, Ye L, Niu J
and Zhang X: Hepatitis B virus X protein mutant HBxΔ127 promotes
proliferation of hepatoma cells through up-regulating miR-215
targeting PTPRT. Biochem Biophys Res Commun. 444:128–134. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yip WK, Cheng AS, Zhu R, Lung RW, Tsang
DP, Lau SS, Chen Y, Sung JG, Lai PB, Ng EK, et al:
Carboxyl-terminal truncated HBx regulates a distinct microRNA
transcription program in hepatocellular carcinoma development. PLoS
One. 6:e228882011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen JJ, Tang YS, Huang SF, Ai JG, Wang HX
and Zhang LP: HBx protein-induced upregulation of microRNA-221
promotes aberrant proliferation in HBV-related hepatocellular
carcinoma by targeting estrogen receptor-α. Oncol Rep. 33:792–798.
2015.PubMed/NCBI
|
|
52
|
Zhang T, Zhang J, Cui M, Liu F, You X, Du
Y, Gao Y, Zhang S, Lu Z, Ye L, et al: Hepatitis B virus X protein
inhibits tumor suppressor miR-205 through inducing hypermethylation
of miR-205 promoter to enhance carcinogenesis. Neoplasia.
15:1282–1291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cui M, Xiao Z, Sun B, Wang Y, Zheng M, Ye
L and Zhang X: Involvement of cholesterol in hepatitis B virus X
protein-induced abnormal lipid metabolism of hepatoma cells via
up-regulating miR-205-targeted ACSL4. Biochem Biophys Res Commun.
445:651–655. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cui M, Wang Y, Sun B, Xiao Z, Ye L and
Zhang X: MiR-205 modulates abnormal lipid metabolism of hepatoma
cells via targeting acyl-CoA synthetase long-chain family member 1
(ACSL1) mRNA. Biochem Biophys Res Commun. 444:270–275. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ,
Tsai TF, Lin YJ, Wu CT and Liu HS: Autophagy suppresses
tumorigenesis of hepatitis B virus-associated hepatocellular
carcinoma through degradation of microRNA-224. Hepatology.
59:505–517. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cao Y, Chen J, Wang D, Peng H, Tan X,
Xiong D, Huang A and Tang H: Upregulated in hepatitis B
virus-associated hepatocellular carcinoma cells, miR-331-3p
promotes proliferation of hepatocellular carcinoma cells by
targeting ING5. Oncotarget. 6:38093–38106. 2015.PubMed/NCBI
|
|
57
|
Arzumanyan A, Friedman T, Kotei E, Ng IO,
Lian Z and Feitelson MA: Epigenetic repression of E-cadherin
expression by hepatitis B virus × antigen in liver cancer.
Oncogene. 31:563–572. 2012.PubMed/NCBI
|
|
58
|
Zhao Q, Li T, Qi J, Liu J and Qin C: The
miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in
HBV-related hepatocellular carcinoma and promotes tumorigenesis and
tumor progression. PLoS One. 9:e1097822014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
You X, Liu F, Zhang T, Li Y, Ye L and
Zhang X: Hepatitis B virus X protein upregulates oncogene Rab18 to
result in the dysregulation of lipogenesis and proliferation of
hepatoma cells. Carcinogenesis. 34:1644–1652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bui-Nguyen TM, Pakala SB, Sirigiri DR,
Martin E, Murad F and Kumar R and Kumar R and Kumar R: Stimulation
of inducible nitric oxide by hepatitis B virus transactivator
protein HBx requires MTA1 coregulator. J Biol Chem. 291:11982016.
View Article : Google Scholar :
|
|
61
|
Yang L, Ma Z, Wang D, Zhao W, Chen L and
Wang G: MicroRNA-602 regulating tumor suppressive gene RASSF1A is
overexpressed in hepatitis B virus-infected liver and
hepatocellular carcinoma. Cancer Biol Ther. 9:803–808. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kew MC: Hepatitis B virus × protein in the
pathogenesis of hepatitis B virus-induced hepatocellular carcinoma.
J Gastroenterol Hepatol. 26:(Suppl 1). 144–152. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lizzano RA, Yang B, Clippinger AJ and
Bouchard MJ: The C-terminal region of the hepatitis B virus X
protein is essential for its stability and function. Virus Res.
155:231–239. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Schuster R, Gerlich WH and Schaefer S:
Induction of apoptosis by the transactivating domains of the
hepatitis B virus X gene leads to suppression of oncogenic
transformation of primary rat embryo fibroblasts. Oncogene.
19:1173–1180. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tu H, Bonura C, Giannini C, Mouly H,
Soussan P, Kew M, Paterlini-Bréchot P, Bréchot C and Kremsdorf D:
Biological impact of natural COOH-terminal deletions of hepatitis B
virus X protein in hepatocellular carcinoma tissues. Cancer Res.
61:7803–7810. 2001.PubMed/NCBI
|
|
66
|
Sirma H, Giannini C, Poussin K, Paterlini
P, Kremsdorf D and Bréchot C: Hepatitis B virus X mutants, present
in hepatocellular carcinoma tissue abrogate both the
antiproliferative and transactivation effects of HBx. Oncogene.
18:4848–4859. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J,
Wang Y, Wu MC, Fung J, Bai X, et al: COOH-terminal truncated HBV X
protein plays key role in hepatocarcinogenesis. Clin Cancer Res.
14:5061–5068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sze KM, Chu GK, Lee JM and Ng IO:
C-terminal truncated hepatitis B virus × protein is associated with
metastasis and enhances invasiveness by C-Jun/matrix
metalloproteinase protein 10 activation in hepatocellular
carcinoma. Hepatology. 57:131–139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang H, Shan CL, Li N, Zhang X, Zhang XZ,
Xu FQ, Zhang S, Qiu LY, Ye LH and Zhang XD: Identification of a
natural mutant of HBV X protein truncated 27 amino acids at the
COOH terminal and its effect on liver cell proliferation. Acta
Pharmacol Sin. 29:473–480. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
McDaniel K, Herrera L, Zhou T, Francis H,
Han Y, Levine P, Lin E, Glaser S, Alpini G and Meng F: The
functional role of microRNAs in alcoholic liver injury. J Cell Mol
Med. 18:197–207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ding D, Lou X, Hua D, Yu W, Li L, Wang J,
Gao F, Zhao N, Ren G, Li L, et al: Recurrent targeted genes of
hepatitis B virus in the liver cancer genomes identified by a
next-generation sequencing-based approach. PLoS Genet.
8:e10030652012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lau CC, Sun T, Ching AK, He M, Li JW, Wong
AM, Co NN, Chan AW, Li PS, Lung RW, et al: Viral-human chimeric
transcript predisposes risk to liver cancer development and
progression. Cancer Cell. 25:335–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lanford RE, Hildebrandt-Eriksen ES, Petri
A, Persson R, Lindow M, Munk ME, Kauppinen S and Ørum H:
Therapeutic silencing of microRNA-122 in primates with chronic
hepatitis C virus infection. Science. 327:198–201. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Shimakami T, Yamane D, Jangra RK, Kempf
BJ, Spaniel C, Barton DJ and Lemon SM: Stabilization of hepatitis C
virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA.
109:941–946. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bandiera S, Pfeffer S, Baumert TF and
Zeisel MB: miR-122 - a key factor and therapeutic target in liver
disease. J Hepatol. 62:448–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Luna JM, Scheel TK, Danino T, Shaw KS,
Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP, et
al: Hepatitis C virus RNA functionally sequesters miR-122. Cell.
160:1099–1110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Romito A and Rougeulle C: Origin and
evolution of the long non-coding genes in the X-inactivation
center. Biochimie. 93:1935–1942. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Astronomo RD and Burton DR: Carbohydrate
vaccines: Developing sweet solutions to sticky situations? Nat Rev
Drug Discov. 9:308–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kim YC, Song KS, Yoon G, Nam MJ and Ryu
WS: Activated ras oncogene collaborates with HBx gene of hepatitis
B virus to transform cells by suppressing HBx-mediated apoptosis.
Oncogene. 20:16–23. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhang W, Kong G, Zhang J, Wang T, Ye L and
Zhang X: MicroRNA-520b inhibits growth of hepatoma cells by
targeting MEKK2 and cyclin D1. PLoS One. 7:e314502012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Melegari M, Scaglioni PP and Wands JR:
Cloning and characterization of a novel hepatitis B virus × binding
protein that inhibits viral replication. J Virol. 72:1737–1743.
1998.PubMed/NCBI
|
|
82
|
Naugler WE, Sakurai T, Kim S, Maeda S, Kim
K, Elsharkawy AM and Karin M: Gender disparity in liver cancer due
to sex differences in MyD88-dependent IL-6 production. Science.
317:121–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhao JJ, Lin J, Yang H, Kong W, He L, Ma
X, Coppola D and Cheng JQ: MicroRNA-221/222 negatively regulates
estrogen receptor alpha and is associated with tamoxifen resistance
in breast cancer. J Biol Chem. 283:31079–31086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Pike MC and Spicer DV: Hormonal
contraception and chemoprevention of female cancers. Endocr Relat
Cancer. 7:73–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Dutertre M and Smith CL: Molecular
mechanisms of selective estrogen receptor modulator (SERM) action.
J Pharmacol Exp Ther. 295:431–437. 2000.PubMed/NCBI
|
|
86
|
Chou YT, Lin HH, Lien YC, Wang YH, Hong
CF, Kao YR, Lin SC, Chang YC, Lin SY, Chen SJ, et al: EGFR promotes
lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc
pathway that targets the Ets2 transcriptional repressor ERF. Cancer
Res. 70:8822–8831. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lubbers J, Lewis S, Harper E, Hledin MP,
Marquez GA, Johnson AE, Graves DR and Burnatowska-Hledin MA:
Resveratrol enhances anti-proliferative effect of VACM-1/cul5 in
T47D cancer cells. Cell Biol Toxicol. 27:95–105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Frisch SM and Francis H: Disruption of
epithelial cell-matrix interactions induces apoptosis. J Cell Biol.
124:619–626. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Taddei ML, Giannoni E, Fiaschi T and
Chiarugi P: Anoikis: An emerging hallmark in health and diseases. J
Pathol. 226:380–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zou Z, Anisowicz A, Hendrix MJ, Thor A,
Neveu M, Sheng S, Rafidi K, Seftor E and Sager R: Maspin, a serpin
with tumor-suppressing activity in human mammary epithelial cells.
Science. 263:526–529. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Bailey CM, Khalkhali-Ellis Z, Seftor EA
and Hendrix MJ: Biological functions of maspin. J Cell Physiol.
209:617–624. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang H, Ozaki I, Mizuta T, Hamajima H,
Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K and Matsuhashi S:
Involvement of programmed cell death 4 in transforming growth
factor-beta1-induced apoptosis in human hepatocellular carcinoma.
Oncogene. 25:6101–6112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tanaka Y, Kanai F, Ichimura T, Tateishi K,
Asaoka Y, Guleng B, Jazag A, Ohta M, Imamura J, Ikenoue T, et al:
The hepatitis B virus X protein enhances AP-1 activation through
interaction with Jab1. Oncogene. 25:633–642. 2006.PubMed/NCBI
|
|
94
|
Talotta F, Cimmino A, Matarazzo MR,
Casalino L, De Vita G, DEsposito M, Di Lauro R and Verde P: An
autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1
activity in RAS transformation. Oncogene. 28:73–84. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yang HS, Jansen AP, Nair R, Shibahara K,
Verma AK, Cmarik JL and Colburn NH: A novel transformation
suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB
or ODC transactivation. Oncogene. 20:669–676. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Guo Y, Chen Y, Ito H, Watanabe A, Ge X,
Kodama T and Aburatani H: Identification and characterization of
lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene.
384:51–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Terradillos O, Pollicino T, Lecoeur H,
Tripodi M, Gougeon ML, Tiollais P and Buendia MA: p53-independent
apoptotic effects of the hepatitis B virus HBx protein in vivo and
in vitro. Oncogene. 17:2115–2123. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ng SA and Lee C: Hepatitis B virus X gene
and hepatocarcinogenesis. J Gastroenterol. 46:974–990. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Kim SY, Kim JC, Kim JK, Kim HJ, Lee HM,
Choi MS, Maeng PJ and Ahn JK: Hepatitis B virus X protein enhances
NFkappaB activity through cooperating with VBP1. BMB Rep.
41:158–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Dasari VR, Kaur K, Velpula KK, Gujrati M,
Fassett D, Klopfenstein JD, Dinh DH and Rao JS: Upregulation of
PTEN in glioma cells by cord blood mesenchymal stem cells inhibits
migration via downregulation of the PI3K/Akt pathway. PLoS One.
5:e103502010. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
van Roy F and Berx G: The cell-cell
adhesion molecule E-cadherin. Cell Mol Life Sci. 65:3756–3788.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Seftor RE, Seftor EA, Sheng S, Pemberton
PA, Sager R and Hendrix MJ: maspin suppresses the invasive
phenotype of human breast carcinoma. Cancer Res. 58:5681–5685.
1998.PubMed/NCBI
|
|
103
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang SC, Lin XL, Li J, Zhang TT, Wang HY,
Shi JW, Yang S, Zhao WT, Xie RY, Wei F, et al: MicroRNA-122
triggers mesenchymal-epithelial transition and suppresses
hepatocellular carcinoma cell motility and invasion by targeting
RhoA. PLoS One. 9:e1013302014. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Tian Y, Sir D, Kuo CF, Ann DK and Ou JH:
Autophagy required for hepatitis B virus replication in transgenic
mice. J Virol. 85:13453–13456. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li
F, Wang Y, Tiollais P, Li T, et al: Hepatitis B virus X protein
sensitizes cells to starvation-induced autophagy via up-regulation
of beclin 1 expression. Hepatology. 49:60–71. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Bhogal RH, Weston CJ, Curbishley SM, Adams
DH and Afford SC: Autophagy: A cyto-protective mechanism which
prevents primary human hepatocyte apoptosis during oxidative
stress. Autophagy. 8:545–558. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Kumar S, Gupta P, Khanal S, Shahi A, Kumar
P, Sarin SK and Venugopal SK: Overexpression of microRNA-30a
inhibits hepatitis B virus X protein-induced autophagosome
formation in hepatic cells. FEBS J. 282:1152–1163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Huo TI, Lee SD and Wu JC: Is diabetes a
risk factor for hepatocellular carcinoma? Gastroenterology.
127:360–361; author reply 361–362. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Hsieh JL, Wu CL, Lee CH and Shiau AL:
Hepatitis B virus X protein sensitizes hepatocellular carcinoma
cells to cytolysis induced by E1B-deleted adenovirus through the
disruption of p53 function. Clin Cancer Res. 9:338–345.
2003.PubMed/NCBI
|
|
112
|
Su F, Theodosis CN and Schneider RJ: Role
of NF-kappaB and myc proteins in apoptosis induced by hepatitis B
virus HBx protein. J Virol. 75:215–225. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Majano P, Lara-Pezzi E, López-Cabrera M,
Apolinario A, Moreno-Otero R and García-Monzón C: Hepatitis B virus
X protein transactivates inducible nitric oxide synthase gene
promoter through the proximal nuclear factor kappaB-binding site:
Evidence that cytoplasmic location of X protein is essential for
gene transactivation. Hepatology. 34:1218–1224. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Jahani-Asl A and Bonni A: iNOS: A
potential therapeutic target for malignant glioma. Curr Mol Med.
13:1241–1249. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chisari FV, Isogawa M and Wieland SF:
Pathogenesis of hepatitis B virus infection. Pathol Biol (Paris).
58:258–266. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Fattovich G, Stroffolini T, Zagni I and
Donato F: Hepatocellular carcinoma in cirrhosis: Incidence and risk
factors. Gastroenterology. 127:(Suppl 1). S35–S50. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Huang R and Rofstad EK: Cancer stem cells
(CSCs), cervical CSCs and targeted therapies. Oncotarget.
8:35351–35367. 2017.PubMed/NCBI
|
|
118
|
Schmelzer E, Wauthier E and Reid LM: The
phenotypes of pluripotent human hepatic progenitors. Stem Cells.
24:1852–1858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Munz M, Baeuerle PA and Gires O: The
emerging role of EpCAM in cancer and stem cell signaling. Cancer
Res. 69:5627–5629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Ji J, Zheng X, Forgues M, Yamashita T,
Wauthier EL, Reid LM, Wen X, Song Y, Wei JS, Khan J, et al:
Identification of microRNAs specific for epithelial cell adhesion
molecule-positive tumor cells in hepatocellular carcinoma.
Hepatology. 62:829–840. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhang F and Du G: Dysregulated lipid
metabolism in cancer. World J Biol Chem. 3:167–174. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Rysman E, Brusselmans K, Scheys K,
Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D,
Daniëls VW, Machiels J, et al: De novo lipogenesis protects cancer
cells from free radicals and chemotherapeutics by promoting
membrane lipid saturation. Cancer Res. 70:8117–8126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Chia WJ and Tang BL: Emerging roles for
Rab family GTPases in human cancer. Biochim Biophys Acta.
1795:110–116. 2009.PubMed/NCBI
|
|
124
|
Pulido MR, Diaz-Ruiz A, Jiménez-Gómez Y,
Garcia-Navarro S, Gracia-Navarro F, Tinahones F, López-Miranda J,
Frühbeck G, Vázquez-Martínez R and Malagón MM: Rab18 dynamics in
adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS
One. 6:e229312011. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Morales A, Mari M, Garcia-Ruiz C, Colell A
and Fernandez-Checa JC: Hepatocarcinogenesis and
ceramide/cholesterol metabolism. Anticancer Agents Med Chem.
12:364–375. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Parkes HA, Preston E, Wilks D, Ballesteros
M, Carpenter L, Wood L, Kraegen EW, Furler SM and Cooney GJ:
Overexpression of acyl-CoA synthetase-1 increases lipid deposition
in hepatic (HepG2) cells and rodent liver in vivo. Am J Physiol
Endocrinol Metab. 291:E737–E744. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Li LO, Ellis JM, Paich HA, Wang S, Gong N,
Altshuller G, Thresher RJ, Koves TR, Watkins SM, Muoio DM, et al:
Liver-specific loss of long chain acyl-CoA synthetase-1 decreases
triacylglycerol synthesis and beta-oxidation and alters
phospholipid fatty acid composition. J Biol Chem. 284:27816–27826.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Duarte A, Poderoso C, Cooke M, Soria G,
Maciel Cornejo F, Gottifredi V and Podestá EJ: Mitochondrial fusion
is essential for steroid biosynthesis. PLoS One. 7:e458292012.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wen J and Friedman JR: miR-122 regulates
hepatic lipid metabolism and tumor suppression. J Clin Invest.
122:2773–2776. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
DAmbrogio A, Gu W, Udagawa T, Mello CC and
Richter JD: Specific miRNA stabilization by Gld2-catalyzed
monoadenylation. Cell Rep. 2:1537–1545. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Liu WH, Yeh SH and Chen PJ: Role of
microRNAs in hepatitis B virus replication and pathogenesis.
Biochim Biophys Acta. 1809:678–685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Li J, Xu Z, Zheng Y, Johnson DL and Ou JH:
Regulation of hepatocyte nuclear factor 1 activity by wild-type and
mutant hepatitis B virus X proteins. J Virol. 76:5875–5881. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Piriyapongsa J and Jordan IK: A family of
human microRNA genes from miniature inverted-repeat transposable
elements. PLoS One. 2:e2032007. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Yuan Z, Sun X, Liu H and Xie J: MicroRNA
genes derived from repetitive elements and expanded by segmental
duplication events in mammalian genomes. PLoS One. 6:e176662011.
View Article : Google Scholar : PubMed/NCBI
|