Introduction
Head and neck cancer (HNC), including cancers of the
oropharynx, larynx, hypopharynx, and sinonasal tract, is the sixth
most common cancer worldwide (1).
In addition, HNC is notoriously difficult to treat, and treatment
is often associated with serious side effects (2). Although these cancers are linked by
common clinicopathological findings, including male predominance,
heavy smoking, habitual alcohol use, and common histological
features (squamous cell carcinoma), some types of HNC are known to
have specific etiologies (3).
Oropharyngeal squamous cell carcinoma (OPSCC) is a specific type of
HNCs often caused by human papilloma virus (HPV) infection, as
demonstrated in recent studies (1,4–6).
Moreover, of HNCs, the carcinogenesis of OPSCC has been well
characterized (4–7).
Although most studies have demonstrated that OPSCC
can be caused by factors known to cause various types of HNCs
(3,4), HPV infection has been increasingly
recognized as a major etiological factor for OPSCC (4–7).
Additionally, HPV-induced OPSCC shows distinct epidemiologic,
clinical, and molecular alterations (8–10).
Thus, the presence/absence of HPV may influence therapy or outcomes
in patients with OPSCC (11). In
addition, this observation suggests that eradication of HPV could
prevent OPSCC (11). Accordingly,
detection of HPV is the first step for evaluation of the
pathological role of HPV infection in OPSCC. In situ
hybridization (ISH)- and polymerase chain reaction (PCR)-based
assays have been used to detect HPV in tumor cells in most previous
studies (12–14). However, the results of ISH-based
tests are not always consistent with those of PCR-based tests
(1). Thus, determining the
concordance between these methods will be important for identifying
the molecular role of HPV infection in tumor cells.
Cancer is a disease of uncontrolled cell division
(15–17). Its development and progression are
usually linked to a series of changes in the activities of cell
cycle regulators, which can be classified as positive or negative
regulators (15–17). Common positive regulators of the
cell cycle include cyclin A and cyclin D1, which promote the
progression of the cell cycle to the next phase; in contrast, Rb,
p16, p53, p21, and p27 are negative regulators that inhibit the
progression of the cell cycle to the S phase (16–18).
In addition to these proteins, SKP2 has also been identified as a
novel factor controlling the cell cycle (18,19).
Abnormal expression of these cell cycle-related proteins is
frequently found in various cancers, including not only OPSCC but
also gastric, breast, lung, and colorectal cancers (20–24).
However, the results of immunostaining for each of these cell
cycle-related proteins vary within tumor tissues (20–24).
For example, although positive regulators (e.g., cyclin A, cyclin
D1, and SKP2) are usually overexpressed in cancer cells, negative
regulators can exhibit either low expression (e.g., p21 and p27) or
overexpression [e.g., phospho (p)-Rb and p53] (20–24).
Ki-67 and p-c-myc proteins have been used as traditional markers of
proliferative activity in tumor cells (25–27).
Previous studies have shown that rates of Ki-67 and p-c-myc
expression in tumor cells are correlated with the S-phase fraction,
indicating that the expression levels of Ki-67 and p-c-myc are
closely associated with the expression of cell cycle-related
proteins (25–27). However, the roles of these cell
cycle proteins have not been determined in HPV-positive or
HPV-negative OPSCC.
The oncogenic nature of HPV can be explained by the
transforming properties of HPV oncoproteins E6 and E7, which target
the p53 and p-Rb tumor-suppressor pathways, respectively (1,4). This
finding suggests that E6 and E7 may render infected cells
susceptible to mutations and cancer development. In addition, the
oncogenic nature of HPV may be associated with abnormalities in
cell cycle-related proteins. Our aim is to determine the possible
association of OPSCC with HPV and to better characterize the
immunoprofile, with special emphasis on the expression of cell
cycle-related proteins.
Materials and methods
Patients
We included specimens of OPSCC (n=98) collected at
Iwate Medical University Hospital and Miyagi Prefectural Miyagi
Cancer Center from 1996 to 2016. The tumor specimens were
retrospectively obtained from pathology laboratories, and
histological diagnosis was performed based on the General Rules for
Clinical Studies on Head and Neck Cancer (28). Histological classification was not
examined according to HPV status. Clinicopathological data are
shown in Table I. Our study was
approved by the ethics committees of Iwate Medical University
(approval no. H27-126) and Miyagi Cancer Center (approval no.
27-85). All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
institutional and national research committee and with the 1964
Declaration of Helsinki and its later amendments or comparable
ethical standards. Informed consent was obtained from all
individual participants included in the study.
 | Table I.Clinicopathological findings of
oropharyngeal carcinoma examined. |
Table I.
Clinicopathological findings of
oropharyngeal carcinoma examined.
|
Characteristics | Number (%) |
|---|
| Total | 98 (100) |
| Sex |
|
|
Male | 86 (87.8) |
|
Female | 12 (12.2) |
| Age (years) |
|
| Median
(range) | 65 (33–83) |
| Tumor subsites |
|
| LW | 63 (64.3) |
| AW | 19 (19.4) |
| SW | 11 (11.2) |
| PW | 5 (5.1) |
|
Differentiation |
|
|
Well | 26 (26.5) |
|
Moderate | 47 (48.0) |
|
Poor | 25 (25.5) |
| Nodal status |
|
| N0 | 45 (45.9) |
|
N1-3 | 53 (54.1) |
| Tumor stage |
|
| I | 16 (16.3) |
| II | 22 (22.5) |
|
III | 12 (12.2) |
| IV | 48 (49.0) |
Sample preparation of pathological
tissue
Serial sections were cut to a thickness of 3 µm and
mounted on positively charged glass slides for ISH. Paraffin
sections were cut from each block (3 sections, 10 µm) for DNA
extraction. The extra sections cut before and after each tissue
section were stained with hematoxylin and eosin (H&E) and used
to determine specimen quality for testing. To avoid
cross-contamination, the microtome was cleaned and a new blade was
used for each sample.
Preparation of digital slides for
OPSCC specimens
Digital images of the OPSCC specimens were acquired
(Aperio ScanScope; Leica Biosystem Imaging, Wetzlar, Germany).
Detection of HPV was carried out using digital image slides.
Assessments of ISH for HPV and immunohistochemical examination for
cell cycle-related proteins were performed using digital images.
The positive rates (PRs) in tumor tissues were measured within the
hot spot area on the slide. At the hot spot area, the PR was
calculated as the percentage using automated measuring software
(Aperio image scope 12.0.1).
ISH for HPV
Probe sets able to detect 12 types of oncogenic HPV
(16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66) were obtained
from Roche Medical Systems (INFORM®, 12 High-risk HPV
Genotype kit). ISH was performed according to the manufacturer's
guidelines using a Roche automated slide staining system (Roche
Medical System) (12). HPV control
slides consisted of formalin-fixed, paraffin-embedded sections
containing three separate collections of cells on a single slide
(Roche Medical Systems). These cells consisted of the CaSki
cervical cancer cell line (containing 200–400 copies of HPV16 per
cell); the HeLa cervical cancer cell line (containing 10–50 copies
of HPV18 per cell); and the C-33A cell line, which served as a
negative control. The negative control was set using negative
control probes provided by Ventana Medical Systems.
Nuclear staining was considered a
positive result for HPV DNA
Two nuclear staining patterns, diffuse staining and
dot-like staining, were primarily observed in the tumor tissues.
All staining patterns were defined as positive signals. HPV
positivity was determined using a 10% cutoff (29).
Two pathologists independently
reviewed the INFORM slides
In some cases in which the evaluation provided
different results, a consensus interpretation was reached after
re-examination.
DNA extraction from paraffin-embedded
sections
DNA from normal and tumor tissue was extracted by
standard SDS proteinase K treatment. DNA extracted from the samples
was resuspended in TE buffer [10 mM Tris-HCl, 1 mM EDTA (pH
8.0)].
Detection of HPV by Cobas 4800
assays
Cobas tests were carried out according to the
manufacturer's protocol (31). DNA
extraction was accomplished using a fully automated Cobas × 480
instrument. Briefly, specimens were digested under denaturing
conditions at elevated temperatures and then lysed in the presence
of chaotropic reagent. Released HPV nucleic acids, along with
β-globin DNA as a process control, were purified through adsorption
to magnetic glass particles, washed, separated from the particles,
and subjected to PCR amplification and detection.
The amplification plates were then manually
transferred to the Cobas z 480 analyzer for real-time PCR
amplification of high-risk HPV and β-globin DNA. The Cobas HPV test
uses primers that define a sequence of approximately 200
nucleotides within the polymorphic L1 region of the HPV genome, as
previously described. A pool of HPV primers present in the master
mix was designed to amplify HPV DNA from 14 high-risk types (16,
18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68).
Fluorescent oligonucleotide probes were used to bind to the
polymorphic regions within the sequence defined by the primers. An
additional primer pair and probe targeting the human β-globin gene
(330-bp amplicon) was included as an internal control to provide a
measure of specimen adequacy and to monitor the quality of the
extraction and amplification process. Interpretation of
amplification results was performed using proprietary software
provided with the Cobas z 480 analyzer. The cycle threshold cutoffs
were set at 40.5 for HPV16 and 40 for HPV18 and for the remaining
12 high-risk HPV genotypes. Positive and negative controls were
included in each run.
Assessment of HPV positivity in tumor
cells
HPV was defined as positive if either the ISH-based
test or the PCR-based test was positive or if both tests were
positive within the given section. HPV was defined as negative if
both tests were negative.
Immunohistochemistry of tumor tissue
sections
Tissues for analysis were fixed for several hours in
10% neutral buffered formalin and then embedded in paraffin.
Paraffin sections (3 µm) were routinely dewaxed and rehydrated,
then subjected to heat-induced epitope retrieval in either High pH
Target Retrieval solution (Dako, Carpinteria, CA, USA) or Reveal
Decloaking solution (BioCare Medical, Concord, MA, USA) for 8 min,
and then incubated with primary antibody for 90 min (21). The following antibodies were used in
this analysis: anti-p16 (clone E6H4, ready-to-use; Roche
Diagnostics, Basel, Switzerland), anti-p53 (clone DO7, 1:100
dilution; Neomarkers Inc., Fremont, CA, USA), anti-p-Rb
(polyclonal, 1:300 dilution; Cell Signaling Technology, Beverly,
MA, USA), anti-p21 (clone SX118, 1:50 dilution; Dako), anti-p27
(clone SX53G8, 1:50 dilution; Dako), anti-SKP2 (polyclonal, 1:50
dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA),
anti-cyclin D1 (clone SP4, 1:100; Nichirei, Tokyo, Japan),
anti-cyclin A (clone, 6E8, 1:100; Novocastra, Newcastle, UK),
anti-p-c-myc (clone, 33A12E10, 1:50; Abcam, Cambridge, UK), and
anti-Ki-67 (clone MIB1, ready-to-use; Dako). After primary antibody
treatment, sections were analyzed using the EnVision Plus Mouse HRP
detection system (Dako), and antigen binding was detected using
DAB+ liquid chromogen (Dako). Sections were then counterstained
with hematoxylin before mounting. Assessment of the expression of
each marker was carried out within the hot spot of each marker.
Statistical analysis
Clinicopathological findings were analyzed using
Chi-square tests (Stat Mate for Windows version 3.07; Atom, Tokyo,
Japan). Differences in the frequencies of immunohistochemical
expression of cell cycle-related proteins between HPV-positive and
HPV-negative OPSCC were analyzed using Mann-Whitney U tests
(GraphPad Prism 6; MDF, Tokyo, Japan). Differences with P-values of
<0.05 were considered to indicate a statistically significant
difference.
Results
Staining patterns of HPV and INFORM
for detection of HPV
Positive staining for HPV varied within the same
tumor (i.e., exhibiting a heterogeneous staining pattern). Although
diffuse HPV staining was observed in 12 of 30 tumors (40%),
heterogeneity of staining was found in 18 of 30 tumors (60%).
Overall, HPV was positive in 30 of 98 tumors (30.6%) when using ISH
as the detection method.
Detection of HPV using a PCR-based
assay
Although the most frequent HPV subtype detected by
PCR-based assay was HPV subtype 16 (41 of 46 HPV-positive tumors,
89.1%), HPV subtype 18 was rarely detected in OPSCCs (1 of 46
HPV-positive tumors, 2.2%). The frequency of HPV for subtypes other
than 16 and 18 was 8.7% (4 of 46 HPV-positive tumors).
Comparison of ISH- and PCR-based
assays
The concordance between ISH- (INFORM) and PCR-based
assays in the detection of HPV DNA is shown in Table II. The two tests were concordant in
81.6% (80 of 98) of cases (29 positive cases and 51 negative
cases). The INFORM and PCR-based assays had good agreement (Kappa
coefficient of 0.62) in detecting HPV DNA in OPSCC. HPV positivity
in OPSCC, as determined by combined ISH- and PCR-based assays, was
48.0% (47/98; positive in either ISH- or PCR-based assays). This
positivity was regarded as overall HPV positivity in this study.
These results are summarized in Table
II.
 | Table II.Concordance of ISH- and PCR-based
assays in OPSCC. |
Table II.
Concordance of ISH- and PCR-based
assays in OPSCC.
|
| PCR |
|
|
|---|
|
|
|
|
|
|---|
|
| Total | Positive | Negative | P-value | κ |
|---|
| HPV-ISH |
|
Total | 98 | 46 | 52 |
|
|
|
Positive | 30 | 29 | 1 |
|
|
|
Negative | 68 | 17 | 51 | <0.001 | 0.62 |
The OPSCC specimens examined in this study were
classified as HPV positive (+) or HPV negative (−). There were
significant differences in median age (P<0.01), tumor location
(P<0.01), tumor differentiation (P<0.05), and lymph node
metastasis (P<0.05) between HPV (+) and HPV (−) samples. No
differences in histological features were observed in the OPSCC
specimens examined. Clinicopathological findings for these two
groups are listed in Table
III.
 | Table III.Clinicopathological findings of
oropharyngeal carcinoma according to the HPV status. |
Table III.
Clinicopathological findings of
oropharyngeal carcinoma according to the HPV status.
|
|
| HPV-status |
|
|---|
|
|
|
|
|
|---|
| Factors | Total (%) | Positive (%) | Negative (%) | P-value |
|---|
| No. of cases | 98 (100) | 47 (100) | 51 (100) |
|
| Sex |
|
|
| NS |
|
Male | 86 (87.8) | 40 (85.1) | 46 (90.2) |
|
|
Female | 12 (12.2) | 7
(14.9) | 5 (9.8) |
|
| Age (years) |
| Median
(range) | 65
(33–83) | 62
(36–83) | 67
(33–83) | <0.01 |
| Tumor subsites |
| LW | 63 (64.3) | 39 (83.0) | 24 (47.1) | <0.001 |
| AW | 19 (19.4) | 3 (6.4) | 16 (31.4) | <0.01 |
| SW | 11 (11.2) | 4 (8.5) | 7
(13.7) | NS |
| PW | 5 (5.1) | 1 (2.1) | 4 (7.8) | NS |
|
Differentiation |
|
Well | 26 (26.5) | 8
(17.0) | 18 (35.3) | <0.05 |
|
Moderate | 47 (48.0) | 25 (53.2) | 22 (43.1) | NS |
|
Poor | 25 (25.5) | 14 (29.8) | 11 (21.6) | NS |
| Nodal status |
|
|
| <0.05 |
| N0 | 45 (45.9) | 16 (34.0) | 29 (56.9) |
|
|
N1-3 | 53 (54.1) | 31 (66.0) | 22 (43.1) |
|
| Tumor stage |
|
|
| NS |
| I | 16 (16.3) | 3 (6.4) | 10 (19.6) |
|
| II | 22 (22.5) | 11 (23.4) | 13 (25.5) |
|
|
III | 12 (12.2) | 6
(12.8) | 6
(11.7) |
|
| IV | 48 (49.0) | 27 (57.4) | 21 (41.2) |
|
Measurement of viral load (HPV)
between PCR+/ISH− and
PCR+/ISH+
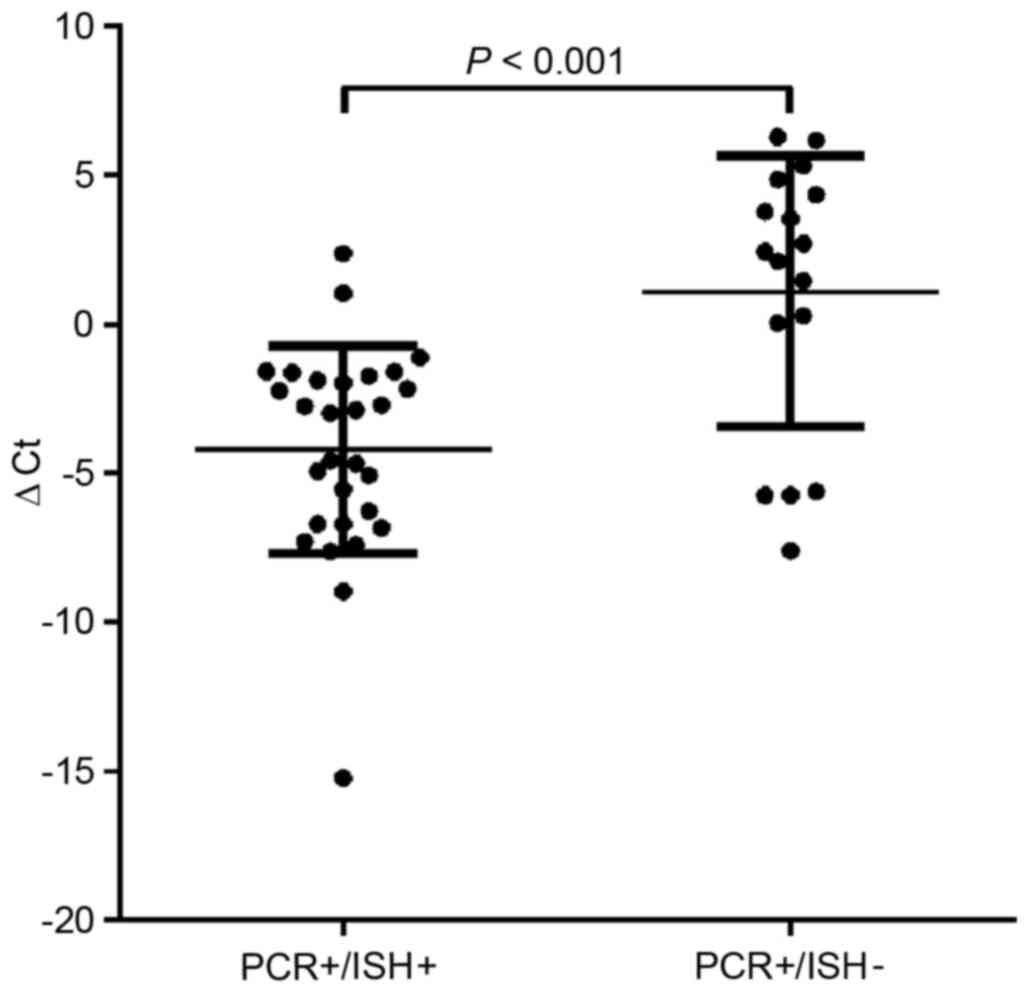
We next examined the viral load (HPV) using
real-time PCR in PCR+/ISH− and
PCR+/ISH+ tumor cells using the ∆Ct. The
results showed that the viral loads in tumor cells with
PCR+/ISH+ increased compared with those with
PCR+/ISH− (p<0.001; Fig. 1). In addition, we examined the
clinicopathological differences between
PCR+/ISH− and PCR+/ISH+
cases. In this study, oropharyngeal squamous cell carcinoma with
PCR+/ISH+ was preferentially located at the
lateral wall of the oropharyngeal region compared with that of
PCR+/ISH−. Finally, the frequency of lymph
node metastasis was significantly higher in
PCR+/ISH+ tumors than in
PCR+/ISH− tumors. These findings are
summarized in Table IV.
 | Table IV.Clinicopathological differences
between PCR+/ISH− and
PCR+/ISH+ cases. |
Table IV.
Clinicopathological differences
between PCR+/ISH− and
PCR+/ISH+ cases.
|
| HPV type16 +
OPSCC |
|
|---|
|
|
|
|
|---|
| Factors |
PCR+/ISH+ |
PCR+/ISH− | P-value |
|---|
| Cases | 27 | 14 |
|
| Sex |
|
| NS |
|
Male | 24 | 12 |
|
|
Female | 3 | 2 |
|
| Age |
|
| NS |
|
Median | 60 (36–83) | 63.5 (43–76) |
|
| Tumorsubsites |
| LW | 26 | 8 | <0.01 |
| AW | 1 | 1 |
|
| SW | 0 | 4 | <0.05 |
| PW | 0 | 1 |
|
|
Differentiation |
|
| NS |
|
Well | 2 | 5 |
|
|
Moderate | 15 | 8 |
|
|
Poor | 10 | 1 |
|
| Nodal status |
|
| <0.01 |
| N0 | 5 | 10 |
|
|
N1-3 | 22 | 4 |
|
| Stage |
|
| NS |
| I | 0 | 3 |
|
| II | 4 | 5 |
|
|
III | 4 | 1 |
|
| IV | 19 | 5 |
|
Association of p16 expression with HPV
status
Most tumors exhibited intense, diffuse p16
expression in both the nucleus and cytoplasm. The positive cutoff
value for p16 expression was defined as >30% of tumor cells. p16
expression and HPV status were both positive in 37 of 98 tumors
(37.8%) by PCR-based assays, whereas that by ISH-based assays was
27 of 98 (27.6%), as shown in Table V-A
and V-B. The overall concordance of p16 expression and HPV
status (as determined by both ISH- and PCR-based tests) was 38 of
98 (38.8%), with 38 HPV (+) cases and 41 HPV (−) cases, as shown in
Table V-C. The concordance of p16
expression and HPV positivity by PCR-based and ISH assays was
moderate (κ values of 0.51 and 0.59, respectively), consistent with
the combined HPV DNA positivity observed by INFORM and PCR-based
assays (κ value, 0.61). The HPV-positive area, as determined using
ISH, was observed within the p16 expression area. Therefore, the
area of p16 staining was larger than that of HPV positivity.
 | Table V.Concordance between HPV positivity
and p16 expression in OPSCC. |
Table V.
Concordance between HPV positivity
and p16 expression in OPSCC.
|
| (A) HPV-ISH |
|
|
|---|
|
|
|
|
|
|---|
|
| Total | Positive | Negative | P-value | κ |
|---|
| p16 |
|
Total | 98 | 30 | 68 |
|
|
|
Positive | 48 | 27 | 21 |
|
|
|
Negative | 50 | 3 | 47 | <0.001 | 0.51 |
|
|
| (B) PCR |
|
|
|
|
|
|
|
|
| Total | Positive | Negative | P-value | κ |
|
| p16 |
|
Total | 98 | 46 | 52 |
|
|
|
Positive | 48 | 37 | 11 |
|
|
|
Negative | 50 | 9 | 41 | <0.001 | 0.59 |
|
|
| (C) HPV |
|
|
|
|
|
|
|
|
| Total | Positive | Negative | P-value | κ |
|
| p16 |
|
Total | 98 | 47 | 51 |
|
|
|
Positive | 48 | 38 | 10 |
|
|
|
Negative | 50 | 9 | 41 | <0.001 | 0.61 |
Assessment of Ki-67, p-c-myc, SKP2,
and cell cycle-related proteins (p53, p21, p27, p-Rb, cyclin D1,
and cyclin A) in tumor tissue sections based on HPV status
Although p16 expression was observed in both the
nucleus and cytoplasm, as mentioned above, Ki-67, p-c-myc, SKP2,
p53, p21, p27, p-Rb, cyclin A, and cyclin D1 were only expressed in
the nucleus in tumor cells. There were no significant differences
in the PRs of Ki-67 between HPV (+) and HPV (−) cases of OPSCC.
Although the PR of p27 expression was significantly higher in HPV
(+) samples than in HPV (−) samples, the PRs of p53 overexpression
and p-Rb, p-c-myc, and cyclin D1 levels were significantly higher
in HPV (−) samples than in HPV (+) samples. In addition, the PR of
SKP2 was significantly higher in HPV (−) OPSCC than in HPV (+)
OPSCC. In contrast, the PRs of p21 and cyclin A expression did not
differ significantly between HPV (+) and HPV (−) samples. These
results are shown in Fig. 2.
Representative immunohistochemical features based on HPV status are
depicted in Figs. 3 and 4.
Discussion
The majority of studies have utilized HPV DNA ISH-
or PCR-based methods for detection of HPV infection (12,30,31).
Although ISH- and PCR-based assays are generally concordant for HPV
infection, it is important to assess commonly used methods for HPV
detection. In this study, we observed that ISH assays using the
INFORM probe were comparable to PCR-based assays with regard to
detection of HPV DNA in formalin-fixed, paraffin-embedded tissues
from patients with OPSCC. However, PCR-based assays could not be
used to determine whether the HPV originated from the tumor cells
or from adjacent non-tumor cells, leading to the concern that
PCR-based assays may yield false-positive results (32). Therefore, the use of ISH-based
assays could overcome this limitation of PCR-based assays and
provide more accurate results. Moreover, an important advantage of
ISH is the ability to determine the cellular localization of HPV
(32). However, both HPV DNA PCR-
and ISH-based methods only evaluate the presence of HPV DNA in the
sample and do not indicate whether the virus is transcriptionally
active (32). Based on our
analysis, we suggest that a combination of PCR- and ISH-based
assays is required to accurately detect HPV infection in tumor
cells, although active HPV may not be detected. Although
false-positive results for HPV status between PCR-based and ISH
assays may exist, we think that our definition of HPV positivity as
positive in either ISH- or PCR-based assays is an appropriate
criterion for determination of HPV status. This finding is
supported by a previous study in which the results of PCR-based
assays were found to be correlated with those of ISH to measure the
HPV viral load (33).
Heterogeneity within the same tumor is an important
issue for tumor development. Therefore, intratumoral differences in
HPV status within the same tumor are of interest. One explanation
is that clonal differences occur within the tumor due to
differences in time when viral infection occurs within the tumor.
An alternative explanation is that there may be differences in
biological sensitivity (e.g., immunosensitivity) between HPV (+)
and HPV (−) cases, even within the same tumor. However, the reason
for the heterogeneity of HPV status within OPSCC remains
unknown.
Cell cycle progression can be accurately controlled
to regulate cell proliferation (15,16).
Cellular proliferation is defined by regulation of cell
cycle-related proteins, including the negative regulators p53, p21,
p27, p16, and p-Rb and the positive regulators cyclin A and cyclin
D1 (15,16,34).
Thus, proliferative activity can be measured by determining the
expression levels of Ki-67 and p-c-myc (25–27).
In human cancer cells, expression of p53, p-Rb, cyclin D1, and
cyclin A is thought to indicate a high rate of proliferation,
whereas expression of p21, p27, and p16 is thought to indicate a
low rate of proliferation (20).
The present results suggested that low expression of p53, p-Rb,
p-c-myc, and cyclin D1, combined with high expression of p27 and
p16, characterized the HPV-positive status and that opposite
results characterized the HPV-negative status. Additionally, the
expression of SKP2, which causes multi-ubiquitylation of p27, was
high in HPV-negative tumors. However, the PR of Ki-67 in
HPV-positive tumors was not different from that in HPV-negative
tumors (35). In addition, in the
present study, the PRs of cyclin A and p21 were not different
between HPV-positive and HPV-negative tumors. The present results
supported the notion that patients with OPSCC having HPV-positive
tumors had better prognoses than patients with OPSCC having
HPV-negative tumors. However, to the best of our knowledge, this is
the first study reporting an extensive analysis of the association
between cell cycle-related proteins and HPV status in patients with
OPSCC.
Despite its function as a tumor suppressor, p16
expression is increased in HPV-positive OPSCC specimens, and there
are strong correlations between HPV positivity and p16 expression
in OPSCC specimens (36,37). In the present study, the frequency
of HPV positivity was concordant with p16 expression in OPSCC
specimens. This result suggested that immunohistochemical
expression of p16 could be a surrogate biomarker for prediction of
HPV infection. p16 is thought to reflect the activity of the E7
oncoprotein of HPV, which disrupts the Rb pathway. HPV E7
oncoprotein binds to p-Rb, releasing E2F transcription factor,
which activates DNA synthesis by promoting the transcription of
genes involved in this process (38). As a consequence, p16 is strongly
activated by these events, and immunoexpression of p16 has been
used in the clinical setting as a surrogate for transcriptionally
active HPV infection because the tumor-suppressive p16 protein is
usually inactivated in HPV-negative OPSCC. Recent studies have
shown that HPV infection is associated with survival in patients
with OPSCC and that this factor can predict patient prognosis in
OPSCC (36,37). One possible explanation for the
improved prognoses in patients with HPV-positive OPSCC is the high
expression of p16 in these tumors (36,37).
Thus, we suggest that combined detection of p16 protein expression
and the presence of HPV DNA is necessary for treating OPSCC.
A previous study showed that p16 expression is a
powerful prognostic factor in patients with OPSCC. p16 positivity
is associated with a survival benefit in these patients,
independent of clinicopathological parameters such as TNM
classification (39). HPV-induced
p16-positive OPSCC is a distinct type of HNSCC with a generally
favorable outcome compared with p16-negative OPSCC, which may be
independent of the treatment modality chosen. In the present study,
we did not examine the association of p16-positive OPSCC with
survival. Further studies are needed to assess these
associations.
In previous studies, p53 overexpression, which
reflects mutations in p53, has been shown to be associated with
aggressive clinical course in several types of cancer, including
OPSCC, gastric cancer, and colorectal cancer (20,40,41).
In the present study, p53 was frequently found to have a high PR
(almost equivalent to overexpression) in HPV-negative OPSCC
compared with that in HPV-positive OPSCC. A previous study showed
that p53 overexpression was a predictive and prognostic factor in
locally advanced pharyngeal cancer treated with induction
chemotherapy (42). Therefore, the
present results suggested that patients with HPV-positive OPSCC had
better prognoses than patients with HPV-negative OPSCC. In
addition, Shinohara et al indicated that a group of patients
with p16-positive/p53-negative tumors had better overall survival
and disease-specific survival (43). This finding was supported by our
results showing that HPV-negative OPSCC showed higher expression of
p53 and lower expression of p16 than HPV-positive OPSCC, supporting
the poor prognosis.
The cyclin kinase inhibitor p27 is a central
regulator of the cell cycle (15,16).
Overexpression of p27 arrests cells in the G1 phase, whereas loss
of p27 leads to an increase in cell proliferation (15,16).
The degradation of p27 at the G1-S transition is mediated by SKP2,
which shows an inverse expression pattern relative to that of p27
in cells (18). Previous studies
have shown that low expression of p27 is correlated with poor
prognosis in patients with gastric, breast, and prostate cancers
(44–46). Thus, this finding suggested that
high expression of p27 predicted a favorable prognosis in
HPV-positive OPSCC. Additionally, we examined the relationship
between p21 expression and HPV-positive OPSCC. Low expression of
p21 is found in various cancers, including gastric and colorectal
cancers (20,47). In colorectal cancer, low expression
of p21 is closely associated with tumor progression. However, the
present results showed that there were no differences in p21
expression levels between HPV-positive and -negative OPSCC,
suggesting that low p21 expression may be a common tumorigenic
mechanism in both HPV-positive and -negative OPSCC.
Previous studies have shown that overexpression of
cyclin D1 is a potential prognostic marker in OPSCC (34). Indeed, cyclin D1 is commonly
upregulated in various cancers, including esophageal and colorectal
cancers (48,49). In the present study, our findings
suggested that cyclin D1 expression was downregulated in
HPV-positive OPSCC, possibly as a result of p-Rb suppression
(34). In addition, a previous
study showed that cyclin D1 negativity was associated with p16
overexpression in OPSCC. This inverse relationship is consistent
with the hypothesis that HPV positivity and subsequent p16
upregulation leads to suppression of cyclin D1 expression (34). In contrast, the expression of cyclin
A, which promotes tumor proliferation, did not differ between
HPV-positive and HPV-negative OPSCC. This finding suggested that
overexpression of cyclin A may play a common role in the
development of HPV-positive and HPV-negative OPSCC.
In conclusion, HPV detection was performed in 98
OPSCC samples using ISH- and PCR-based assays. Although there are
some concerns that must be considered when interpreting HPV
detection data, we did not find evidence of differences in HPV
detection using ISH- and PCR-based assays in the present study.
Moreover, while overexpression of p16 may be a useful marker for
detection HPV infection in tumor cells, we suggest that the
combination of ISH- and PCR-based assays may be most effective for
accurate detection of HPV positivity. Overexpression of p16 is an
important factor that may provide an explanation for the improved
prognosis in patients with HPV-positive OPSCC. Furthermore, cell
cycle-related proteins may also be effective for identification of
the molecular mechanisms of OPSCC pathogenesis according to HPV
status.
Acknowledgements
We gratefully acknowledge the technical assistance
of members of the Department of Molecular Diagnostic Pathology,
Iwate Medical University. We also thank members of the Department
of Pathology, Miyagi Cancer Center, the Department of Head and Neck
Surgery, Iwate Medical University and Miyagi Cancer Center for
their support.
References
|
1
|
Elrefaey S, Massaro MA, Chiocca S, Chiesa
F and Ansarin M: HPV in oropharyngeal cancer: The basics to know in
clinical practice. Acta Otorhinolaryngol Ital. 34:299–309.
2014.PubMed/NCBI
|
|
2
|
Ausweger C, Burgschwaiger E, Kugler A,
Schmidbauer R, Steinek I, Todorov Y, Thurnher D and Krapfenbauer K:
Economic concerns about global healthcare in lung, head and neck
cancer: Meeting the economic challenge of predictive, preventive
and personalized medicine. EPMA J. 1:627–631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gale N, Michaels L, Luzar B, Poljak M,
Zidar N, Fischinger J and Cardesa A: Current review on squamous
intraepithelial lesions of the larynx. Histopathology. 54:639–656.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kreimer AR, Clifford GM, Boyle P and
Franceschi S: Human papillomavirus types in head and neck squamous
cell carcinomas worldwide: A systematic review. Cancer Epidemiol
Biomarkers Prev. 14:467–475. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Monsjou HS, Balm AJ, van den Brekel MM
and Wreesmann VB: Oropharyngeal squamous cell carcinoma: A unique
disease on the rise? Oral Oncol. 46:780–785. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mignogna MD, Fedele S and Russo Lo L: The
World Cancer Report and the burden of oral cancer. Eur J Cancer
Prev. 13:139–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sedrak M and Rizzolo D: Understanding the
link between HPV and oropharyngeal cancers. JAAPA. 22:42–46.
2009.PubMed/NCBI
|
|
8
|
Gillison ML: Human
papillomavirus-associated head and neck cancer is a distinct
epidemiologic, clinical, and molecular entity. Semin Oncol.
31:744–754. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Adelstein DJ and Rodriguez CP: Human
papillomavirus: Changing paradigms in oropharyngeal cancer. Curr
Oncol Rep. 12:115–120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stransky N, Egloff AM, Tward AD, Kostic
AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C,
McKenna A, et al: The mutational landscape of head and neck
squamous cell carcinoma. Science. 333:1157–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ang KK, Harris J, Wheeler R, Weber R,
Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C,
et al: Human papillomavirus and survival of patients with
oropharyngeal cancer. N Engl J Med. 363:24–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo M, Gong Y, Deavers M, Silva EG, Jan
YJ, Cogdell DE, Luthra R, Lin E, Lai HC, Zhang W, et al: Evaluation
of a commercialized in situ hybridization assay for detecting human
papillomavirus DNA in tissue specimens from patients with cervical
intraepithelial neoplasia and cervical carcinoma. J Clin Microbiol.
46:274–280. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pannone G, Rodolico V, Santoro A, Muzio Lo
L, Franco R, Botti G, Aquino G, Pedicillo MC, Cagiano S, Campisi G,
et al: Evaluation of a combined triple method to detect causative
HPV in oral and oropharyngeal squamous cell carcinomas: p16
immunohistochemistry, consensus PCR HPV-DNA, and in situ
hybridization. Infect Agent Cancer. 7:42012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stein AP, Saha S, Kraninger JL, Swick AD,
Yu M, Lambert PF and Kimple RJ: Prevalence of human papillomavirus
in oropharyngeal cancer: A systematic review. Cancer J. 21:138–146.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Williams GH and Stoeber K: The cell cycle
and cancer. J Pathol. 226:352–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Williams GH and Stoeber K: Cell cycle
markers in clinical oncology. Curr Opin Cell Biol. 19:672–679.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee Y and Lim HS: Skp2 inhibitors: Novel
anticancer strategies. Curr Med Chem. 23:2363–2379. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiu L, Lv J, Chen Y, Wang J and Wu R:
Expression of Skp2 and p27kip1 proteins in
hypopharyngeal squamous cell carcinoma and its clinical
significance. Oncol Lett. 10:3756–3760. 2015.PubMed/NCBI
|
|
20
|
Sugai T, Tsukahara M, Endoh M, Shioi Y,
Takebe N, Mue Y, Matsushita H, Toyota M and Suzuki K: Analysis of
cell cycle-related proteins in gastric intramucosal
differentiated-type cancers based on mucin phenotypes: A novel
hypothesis of early gastric carcinogenesis based on mucin
phenotype. BMC Gastroenterol. 10:552010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Juríková M, Danihel Ľ, Polák Š and Varga
I: Ki67, PCNA, and MCM proteins: Markers of proliferation in the
diagnosis of breast cancer. Acta Histochem. 118:544–552. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ko E, Kim Y, Park SE, Cho EY, Han J, Shim
YM, Park J and Kim DH: Reduced expression of cyclin D2 is
associated with poor recurrence-free survival independent of cyclin
D1 in stage III non-small cell lung cancer. Lung Cancer.
77:401–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bahnassy AA, Zekri AR, El-Houssini S,
El-Shehaby AM, Mahmoud MR, Abdallah S and El-Serafi M: Cyclin A and
cyclin D1 as significant prognostic markers in colorectal cancer
patients. BMC Gastroenterol. 4:222004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Boscolo-Rizzo P, Pawlita M and Holzinger
D: From HPV-positive towards HPV-driven oropharyngeal squamous cell
carcinomas. Cancer Treat Rev. 42:24–29. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miller DM, Thomas SD, Islam A, Muench D
and Sedoris K: c-Myc and cancer metabolism. Clin Cancer Res.
18:5546–5553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Inwald EC, Klinkhammer-Schalke M,
Hofstädter F, Zeman F, Koller M, Gerstenhauer M and Ortmann O:
Ki-67 is a prognostic parameter in breast cancer patients: Results
of a large population-based cohort of a cancer registry. Breast
Cancer Res Treat. 139:539–552. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li LT, Jiang G, Chen Q and Zheng JN: Ki67
is a promising molecular target in the diagnosis of cancer
(Review). Mol Med Rep. 11:1566–1572. 2015.PubMed/NCBI
|
|
28
|
Kuo KT, Hsiao CH, Lin CH, Kuo LT, Huang SH
and Lin MC: The biomarkers of human papillomavirus infection in
tonsillar squamous cell carcinoma-molecular basis and predicting
favorable outcome. Mod Pathol. 21:376–386. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Izumo T, Kirita T, Ariji E, Ozeki S, Okada
N, Okabe S, Okazaki Y, Omura K, Kusama M, Sato T, et al: Working
Group 1 on the ‘Guidelines for Clinical and Pathological Studies of
Oral Cancer’, Scientific Committee, Japan Society for Oral Tumors:
General rules for clinical and pathological studies on oral cancer:
A synopsis. Jpn J Clin Oncol. 42:1099–1109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cui M, Chan N, Liu M, Thai K, Malaczynska
J, Singh I, Zhang D and Ye F: Clinical performance of Roche Cobas
4800 HPV Test. J Clin Microbiol. 52:2210–2211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gage JC, Sadorra M, Lamere BJ, Kail R,
Aldrich C, Kinney W, Fetterman B, Lorey T, Schiffman M and Castle
PE: PaP Cohort Study Group: Comparison of the cobas Human
Papillomavirus (HPV) test with the hybrid capture 2 and linear
array HPV DNA tests. J Clin Microbiol. 50:61–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stein AP, Saha S, Yu M, Kimple RJ and
Lambert PF: Prevalence of human papillomavirus in oropharyngeal
squamous cell carcinoma in the United States across time. Chem Res
Toxicol. 27:462–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schache AG, Liloglou T, Risk JM, Filia A,
Jones TM, Sheard J, Woolgar JA, Helliwell TR, Triantafyllou A,
Robinson M, et al: Evaluation of human papilloma virus diagnostic
testing in oropharyngeal squamous cell carcinoma: Sensitivity,
specificity, and prognostic discrimination. Clin Cancer Res.
17:6262–6271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin RJ, Lubpairee T, Liu KY, Anderson DW,
Durham S and Poh CF: Cyclin D1 overexpression is associated with
poor prognosis in oropharyngeal cancer. J Otolaryngol Head Neck
Surg. 42:232013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu J, Zhang M, Rose B, Veillard AS, Jones
D, Zhang X, Lee Soon C, Milross C and Hong A: Ki67 expression has
prognostic significance in relation to human papillomavirus status
in oropharyngeal squamous cell carcinoma. Ann Surg Oncol.
22:1893–1900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lai S, Wenaas AE, Sandulache VC, Hartman
C, Chiao E, Kramer J and Zevallos JP: Prognostic significance of
p16 cellular localization in oropharyngeal squamous cell carcinoma.
Ann Clin Lab Sci. 46:132–139. 2016.PubMed/NCBI
|
|
37
|
Saber CN, Larsen Grønhøj C, Dalianis T and
von Buchwald C: Immune cells and prognosis in HPV-associated
oropharyngeal squamous cell carcinomas: Review of the literature.
Oral Oncol. 58:8–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yim EK and Park JS: The role of HPV E6 and
E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer
Res Treat. 37:319–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fischer CA, Zlobec I, Green E, Probst S,
Storck C, Lugli A, Tornillo L, Wolfensberger M and Terracciano LM:
Is the improved prognosis of p16 positive oropharyngeal squamous
cell carcinoma dependent of the treatment modality? Int J Cancer.
126:1256–1262. 2010.PubMed/NCBI
|
|
40
|
Friedman JM, Stavas MJ and Cmelak AJ:
Clinical and scientific impact of human papillomavirus on head and
neck cancer. World J Clin Oncol. 5:781–791. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sugai T, Nakamura SI, Habano W, Uesugi N,
Sato H, Yoshida T and Orii S: Usefulness of proliferative activity,
DNA ploidy pattern and p53 products as diagnostic adjuncts in
colorectal adenomas and intramucosal carcinomas. Pathol Int.
49:617–625. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lassaletta L, Brandáriz JA, Benito A, de
la Cruz J, Gómez C, Ballestín C, Hitt R, Colomer R and
Alvarez-Vicent JJ: p53 expression in locally advanced pharyngeal
squamous cell carcinoma. Arch Otolaryngol Head Neck Surg.
125:1356–1359. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shinohara S, Kikuchi M, Tona R, Kanazawa
Y, Kishimoto I, Harada H, Imai Y and Usami Y: Prognostic impact of
p16 and p53 expression in oropharyngeal squamous cell carcinomas.
Jpn J Clin Oncol. 44:232–240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang RM, Naitoh J, Murphy M, Wang HJ,
Phillipson J, deKernion JB, Loda M and Reiter RE: Low p27
expression predicts poor disease-free survival in patients with
prostate cancer. J Urol. 159:941–945. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hayashi H, Ito T, Yazawa T, Ikeda M,
Inayama Y, Nakatani Y, Kameda Y, Nakamura N and Kitamura H: Reduced
expression of p27/Kip1 is associated with the development of
pulmonary adenocarcinoma. J Pathol. 192:26–31. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yasui W, Kudo Y, Semba S, Yokozaki H and
Tahara E: Reduced expression of cyclin-dependent kinase inhibitor
p27Kip1 is associated with advanced stage and invasiveness of
gastric carcinomas. Jpn J Cancer Res. 88:625–629. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mitomi H, Ohkura Y, Fukui N, Kanazawa H,
Kishimoto I, Nakamura T, Yokoyama K, Sada M, Kobayashi K, Tanabe S,
et al: P21WAF1/CIP1 expression in colorectal carcinomas is related
to Kras mutations and prognosis. Eur J Gastroenterol Hepatol.
19:883–889. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang X-P, Rong T-H, Lin P, Wu Q-L, Yao
G-Y, Hou J-H, Su X-D, Li X-D, Li B-J, Zhang P-Y, et al: Cyclin D1
overexpression in esophageal cancer from southern China and its
clinical significance. Cancer Lett. 231:94–101. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Y, Wei J, Xu C, Zhao Z and You T:
Prognostic significance of cyclin D1 expression in colorectal
cancer: A meta-analysis of observational studies. PLoS One.
9:e945082014. View Article : Google Scholar : PubMed/NCBI
|