Introduction
In adults, glioma is the most common type of
malignant tumor of the brain and central nervous system,
characterized by rapid progression, aggressive behavior, frequent
recurrence and poor prognosis (1,2).
Despite advances in diagnosis and treatment, frequent recurrence
and a low cure rate of glioma remain due to the rapid proliferation
and high invasiveness of this disease, particularly for
glioblastoma multiforme patients (3,4). It is
in essence a genetic disease with overexpression of oncogenes and
deletions or mutations of tumor-suppressor genes, which lead to the
uncontrolled proliferation of cancer cells (5,6).
Therefore, it is essential to investigate the underlying mechanisms
of this disease.
A human cDNA encoding the novel protein chondroitin
polymerizing factor (CHPF) which was initially discovered by
Kitagawa et al in 2003, is essential for chondroitin
polymerizing activity (7). In
addition, knockdown of the expression of CHPF leads to specific
elimination of chondroitin sulfate (CS) and dermatan sulfate. In
recent years, more and more evidence has revealed the potential
roles of CS in tumor biology. Notably, numerous studies have shown
that the CS expression has been observed significantly increased in
many human tumors, such as colon (8) and ovarian epithelial cancer (9). The gene encoding CHPF has been
identified in humans (10) and mice
(11). Thus, CHPF plays an
important role in tumor progression. However, no studies have been
carried out investigating the functional role of CHPF in human
types of cancer, and particularly in glioma.
In the present study, we demonstrated that CHPF was
highly expressed in human glioma tissues and 4 glioma cell lines.
Consequently, we constructed a lentivirus vector mediating RNAi
targeting of CHPF (LV-shRNA-CHPF). We employed the LV-shRNA-CHPF to
determine whether CHPF affects the progression of human glioma
in vitro.
Materials and methods
Cancer cell lines
The human glioma cell lines A172, U373, U251, U87
and the human renal epithelial 293T cells were purchased from the
Shanghai Cell Bank (Shanghai, China). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM) (Gibco, Carlsbad, CA,
USA) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA,
USA) in an incubator at 37°C with 5% CO2.
Patients and tissue samples
Patients with glioma (n=32) who underwent initial
surgery at The Second Affiliated Hospital of Nanchang University
between 2013 and 2015 were retrospectively selected for the present
study. Normal brain tissues (n=10) were obtained from epileptic
resections. Each patient participated after providing informed
consent, and the use of the samples for research was approved by
the Medical Ethics Committee of The Second Affiliated Hospital of
Nanchang University. All specimens were immediately frozen in
liquid nitrogen after resection and stored at −80°C until RNA
extraction. No patients had received therapy before resection.
Quantitative real-time polymerase
chain reaction (qRT-PCR) assay
Total RNA was isolated from the 4 glioma cell lines
(A172, U373, U251 and U87) using TRIzol Total RNA Reagent
(Invitrogen, Shanghai, China). Complementary DNA (cDNA) synthesis
was performed with 2 µg of total RNA using the RevertAid™ H Minus
First Strand cDNA Synthesis kit (Takara, Otsu, Japan). The CHPF
primers were obtained from GeneChem Co. Ltd. (Shanghai, China), and
GAPDH was used as an internal control. The primers used were as
follows: CHPF forward, 5′-GGA ACG CAC GTA CCA GGAG-3′ and reverse,
5′-CAC CCT GTT GCT GTA GCC AAA-3′; and GAPDH forward, 5′-TGA CTT
CAA CAG CGA CAC CCA-3′ and reverse, 5′-CAC CCT GTT GCT GTA GCC
AAA-3′. Quantitative PCR was performed using the SYBR PrimeScript
RT-PCR kit (Takara) on an Applied Biosystems 7300 fluorescence
quantitative PCR system (Applied Biosystems, Foster City, CA, USA).
The reaction mixtures were incubated at 95°C for 30 sec, followed
by 45 amplification cycles at 95°C for 5 sec and 60°C for 30 sec.
The PCR products of CHPF and GAPDH were 117 and 121 bp,
respectively. The relative expression of the CHPF mRNA was
calculated with the 2−ΔΔCt method, using the GAPDH mRNA
expression level for normalization.
Construction of the shRNA lentiviral
vector and cell transfection
The lentiviral expressing the short hairpin RNA
(shRNA) targeting the sequence of the CHPF gene (CTGGCCATGCT
ACTCTTTG) and negative control (NC) (TTCTCCGAACGT GTCACGT) were
purchased from Shanghai GeneChem Co., Ltd. Then, they were cloned
into the pGCSIL-GFP lentiviral vector with AgeI/EcoRI
sites to form the recombinant lentiviral shRNA expression vector.
Lentiviral vectors and packaging vectors were transfected into 293T
cells using Lipofectamine 2000. Lentiviral particles were purified
using ultracentrifugation and the titer of the lentiviruses was
determined by endpoint dilution assay.
The U251 glioblastoma cells were infected with the
shRNA-CHPF-lentivirus or the NC lentivirus. The U251 cells were
seeded in a 6-well plate at 40% confluence with 4.0×105
cells/well. After 72 h of infection, the cells were observed under
a fluorescence microscope (MicroPublisher 3.3RTV; Olympus, Tokyo,
Japan). After 5 days of infection, the ability of the shRNA-CHPF
vectors to knock down CHPF was investigated using quantitative
PCR.
Western blot analysis
The cells were harvested at 48 h after transfection,
and lysed with RIPA buffer for 30 min at 4°C. The protein
concentration was determined using a BCA protein assay kit (Pierce,
Rockford, IL, USA). Equal amounts of total protein of each
treatment were separated using 12.5% SDS-PAGE according to
Laemmli's method (12), and were
then transferred onto polyvinylidene difluoride (PVDF) membranes.
The first antibodies used were polyclonal mouse anti-CHPF (1:2,000
dilutions; Sigma-Aldrich® Co. LLC., St. Louis, MO, USA),
and the anti-GAPDH antibody (1:2,000 dilution; Santa Cruz
Biotechnology, Santa Cruz, CA, USA). The secondary antibody was
anti-goat IgG-conjugated with horseradish peroxidase at a dilution
of 1:2,000. The bound antibodies were detected using the Enhanced
Chemiluminescence Plus Western Blotting Detection system (GE
Healthcare, Bethesda, MD, USA). GAPDH was used as an internal
control to normalize the CHPF expression levels.
Cell growth and MTT cell proliferation
assays
Cell growth was assessed using multiparametric
high-content screening (HCS). U251 cells at the logarithmic phase
after being infected with either the NC or shRNA-CHPF lentiviruses
were seeded at 2×103 cells/well in 96-well tissue
culture plates, and incubated at 37°C for 1–5 days. The cells in
the plates were counted using the Cellomics ArrayScan™ VT1 HCS
automated reader (Cellomics, Inc., Pittsburgh, PA, USA) for daily
analysis. In each well, at least 800 cells were analyzed. All
experiments were performed in triplicate.
In vitro cell proliferation was determined
using an MTT cell proliferation assay kit as described by the
manufacturer [American Type Culture Collection (ATCC) Manassas, VA,
USA]. At the end of the culture period, the cells were washed with
phosphate-buffered saline, the MTT reagents were then added
according to the manufacturer's recommendations, and the absorbance
was assessed at 570 nm using a microplate reader. Each experiment
was repeated 3 times in duplicate.
Apoptosis and cell cycle analysis
using flow cytometry
Annexin V-FITC/propidium iodide (PI) double labeling
was used to determine the cell cycle distribution or detect
apoptosis. U251 cells were infected with the shRNA-CHPF or NC
plasmids and incubated at 37°C. U251 cells were harvested by
trypsinization and incubated with FITC-conjugated Annexin V and PI
according to the manufacturer's instructions (KeyGen Biotech,
Nanjing, China). FACSCalibur II sorter and Cell Quest Research
Software (BD Biosciences, San Diego, CA, USA) were used for
analysis. To further confirm the changes of DNA content
distribution in the shRNA-CHPF-treated cells, we performed PI
staining. The cells of different groups were collected and fixed in
70% ethanol at 4°C overnight. After incubation with 50 µg/ml of PI
and RNase A (Sigma-Aldrich) for 30 min at 37°C in the dark, the
cells were subjected to FACS analysis as described above. All
experiments were performed in triplicate.
Statistical analysis
All statistical analyses were implemented in the
SPSS 20.0 statistical software package. All data are presented as
the means ± standard deviations (SD) and the experiments were
repeated 3 times. The Student's t-test was used for raw data
analysis. A P-value of <0.05 was considered to indicate a
statistically significant result.
Results
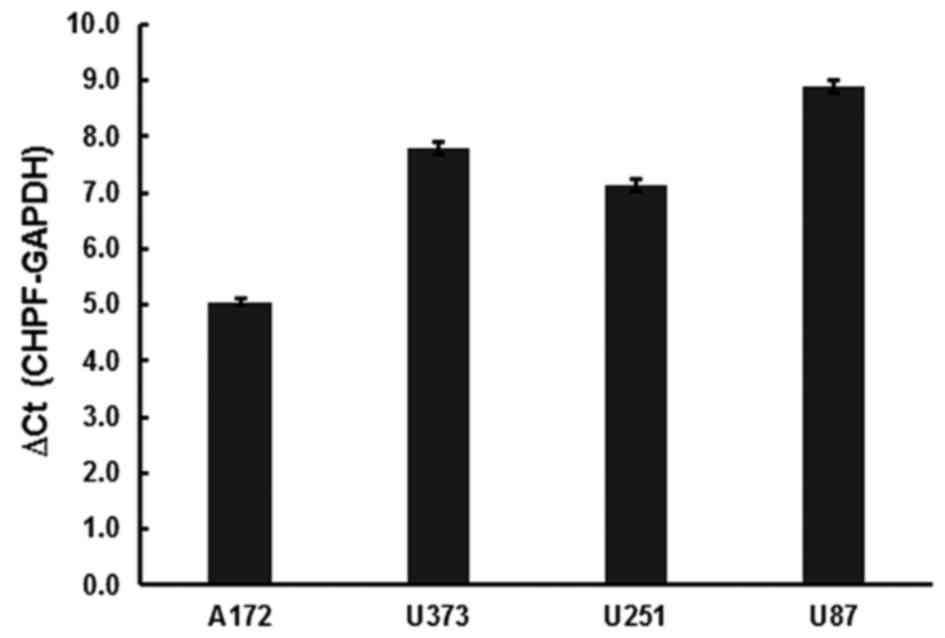
CHPF mRNA detection in 4 human glioma
cell lines
The expression levels of CHPF mRNA were examined in
glioma cell lines A172, U373, U251 and U87 by quantitative RT-PCR.
The results revealed that CHPF mRNA was expressed in all 4 cell
lines (Fig. 1).
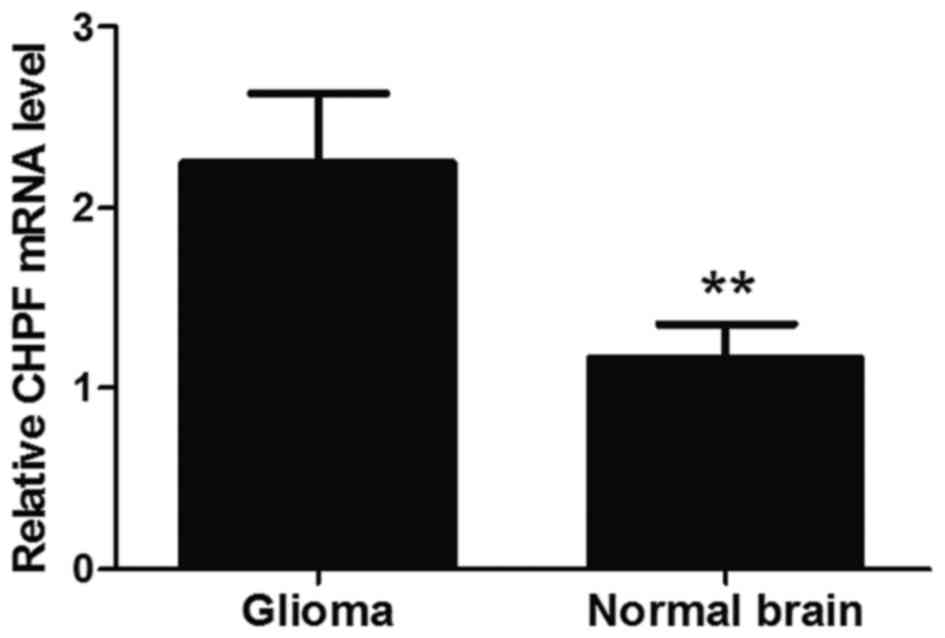
CHPF is overexpressed in human glioma
tissues
In order to determine whether CHPF was involved in
the regulation of tumorigenesis of human glioma we assessed the
CHPF expression level in glioma tissue specimens and normal brain
tissues. Quantitative RT-PCR assays revealed that the mRNA levels
of CHPF in gliomas tissues were significantly higher than those in
normal brain tissues. The average expression level of CHPF in
gliomas tissues was 2.484 and in normal brain tissues it was 1.231
(P<0.01; Fig. 2). This result
demonstrated that CHPF was upregulated in glioma tissues compared
with normal brain tissues.
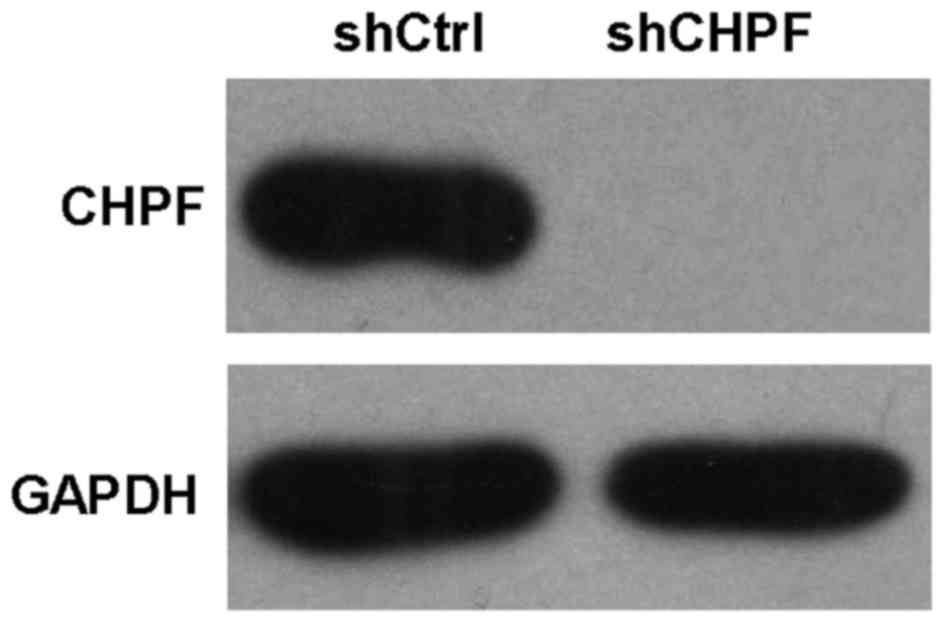
Knockdown efficiency determined by
western blot analysis
Human renal epithelial 293T cells were infected with
shRNA-CHPF or NC lentiviruses. The protein levels of CHPF were
assessed using western blotting in 293T cells. shRNA-CHPF-infected
cultures had a significantly lower level of CHPF compared to the
shRNA-Ctrl-infected cultures (Fig.
3).
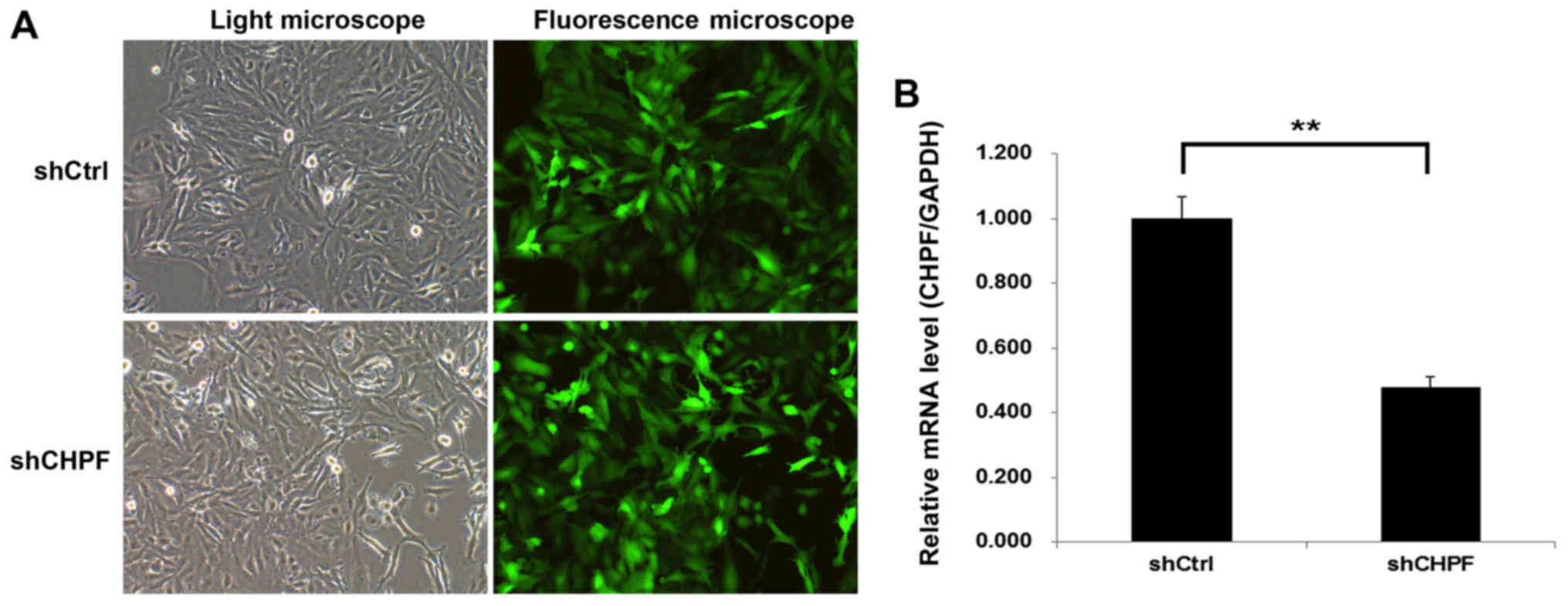
Knockdown of CHPF by the shRNA
lentiviral system in human glioma cell line U251
To explore the role of CHPF in glioma, shRNA-CHPF
and shRNA-Ctrl expressing GFP were generated and infected into
human glioma cell line U251. The concentration dose of the
shRNA-CHPF used was 4×108 TU/ml. More than 80% of the
cells exhibited positive green fluorescence in both the shRNA-CHPF
and shRNA-Ctrl groups, indicating the successful lentiviral
infection (Fig. 4A). The knockdown
effect was analyzed by qRT-PCR. At 5 days post-infection, the CHPF
mRNA level of U251 cells infected with shRNA-CHPF (0.477±0.034) was
significantly lower than that infected with the shRNA-Ctrl
(1.001±0.065) (Fig. 4B).
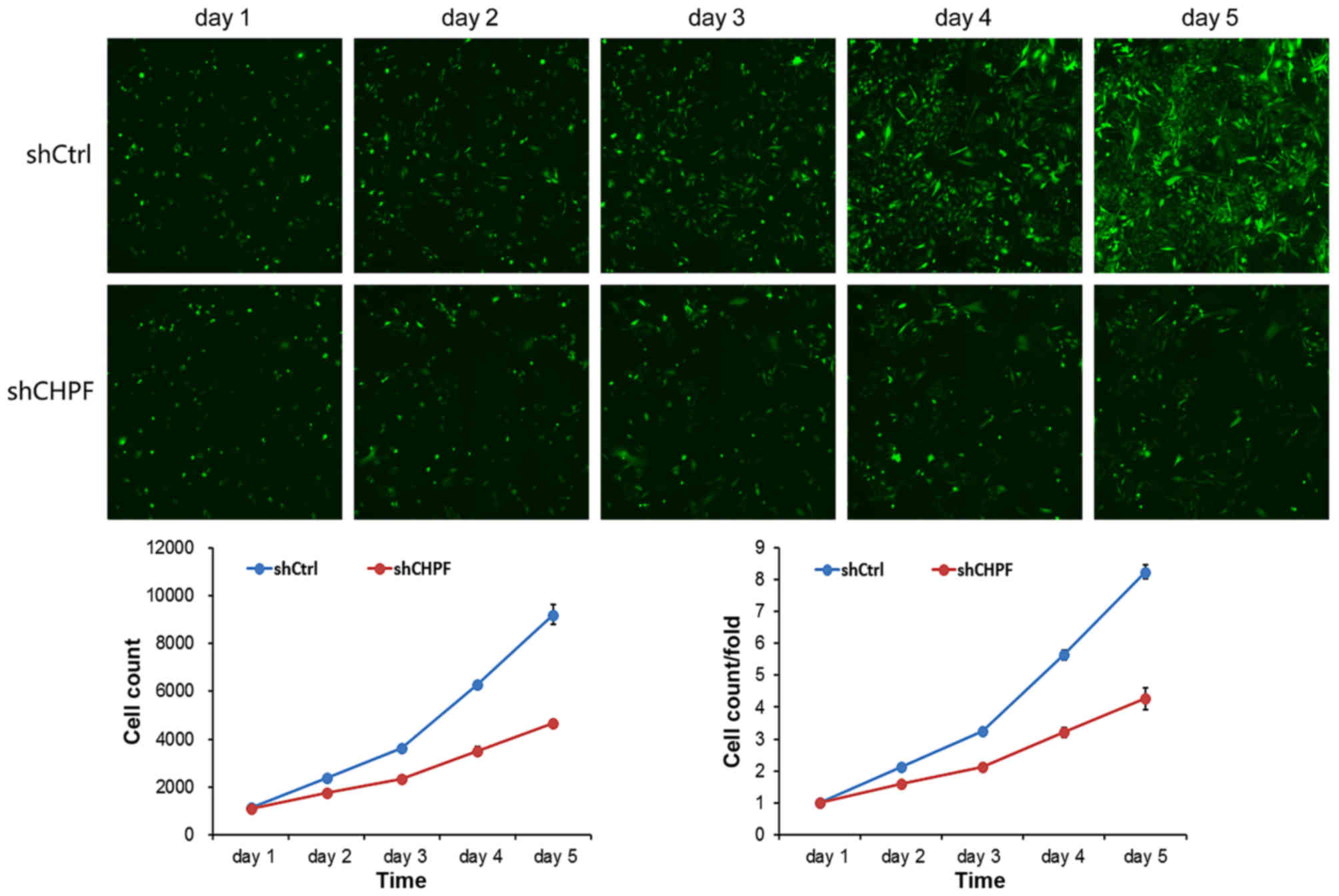
Knockdown of CHPF noticeably inhibits
U251 cell growth
To examine the effect of CHPF on cell growth, U251
cells expressing either the shRNA-CHPF or shRNA-Ctrl were seeded
into 96-well plates and analyzed by Cellomics daily for 5 days. As
shown in Fig. 5, the cell
proliferation level in the shRNA-CHPF group remained lower from the
second day after infection, and the gap between the shRNA-CHPF and
shRNA-Ctrl groups increased with time. The results of the present
study revealed that CHPF knockdown significantly inhibited the
growth of the U251 cells.
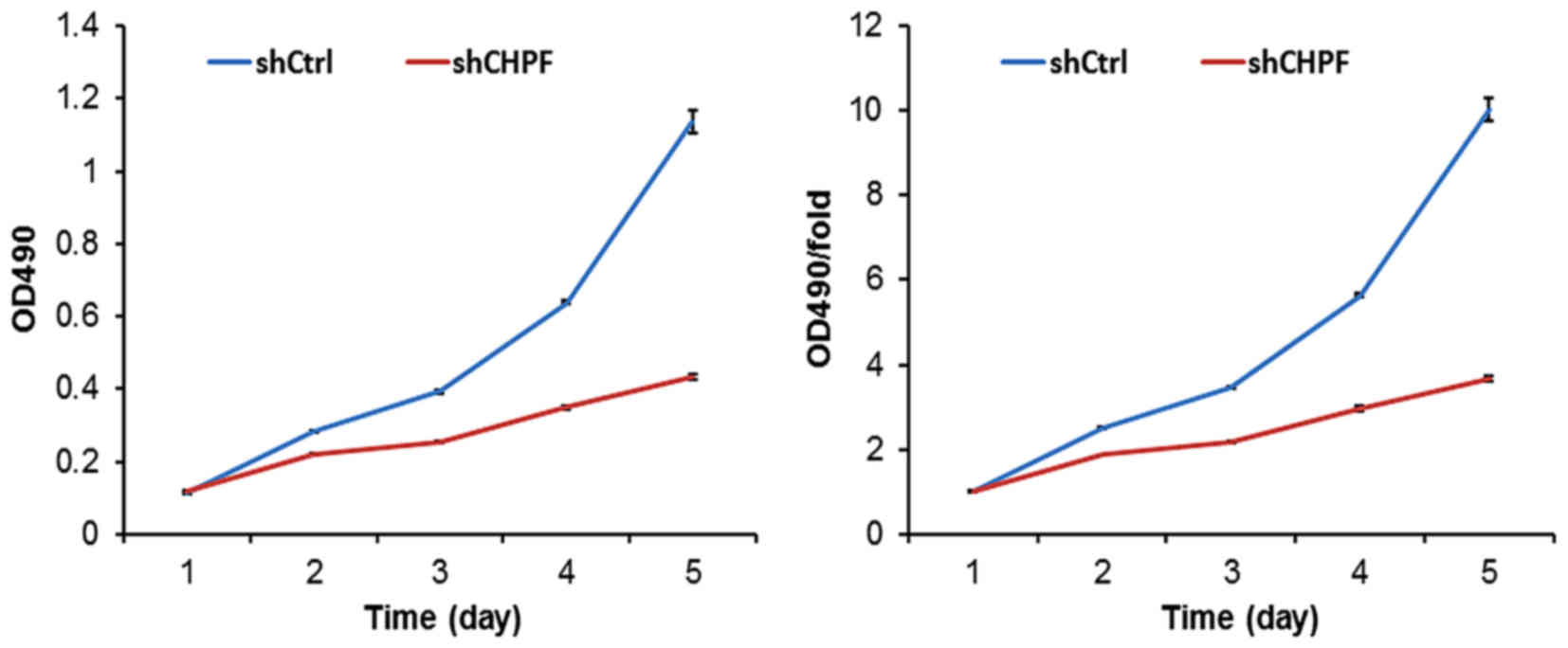
To further evaluate the effect of CHPF on the
regulation of glioma cell proliferation, an MTT assay was applied
in U251 cells. As shown in Fig. 6,
knockdown of CHPF expression markedly decreased the proliferation
potential from day 2 (P<0.01). The growth of U251 cells treated
with the shRNA-CHPF lentivirus was markedly inhibited compared to
the shRNA-Ctrl group. The shRNA-CHPF cells had significantly
decreased in vitro growth on days 4 (shCtrl, 5.64±0.04 vs.
shCHPF 2.96±0.05; P<0.01) and 5 (shCtrl, 10.03±0.28 vs. shCHPF,
3.67±0.07; P<0.01). Based on these data, in vitro U251
cell growth was dependent on CHPF expression.
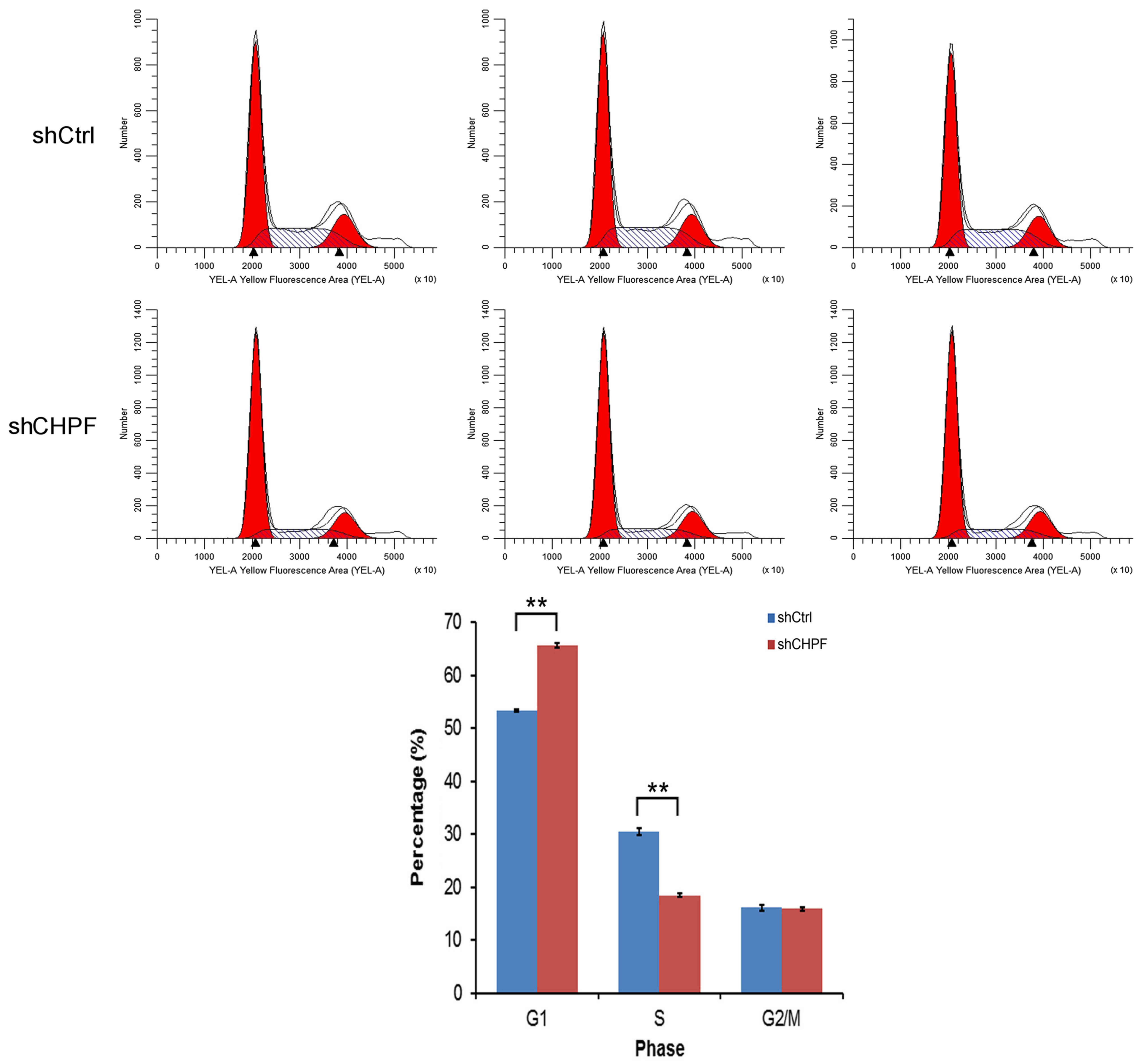
Inhibition of CHPF induces G0/G1 phase
arrest in glioma U251 cells
To ascertain whether CHPF is necessary for cell
cycle progression in U251 cells, we assessed the cell cycle phases
in U251 cells by flow cytometry. As shown in Fig. 7, the shRNA-CHPF group displayed the
following: G0/G1 phase, 65.57±0.42%; S phase, 18.48±0.26%; and G2/M
phase, 15.95±0.32%, and the shRNA-Ctrl group displayed the
following distribution: G0/G1 phase, 53.42±0.23%; S phase,
30.49±0.70%; and G2/M phase, 16.10±0.50%. With the absence of CHPF,
the number of cells entering the G0/G1 phase increased by 22.7%
(P<0.05) and the cells entering the S phase decreased by 39.4%
(P<0.01). In conclusion, these data revealed that CHPF regulated
U251 cell growth and blocked cell cycle progression in the G0/G1
phase.
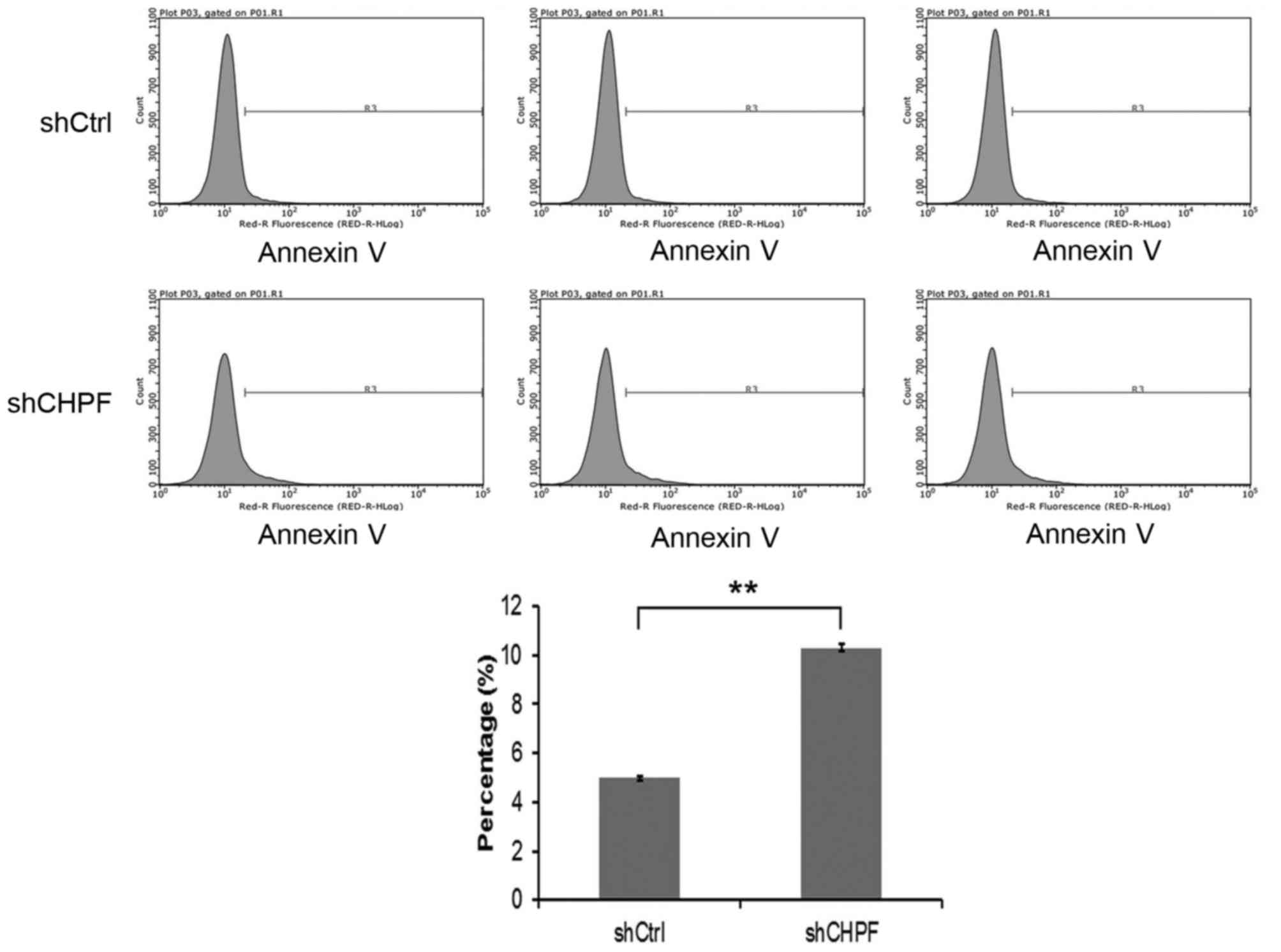
CHPF knockdown increases glioma U251
cell apoptosis
We further examined the effect of knockdown of CHPF
on the cell apoptosis of glioma U251 cells by flow cytometry. The
apoptosis percentage of U251 cells in the shRNA-CHPF group
(10.30±0.16)% was significantly higher (P<0.05) than in the
shRNA-Ctrl group (4.99±0.09)% (Fig.
8). These results indicated that CHPF silencing inhibited the
proliferation of U251 cells by inducing G0/G1 phase cell cycle
arrest and apoptosis.
Discussion
Glioma, particularly glioblastoma, has a poor
prognosis (4,13). It is one of the most common highly
vascularized malignant tumors of the CNS, with a ≤12-month median
survival period (5,14). Therefore, it is essential to
investigate the mechanisms involved in glioma initiation and
progression.
Chondroitin sulfate (CS) is a polysaccharide
consisting of repeating disaccharide units of
N-acetyl-D-galactosamine and D-glucuronic acid residues, modified
with sulfated residues at various positions (11). CS biosynthesis and sulfation balance
are tightly controlled and play important roles in the progression
of the disease (15,16). In addition, the specific sulfation
pattern of CS chains dictates its function and binding affinities
(17,18). Previous studies utilizing inhibitors
or enzymes which degrade CS chains indicate that CS has a
considerable effect on the processes of tumor metastasis,
proliferation and adhesion (19,20).
The identification of CHPF, a unique protein factor required for
chondroitin polymerization activity, may shed light on the
molecular basis of CS. Therefore, CHPF may play an important role
in tumor progression. However, to date, only 3 studies have
revealed the expression level of CHPF in cancer, including
colorectal cancer (10), head and
neck squamous cell carcinoma (21)
and laryngeal cancer (22). To
date, the function and the role of CHPF in human types of cancer
remain unknown. Therefore, we employed LV-shRNA-CHPF to determine
whether CHPF has effects on the progression of human glioma in
vitro.
In the present study, we firstly demonstrated that
CHPF was highly expressed in human glioma tissues and 4 glioma cell
lines. To explore the role of CHPF in glioma, a lentiviral vector
expressing CHPF shRNA was constructed and transfected into the
glioma U251 cells, which stably downregulated the expression levels
of the CHPF gene in U251 cells in vitro. Compared to the
shRNA-Ctrl group of cells, the shRNA-CHPF group of cells exhibited
decreased proliferation and a significant increase in the
proportion of cells in the G0/G1 phase. In addition, we found that
knockdown of the expression of CHPF increased apoptosis in glioma
U251 cells. Therefore, our results confirmed that CHPF promotes
glioma U251 cell growth. Further validation and functional
evaluation are warranted to assess the role of CHPF in other glioma
cell lines and the mechanism underlying this association.
In conclusion, our findings indicate that CHPF plays
an important role in the progression of human glioma. A high level
of expression of CHPF was found in human glioma tissues and 4
glioma cell lines. Downregulation of CHPF expression by the
shRNA-CHPF lentiviral vector inhibited glioma U251 cell
proliferation and induced cell apoptosis. This demonstrated that
CHPF may be a novel target in the clinical treatment of
gliomas.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81660420), the
Construction Plan of the Superior Science and Technology Innovation
Team of Jiangxi Province (grants no. 20152BCB24009), and the
Foreign Science and Technology Cooperation Plan of Jiangxi Province
(grants no. 20151BDH80009).
References
|
1
|
Wu M, Fan Y, Lv S, Xiao B, Ye M and Zhu X:
Vincristine and temozolomide combined chemotherapy for the
treatment of glioma: A comparison of solid lipid nanoparticles and
nanostructured lipid carriers for dual drugs delivery. Drug Deliv.
23:2720–2725. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alves TR, Lima FR, Kahn SA, Lobo D, Dubois
LG, Soletti R, Borges H and Neto VM: Glioblastoma cells: A
heterogeneous and fatal tumor interacting with the parenchyma. Life
Sci. 89:532–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Castro MG, Candolfi M, Kroeger K, King GD,
Curtin JF, Yagiz K, Mineharu Y, Assi H, Wibowo M, Muhammad Ghulam
AK, et al: Gene therapy and targeted toxins for glioma. Curr Gene
Ther. 11:155–180. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiao B, Zhou X, Ye M, Lv S, Wu M, Liao C,
Han L, Kang C and Zhu X: MicroRNA-566 modulates vascular
endothelial growth factor by targeting Von Hippel-Landau in human
glioblastoma in vitro and in vivo. Mol Med Rep.
13:379–385. 2016.PubMed/NCBI
|
|
5
|
Fan YH, Ye MH, Wu L, Lv SG, Wu MJ, Xiao B,
Liao CC, Ji QK, Chai Y and Zhu XG: Overexpression of miR-98
inhibits cell invasion in glioma cell lines via downregulation of
IKKε. Eur Rev Med Pharmacol Sci. 19:3593–3604. 2015.PubMed/NCBI
|
|
6
|
Wu L, Yang L, Xiong Y, Guo H, Shen X,
Cheng Z, Zhang Y, Gao Z and Zhu X: Annexin A5 promotes invasion and
chemoresistance to temozolomide in glioblastoma multiforme cells.
Tumour Biol. 35:12327–12337. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kitagawa H, Izumikawa T, Uyama T and
Sugahara K: Molecular cloning of a chondroitin polymerizing factor
that cooperates with chondroitin synthase for chondroitin
polymerization. J Biol Chem. 278:23666–23671. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalathas D, Theocharis DA, Bounias D,
Kyriakopoulou D, Papageorgakopoulou N, Stavropoulos MS and Vynios
DH: Alterations of glycosaminoglycan disaccharide content and
composition in colorectal cancer: Structural and expressional
studies. Oncol Rep. 22:369–375. 2009.PubMed/NCBI
|
|
9
|
Pothacharoen P, Siriaunkgul S, Ong-Chai S,
Supabandhu J, Kumja P, Wanaphirak C, Sugahara K, Hardingham T and
Kongtawelert P: Raised serum chondroitin sulfate epitope level in
ovarian epithelial cancer. J Biochem. 140:517–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kalathas D, Theocharis DA, Bounias D,
Kyriakopoulou D, Papageorgakopoulou N, Stavropoulos MS and Vynios
DH: Chondroitin synthases I, II, III and chondroitin sulfate
glucuronyltransferase expression in colorectal cancer. Mol Med Rep.
4:363–368. 2011.PubMed/NCBI
|
|
11
|
Ogawa H, Shionyu M, Sugiura N, Hatano S,
Nagai N, Kubota Y, Nishiwaki K, Sato T, Gotoh M, Narimatsu H, et
al: Chondroitin sulfate synthase-2/chondroitin polymerizing factor
has two variants with distinct function. J Biol Chem.
285:34155–34167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu H, Liang S, Yang X, Ji Z, Zhao W, Ye X
and Rui J: RNAi-mediated RPL34 knockdown suppresses the growth of
human gastric cancer cells. Oncol Rep. 34:2267–2272.
2015.PubMed/NCBI
|
|
13
|
Ivo D'Urso P, Fernando D'Urso O, Damiano
Gianfreda C, Mezzolla V, Storelli C and Marsigliante S: miR-15b and
miR-21 as circulating biomarkers for diagnosis of glioma. Curr
Genomics. 16:304–311. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Viswanath P and Ronen SM: Metabolic
reprogramming of pyruvate dehydrogenase is essential for the
proliferation of glioma cells expressing mutant IDH1. Mol Cell
Oncol. 3:e10779222016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prinz RD, Willis CM, van Kuppevelt TH and
Klüppel M: Biphasic role of chondroitin sulfate in cardiac
differentiation of embryonic stem cells through inhibition of
Wnt/β-catenin signaling. PLoS One. 9:e923812014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Willis CM and Klüppel M: Chondroitin
sulfate-E is a negative regulator of a pro-tumorigenic
Wnt/beta-catenin-Collagen 1 axis in breast cancer cells. PLoS One.
9:e1039662014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Klüppel M: The roles of
chondroitin-4-sulfotransferase-1 in development and disease. Prog
Mol Biol Transl Sci. 93:113–132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Karangelis DE, Kanakis I, Asimakopoulou
AP, Karousou E, Passi A, Theocharis AD, Triposkiadis F, Tsilimingas
NB and Karamanos NK: Glycosaminoglycans as key molecules in
atherosclerosis: The role of versican and hyaluronan. Curr Med
Chem. 17:4018–4026. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fthenou E, Zong F, Zafiropoulos A, Dobra
K, Hjerpe A and Tzanakakis GN: Chondroitin sulfate A regulates
fibrosarcoma cell adhesion, motility and migration through JNK and
tyrosine kinase signaling pathways. In Vivo. 23:69–76.
2009.PubMed/NCBI
|
|
20
|
Denholm EM, Lin YQ and Silver PJ:
Anti-tumor activities of chondroitinase AC and chondroitinase B:
Inhibition of angiogenesis, proliferation and invasion. Eur J
Pharmacol. 416:213–221. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Teh MT, Gemenetzidis E, Patel D, Tariq R,
Nadir A, Bahta AW, Waseem A and Hutchison IL: FOXM1 induces a
global methylation signature that mimics the cancer epigenome in
head and neck squamous cell carcinoma. PLoS One. 7:e343292012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kalathas D, Triantaphyllidou IE,
Mastronikolis NS, Goumas PD, Papadas TA, Tsiropoulos G and Vynios
DH: The chondroitin/dermatan sulfate synthesizing and modifying
enzymes in laryngeal cancer: Expressional and epigenetic studies.
Head Neck Oncol. 2:272010. View Article : Google Scholar : PubMed/NCBI
|