Introduction
Lung cancer is a tumor with a highly malignant
phenotype characterized by rapid progression, late diagnosis, and a
limited response to radiotherapy and chemotherapy. In the past 10
years, despite of the FDA approved therapeutic regimens and great
improvements in medical care, we have observed no momentous effect
on lung cancer patient survival (1,2).
Therefore, to identify biomarkers which may promote early diagnosis
and allow personalized treatment strategies for patients at a high
risk of lung cancer has become an urgent need (2).
MicroRNAs (miRNAs) are particularly important in
almost all tumor developmental processes since they can be targets
of genomic lesions, controlled by classic tumor signals, and they
themselves present as a class of oncogenes or tumor suppressors
(3,4). Recently, a close association between
miRNAs and lung cancer tumorigenesis has been suggested (5,6).
Multiple miRNAs, such as the let-7 family (7), miR-200 (8), miR-486 (9) and miR-146a (10) have been identified as tumor
suppressors. Meanwhile, miR-31 (11), miR-212 (12) and miR-196a (13) were found to promote lung cancer
carcinogenesis. miR-140 has been noted since it is involved in the
development and progression of many types of cancers, including
breast cancer (14), osteosarcoma
(15), colon (16) and lung cancer (17). Yuan et al reported that
miR-140 is significantly downregulated in non-small cell lung
cancer (NSCLC) tissues and cell lines; and that miR-140 suppresses
tumor growth and metastasis of NSCLC by targeting insulin-like
growth factor 1 receptor (18).
These findings suggest that miR-140 plays a tumor-suppressor role
in these cancers. However, to the best of our knowledge, its role
and the potential mechanisms in lung cancer remain unclear.
Human genome sequence data indicate that more than
90% of the DNA sequences actively transcribe, but only 2% encode
protein; thus, the majority of the transcripts are referred to as
non-coding RNAs (ncRNAs) (19,20).
Small non-coding RNAs such as miRNAs have been studied extensively
and their roles in gene regulation and cell function have been
elucidated in numerous types of cancers (20). Recent studies have shown that long
non-coding RNAs (lncRNAs) play important roles in both normal
development and diseases including cancer (21). LncRNAs have emerged as new players
in cancer research and several studies have shown that some lncRNAs
function as oncogenes, tumor suppressor genes or both, depending on
the circumstance (22).
The mechanisms by which lncRNAs exert their effect
vary under different conditions; however, the interaction between
lncRNAs and microRNAs plays a major role (23,24).
Zhu et al reported that the lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting transforming
growth factor β-induced protein (TGFBI) (25). The interaction between miR-141 and
lncRNA-H19 has been regarded as an important component in
regulating cell proliferation and migration in gastric cancer
(26).
In the present study, we report an interaction
between X-inactive specific transcript (XIST) and miR-140 which
regulates lung cancer cell growth by directly targeting inhibitor
of apoptosis-stimulating protein of p53 (iASPP). Our findings
provide a novel understanding of the role of XIST and miR-140 in
lung cancer metastasis and the mechanism involved.
Materials and methods
Tissue samples, cell lines and cell
transfection
We collected 28 paired primary lung cancer and
matched adjacent normal tissues. We obtained all samples from
patients who underwent surgical resection at Xiangya Hospital of
Central South University (Changsha, China). The tissues were
snap-frozen in liquid nitrogen, and then stored at −80°C. The
present study was approved by the Ethics Committee of Xiangya
Hospital of Central South University. Informed consent statements
were obtained from all the subjects included.
We purchased human lung cancer cell lines, A549 and
H1299, from the American Type Culture Collection (ATCC; Manassas,
VA, USA). They were cultured in RPMI-1640 medium (Invitrogen,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco,
Carlsbad, CA, USA) at 37°C in a humidified atmosphere with 5%
CO2. The expression of miR-140 was regulated by
transfection of miR-140 mimics or miR-140 inhibitor (GenePharma,
Shanghai, China) using Lipofectamine 2000 (Invitrogen). A
pcDNA3.1/iASPP was used to achieve overexpression of iASPP
(GeneCopoecia, Guangzhou, China). Cells were plated into 6- or
96-well plates, transfected, incubated for 24 or 48 h, and used for
further assays or RNA/protein extraction.
RNA extraction and SYBR-Green
quantitative PCR analysis
We extracted total RNA from cells using TRIzol
reagent (Invitrogen), and detected mature miR-140 expression in
cells using a Hairpin-it™ miRNAs qPCR kit (GenePharma). We used
expression of RNU6B as an endogenous control. iASPP expression was
measured using SYBR-Green qPCR assay (Takara, Dalian, China). Data
were processed using the 2−ΔΔCt method.
MTT assay
A modified MTT assay was used to evaluate cell
viability. After seeding 2×103 transfected cells/well
into 96-well culture plates we assessed the viability of A549 and
H1299 cells transfected with miR-140 or control at five time points
(on day 1–5). In brief, quantification of mitochondrial
dehydrogenase activity was achieved through the enzymatic
conversion of [3-(4,5-dimethyldiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT); Sigma-Aldrich, St. Louis, MO, USA] to a colored
formazan product. MTT (10 µl, 10 mg/ml) was added to the cells,
incubated for 4 h, and the reaction was terminated by removal of
the supernatant and addition of 100 µl dimethyl sulfoxide (DMSO) to
dissolve the formazan product. After 0.5 h, the optical density
(OD) of each well was measured at 570 nm using a plate reader
(ELx808; Bio-Tek Instruments, Inc., Winooski, VT, USA).
BrdU incorporation assay
DNA synthesis in proliferating cells is determined
by measuring 5-bromo-2-deoxyuridine (BrdU) incorporation. BrdU
assays were performed at 24 and 48 h after transfecting A549 and
H1299 cells with miR-140 or the control vector. After seeding the
infected cells in 96-well culture plates at a density of
2×103 cells/well, they were cultured for 24 or 48 h, and
incubated with a final concentration of 10 µM BrdU (BD Pharmingen,
San Diego, CA, USA) for 2–24 h. When the incubation period ended,
we removed the medium, fixed the cells for 30 min at room
temperature (RT), incubated them with peroxidase-coupled anti-BrdU
antibody (Sigma-Aldrich) for 60 min at RT, washed them three times
with phosphate-buffered saline (PBS), incubated the cells with
peroxidase substrate (tetramethylbenzidine) for 30 min, and
measured the absorbance values at 450 nm. Background BrdU
immunofluorescence was determined in cells not exposed to BrdU, but
stained with the BrdU antibody.
Apoptosis cell death detection by
enzyme-linked immunosorbent assay (ELISA)
To measure apoptosis in vitro, a
DNA-fragmentation ELISA was used according to the manufacturer's
instructions (Roche Diagnostics, Indianapolis, IN, USA).
Western blot analysis
The expression of iASPP and XIST in the lung cancer
cells was detected by performing immunoblotting. We lysed cultured
or transfected cells in RIPA buffer with 1% phenylmethanesulfonyl
fluoride (PMSF), and loaded protein onto a SDS-PAGE minigel and
transferred them onto polyvinylidene difluoride (PVDF) membranes.
After being probed with 1:1,000 diluted rabbit polyclonal iASPP and
XIST antibodies (Abcam, Cambridge, MA, USA) at 4°C overnight, the
blots were subsequently incubated with the HRP-conjugated secondary
antibody (1:5,000). ECL substrates were used to visualize signals
(Millipore, Billerica, MA, USA). β-actin was used as an endogenous
protein for normalization.
Luciferase reporter assay
A549 cells were seeded into a 24-well plate. After
being cultured overnight, the cells were co-transfected with the
wild-type and mutated iASPP 3 untranslated region (3′ UTR) reporter
plasmid, and pRL-TK plasmids, or transfected with miR-140 mimics
and miR-140 inhibitor. Luciferase assays were performed 48 h after
transfection using the Dual-Luciferase Reporter Assay System
(Promega, Madison, WI, USA).
Statistical analysis
Data are expressed as the mean ± SD of three
independent experiments and were processed using SPSS 17.0
statistical software (SPSS, Inc., Chicago, IL, USA). Using
Wilcoxon's paired test we compared the expression of miR-140 in
lung cancer and the paired adjacent normal colonic tissues. The
differences between groups were evaluated using the one-way
analysis of variance (ANOVA). P-values of <0.05 were considered
statistically significant.
Results
miR-140 negatively regulates iASPP
expression in the A549 cell line
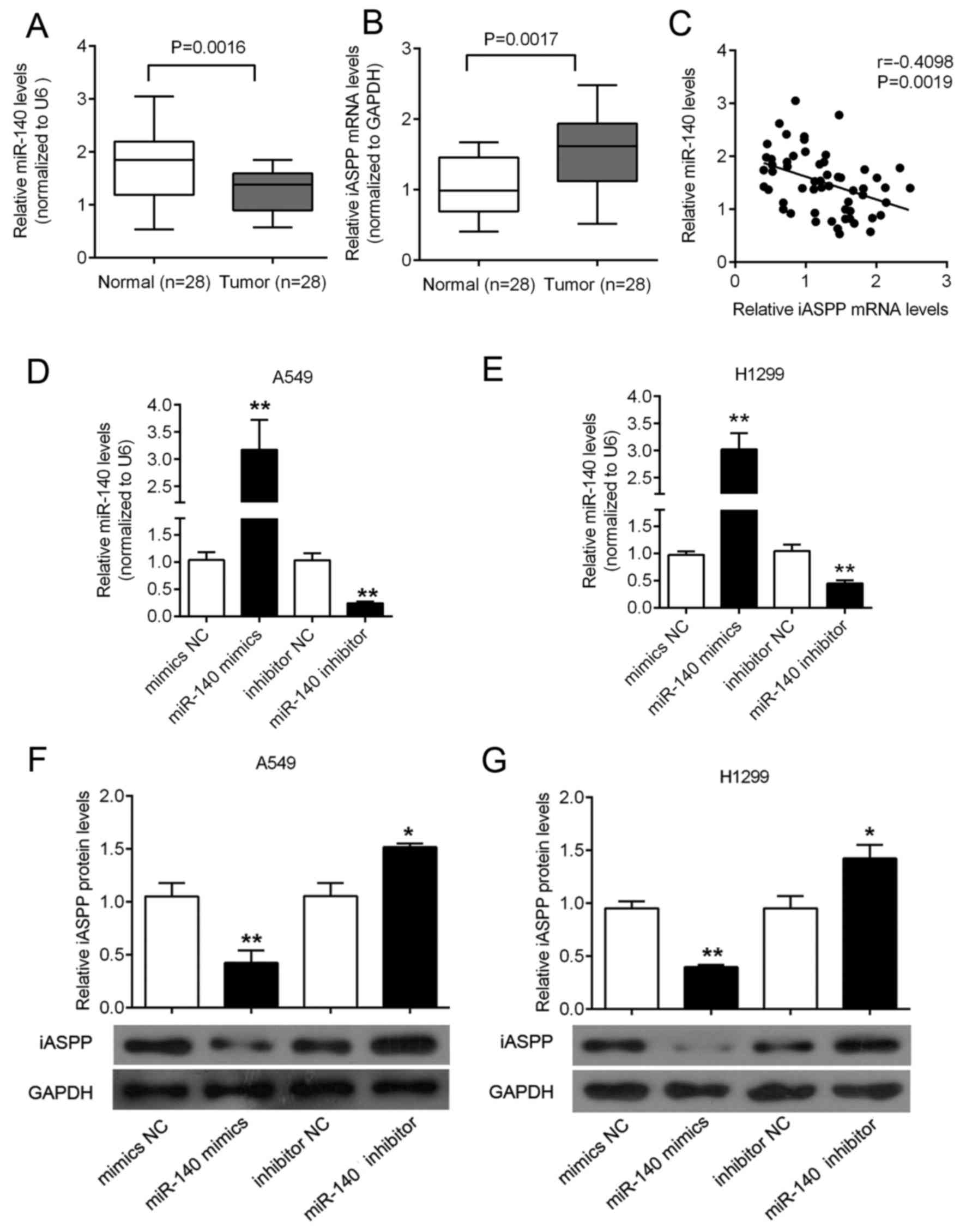
SYBR-Green quantitative PCR analysis was performed
to quantify the expression of miR-140 in lung cancer tissues. In a
large panel of 28 cases of primary lung cancer and the adjacent
normal colonic tissues, the expression of miR-140 was significantly
decreased in the lung cancer tissues, compared with that noted in
the paired adjacent normal tissues (Fig. 1A). In contrast, the expression of
iASPP was upregulated in the lung cancer tissues, compared with
that noted in the paired adjacent normal tissues (Fig. 1B). An inverse correlation between
the expression levels of miR-140 and iASPP was observed (Fig. 1C). To investigate the effect of
miR-140 on iASPP expression, miR-140 mimics were used to achieve
miR-140 overexpression, and miR-140 inhibitor was used to achieve
miR-140 knockdown in the A549 and H1299 cell lines (Fig. 1D and E), and the expression
efficiency was verified using real-time PCR. Next, the protein
expression of iASPP was determined using western blotting in
response to miR-140 overexpression or miR-140 inhibition. Results
showed that miR-140 overexpression suppressed the iASPP protein
expression, while miR-140 inhibition promoted the iASPP protein
expression in both A549 and H1299 cell lines (Fig. 1F and G).
iASPP mRNA is a direct target of
miR-140
It was predicted that miR-140 could bind to and
target the iASPP 3′ UTR using TargetScan, miRanda and
miRWalk (27–29). A wt-iASPP 3′ UTR luciferase
reporter vector (wt-iASPP) was created to confirm this prediction,
as well as a mut-iASPP 3′ UTR luciferase reporter vector
(mut-iASPP) by sequentially mutating the predicted five base pair
miR-140 binding site in the iASPP 3′ UTR (Fig. 2A). The wt-iASPP vector and miR-140
mimics or inhibitor were co-transfected into the A549 cell line. In
the miR-140 mimic-transfected cells, the luciferase activity of the
iASPP 3′ UTR luciferase reporter vector was significantly
reduced while it was induced in the miR-140 inhibitor-transfected
cells, compared to the inhibitor NC group (Fig. 2B and C). However, mutation of the
putative the miR-140 binding site in the iASPP 3′ UTR
abolished miR-140-mediated repression of iASPP 3′ UTR
luciferase reporter activity (Fig. 2B
and C).
miR-140 inhibits the proliferation of
lung cancer cell line through iASPP
To investigate the functional roles of iASPP in lung
cancer cells, we transfected A549 and H1299 cell lines with
pcDNA3.1/iASPP to achieve iASPP overexpression (Fig. 3A). The expression level of iASPP
protein was verified using western blotting (Fig. 3A). Then, pcDNA3.1/iASPP and miR-140
mimics/mimics NC were co-transfected into the A549 and H1299 cell
lines. MTT and BrdU results showed that miR-140 mimic transfection
significantly reduced the cell proliferation, while the forced
expression of iASPP reversed the significant inhibition on cell
proliferation by miR-140 (Fig. 3B and
C). Similarly, miR-140 overexpression notably promoted the DNA
fragmentation of both A549 and H1299 cells, while iASPP
transfection attenuated the promotive effect of miR-140 on DNA
fragmentation of both A549 and H1299 cells (Fig. 3D).
XIST is correlated with miR-140 by
direct targeting
According to previous studies, miR-140 plays a
suppressive role in cancers (30).
Previous studies have reported that XIST is associated with cancers
by regulating miRNAs (31). To
further investigate the mechanism by which miR-140 is regulated in
lung cancer cell lines, we generated a wt-XIST 3′ UTR luciferase
reporter vector (wt-XIST), as well as a mut-XIST3′ UTR luciferase
reporter vector (mut-XIST) by sequentially mutating predicted
miR-140 binding sites in the XIST 3′ UTR (Fig. 4A). We co-transfected the
wt-XIST/mut-XIST vectors and miR-140 NC/miR-140 mimics into A549
cells. The luciferase activity of the XIST luciferase reporter
vector was significantly reduced in the miR-140 mimic-transfected
cells, compared to the mimics NC groups (Fig. 4B). Moreover, the luciferase activity
of the XIST luciferase reporter vector was markedly amplified in
the miR-140 inhibitor-transfected cells, compared to the inhibitor
NC groups (Fig. 4C).
XIST promotes lung cancer cell growth
and inhibits cell apoptosis
Next, we investigated the association of XIST
expression with lung cancer cell proliferation. XIST knockdown was
achieved by si-XIST and the inhibitory efficiency was verified by
real-time PCR (Fig. 5A). Two lung
cancer cell lines, A549 and H1299, were transfected with si-NC or
si-XIST, and then the cell proliferation was determined by MTT and
BrdU assays. MTT assays revealed that knockdown of XIST
significantly attenuated the proliferation of both A549 and H1299
cell lines over time, compared with the si-NC group (Fig. 5B and C). Moreover, it was observed
that DNA fragmentation of both A549 and H1299 cell lines was
significantly promoted (Fig. 5D).
Taken together, these data revealed that lncRNA-XIST promoted lung
cancer cell growth and inhibited cell apoptosis.
XIST promotes iASPP expression through
miR-140
Given that XIST inhibits miR-140 by direct binding,
and that miR-140 inhibits iASPP by binding to its 3′ UTR, we next
investigated the association of XIST expression with iASPP protein
expression in lung cancer cell lines. Results from western blotting
showed that miR-140 knockdown significantly promoted iASPP protein
expression, while XIST knockdown by si-XIST could partly restore
this effect in both A549 and H1299 cell lines (Fig. 6A and B). Taken together, XIST
promoted iASPP expression, most possibly through miR-140.
Discussion
Alterations in the presence of miRNAs are implicated
in almost all fields of cancer biology, including cell growth,
apoptosis, migration and/or invasion, and they function as either
tumor suppressors or oncogenes (28). In the present study, we focused on
miR-140 due to its potential suppressive function in human
malignances. Song et al indicated that cell proliferation in
both osteosarcoma and colon cancer cell lines was inhibited by
overexpression of miR-140 (16).
Recently, Yang et al reported that miR-140-5p was
significantly decreased in HCC tissues and cell lines, and its
overexpression suppressed tumor growth and metastasis by targeting
transforming growth factor β receptor 1 and fibroblast growth
factor 9 (32). To date, however,
the role of miR-140 in lung cancer carcinogenesis and the molecular
mechanisms by which miR-140 exerts its functions remain unclear. In
the present study, the real-time PCR results showed that miR-140
expression was significantly downregulated in lung cancer tissues.
Overexpression of miR-140 significantly inhibited the proliferation
and promoted the cell apoptosis of lung cancer cell. Accordingly,
knockdown of miR-140 promoted cell proliferation and attenuated
cell apoptosis. These results indicate that miR-140 could be a
potential tumor-suppressor miRNA in lung cancer.
The impact of specific miRNAs on cancer biology
depends on their downstream targets (33,34).
Different prediction algorithms were used to predict gene targets
for miR-140, to elucidate the underlying mechanisms involved in the
miR-140-induced inhibition of lung cancer growth and metastasis.
The iASPP oncogene was identified as a critical downstream target
of miR-140, due to its frequent overexpression in many malignancies
and its function as an important regulator of cell proliferation,
survival and metastasis (35–38).
In the present study, miR-140 overexpression significantly
downregulated the protein level of iASPP. Meanwhile, knockdown of
miR-140 upregulated the expression of iASPP. A luciferase reporter
assay validated the binding and repressive effects of miR-140 on
the iASPP 3′ UTR in A549 cells. Western blot analysis also
confirmed that iASPP was downregulated by miR-140 upregulation, and
upregulated by miR-140 downregulation. Moreover, the expression
levels of miR-140 and iASPP in clinical lung cancer samples were
significantly inversely correlated. To further confirm the role of
iASPP in the A549 cell line, we revealed that knockdown of iASPP
inhibited the proliferation and promoted the apoptosis of the A549
cell line. These data regarding the involvement of the
miR-140/iASPP axis in lung cancer suggest that miR-140 may have
potential as a therapeutic target.
To identify the correlation between miRNAs and
lncRNAs and the underlying mechanisms provide potential targets for
tumor treatment. Previous studies have mostly focused on the
targets of miRNAs and the mechanisms by which miRNAs regulate the
targets, thus as to influence tumor initiation and progression. For
example, miRNAs including miR-21, miR-34a, miR-182 (39–41),
have been reported to be involved in tumor growth and progression.
However, few studies have focused on the regulation of miRNAs,
particularly the co-regulation of miRNAs by non-coding RNAs.
Emerging evidence has revealed that the mutual regulation between
miRNAs and lncRNAs play major roles in tumor progression. In the
present study, we revealed the interaction between XIST and miR-140
for the first time. The knockdown of XIST upregulated miR-140,
while forced miR-140 overexpression inhibited the expression of
XIST.
To detect the role of XIST in lung cancer cell
growth regulation, siRNA was transfected into lung cancer cell
lines to knock down XIST. As expected, iASPP was downregulated by
XIST knockdown. In addition, the proliferation of lung cancer cell
lines was reduced by iASPP inhibition. According to previous
studies, iASPP is an oncogene and promotes cancer cell growth
(36,37,42).
To further validate the correlation of XIST, miR-140
and iASPP in lung cancer tissues, the expression levels of XIST,
miR-140 and iASPP were determined. The data showed that the
expression levels of XIST and iASPP were upregulated, while miR-140
expression was downregulated.
In summary, through targeting iASPP, lung cancer
cell viability and proliferation were inhibited while cell
apoptosis was promoted by miR-140. These effects were due to the
regulation of miR-140 by XIST. The XIST/miR-140/iASPP axis
identified in the present study may play a key role in regulating
lung cancer cell proliferation and apoptosis, and may provide a
potential therapeutic strategy for lung cancer.
Acknowledgements
The present study was supported by the National Key
Scientific and Technology Support Program, and the Collaborative
innovation of Clinical Research for Chronic Obstructive Pulmonary
Sisease and Lung Cancer (no. 2013BAI09B09).
References
|
1
|
Neville A: Lung cancer. BMJ Clin Evid.
2009:pii: 1504. 2009.PubMed/NCBI
|
|
2
|
Chirieac LR and Dacic S: Targeted
therapies in lung cancer. Surg Pathol Clin. 3:71–82. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai XL, Huang YH, Li YS, Li GN, Wang LP,
Sun R, Ma YS, Feng SY, Chang ZY, Wang XH, et al: Differential
expression profiling of microRNAs in para-carcinoma, carcinoma and
relapse human pancreatic cancer. Clin Transl Oncol. 17:398–408.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pavlakis E, Papaconstantinou I, Gazouli M,
Theodosopoulos T, Karamanolis G, Genatas K and Ladas SD: MicroRNA
gene polymorphisms in pancreatic cancer. Pancreatology. 13:273–278.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, VandenBoom TG II, Kong D, Wang Z,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-200 and
let-7 by natural agents leads to the reversal of
epithelial-to-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Cancer Res. 69:6704–6712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu J, Ohuchida K, Mizumoto K, Sato N,
Kayashima T, Fujita H, Nakata K and Tanaka M: MicroRNA,
hsa-miR-200c, is an independent prognostic factor in
pancreatic cancer and its upregulation inhibits pancreatic cancer
invasion but increases cell proliferation. Mol Cancer. 9:1692010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mees ST, Mardin WA, Sielker S, Willscher
E, Senninger N, Schleicher C, Colombo-Benkmann M and Haier J:
Involvement of CD40 targeting miR-224 and miR-486 on the
progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol.
16:2339–2350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, VandenBoom TG II, Wang Z, Kong D,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-146a
contributes to the inhibition of invasion of pancreatic cancer
cells. Cancer Res. 70:(Suppl 8). S57032010. View Article : Google Scholar
|
|
11
|
Laurila EM, Sandström S, Rantanen LM,
Autio R and Kallioniemi A: Both inhibition and enhanced expression
of miR-31 lead to reduced migration and invasion of pancreatic
cancer cells. Genes Chromosomes Cancer. 51:557–568. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H,
Wang W, Huang X, Yuan Z and Ai K: miR-212 promotes pancreatic
cancer cell growth and invasion by targeting the hedgehog signaling
pathway receptor patched-1. J Exp Clin Cancer Res. 33:542014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang F, Tang J, Zhuang X, Zhuang Y, Cheng
W, Chen W, Yao H and Zhang S: MiR-196a promotes pancreatic cancer
progression by targeting nuclear factor kappa-B-inhibitor alpha.
PLoS One. 9:e878972014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Q, Yao Y, Eades G, Liu Z, Zhang Y and
Zhou Q: Downregulation of miR-140 promotes cancer stem cell
formation in basal-like early stage breast cancer. Oncogene.
33:2589–2600. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hwang S, Park SK, Lee HY, Kim SW, Lee JS,
Choi EK, You D, Kim CS and Suh N: miR-140-5p suppresses
BMP2-mediated osteogenesis in undifferentiated human mesenchymal
stem cells. FEBS Lett. 588:2957–2963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song B, Wang Y, Xi Y, Kudo K, Bruheim S,
Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al:
Mechanism of chemoresistance mediated by miR-140 in human
osteosarcoma and colon cancer cells. Oncogene. 28:4065–4074. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kong XM, Zhang GH, Huo YK, Zhao XH, Cao
DW, Guo SF, Li AM and Zhang XR: MicroRNA-140-3p inhibits
proliferation, migration and invasion of lung cancer cells by
targeting ATP6AP2. Int J Clin Exp Pathol. 8:12845–12852.
2015.PubMed/NCBI
|
|
18
|
Yuan Y, Shen Y, Xue L and Fan H: miR-140
suppresses tumor growth and metastasis of non-small cell lung
cancer by targeting insulin-like growth factor 1 receptor. PLoS
One. 8:e736042013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou S, Wang J and Zhang Z: An emerging
understanding of long noncoding RNAs in kidney cancer. J Cancer Res
Clin Oncol. 140:1989–1995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing miRNA-lncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu Z, Liu X, Liu L, Deng H, Zhang J, Xu Q,
Cen B and Ji A: Regulation of lncRNA expression. Cell Mol Biol
Lett. 19:561–575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng
R, Wang Y, Huang J, Xu M, Yan J, et al: lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting TGFBI. FEBS J.
281:3766–3775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou X, Ye F, Yin C, Zhuang Y, Yue G and
Zhang G: The interaction between miR-141 and lncRNA-H19 in
regulating cell proliferation and migration in gastric cancer. Cell
Physiol Biochem. 36:1440–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
96:(Suppl). R40–R44. 2007.PubMed/NCBI
|
|
29
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Green D, Dalmay T and Fraser WD: Role of
miR-140 in embryonic bone development and cancer. Clin Sci.
129:863–873. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J,
Chen L, Xi Z, Teng H, Wang Z, et al: Knockdown of long non-coding
RNA XIST exerts tumor-suppressive functions in human glioblastoma
stem cells by up-regulating miR-152. Cancer Lett. 359:75–86. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang H, Fang F, Chang R and Yang L:
MicroRNA-140-5p suppresses tumor growth and metastasis by targeting
transforming growth factor β receptor 1 and fibroblast growth
factor 9 in hepatocellular carcinoma. Hepatology. 58:205–217. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Profumo V, Doldi V, Gandellini P and
Zaffaroni N: Targeting microRNAs to withstand cancer metastasis.
Methods Mol Biol. 1218:415–437. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen J, Sun D, Chu H, Gong Z, Zhang C,
Gong B, Li Y, Li N and Jiang L: Screening of differential microRNA
expression in gastric signet ring cell carcinoma and gastric
adenocarcinoma and target gene prediction. Oncol Rep. 33:2963–2971.
2015.PubMed/NCBI
|
|
35
|
Chen J, Xie F, Zhang L and Jiang WG: iASPP
is over-expressed in human non-small cell lung cancer and regulates
the proliferation of lung cancer cells through a p53 associated
pathway. BMC Cancer. 10:6942010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gillotin S: iASPP, a potential drug target
in cancer therapy. Leuk Res. 33:1175–1177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu T, Li L, Yang W, Jia H, Xu M, Bi J, Li
Z, Liu X, Li Z, Jing H, et al: iASPP is important for bladder
cancer cell proliferation. Oncol Res. 19:125–130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jia Y, Peng L, Rao Q, Xing H, Huai L, Yu
P, Chen Y, Wang C, Wang M, Mi Y, et al: Oncogene iASPP enhances
self-renewal of hematopoietic stem cells and facilitates their
resistance to chemotherapy and irradiation. FASEB J. 28:2816–2827.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Luan S, Sun L and Huang F: MicroRNA-34a: A
novel tumor suppressor in p53-mutant glioma cell line U251. Arch
Med Res. 41:67–74. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li S, Liang Z, Xu L and Zou F:
MicroRNA-21: A ubiquitously expressed pro-survival factor in cancer
and other diseases. Mol Cell Biochem. 360:147–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bai AH, Milde T, Remke M, Rolli CG,
Hielscher T, Cho YJ, Kool M, Northcott PA, Jugold M, Bazhin AV, et
al: MicroRNA-182 promotes leptomeningeal spread of non-sonic
hedgehog-medulloblastoma. Acta Neuropathol. 123:529–538. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Ahmad A and Sarkar FH: ASPP and
iASPP: Implication in cancer development and progression. Cell Mol
Biol. 61:2–8. 2015.PubMed/NCBI
|