Introduction
Glioma, the most common type of malignant tumour in
the brain, is an extremely aggressive and lethal type of brain
tumour that originates from glial cells. In China, glioma has an
annual incidence of 5.26 per 100,000 individuals (1). Gliomas are divided into two groups
according to the 2007 World Health Organization (WHO)
classification: low-grade (grades I and II) and high-grade (grades
III and IV) (2). Despite the
advances in radiotherapy and chemotherapy treatments following
surgical resection, the prognosis of glioma patients still remains
poor (3,4). The median survival of the glioma
patients who received comprehensive therapy is <15 months
(5). Poor glioma prognosis is
partially due to the oncogenic nature and rapid growth of glioma
cell and local invasion of tumour into normal brain tissues
(6,7). Therefore, improving understanding
regarding the underlying mechanisms involved in the formation and
progression of glioma is essential to the identification of more
effective therapeutic strategies for this disease.
MicroRNA (miRNA) is a group of endogenous,
single-stranded and non-protein-coding small RNAs (20–23
nucleotides) without open reading frames (8). miRNAs negatively regulate the
expression of their target genes by undergoing base pairing with
the 3′-untranslated regions (3′-UTRs) of their target messenger
RNAs (mRNAs). The base pairing inhibits mRNA translation or promote
mRNA degradation (9). Notably, a
single miRNA can modulate several targets simultaneously, whereas a
single gene may be regulated by multiple miRNAs (10). Thus, miRNAs are involved in complex
regulatory networks that regulate a multitude of biological
processes, such as cell proliferation, apoptosis, differentiation,
autophagy, angiogenesis, invasion, migration and stem cell renewal
(11–13). Moreover, miRNAs are abnormally
expressed in various cancers and closely related with cancer
initiation and progression (14–16).
They also act as tumour suppressors or promoters by directly
targeting known oncogenes or tumour suppressor genes (17–19).
Therefore, identifying specific miRNAs that play important roles in
tumourigenesis and tumour development might provide therapeutic
biomarkers for cancer diagnosis, prognosis and therapy.
MicroRNA-342 (miR-342) plays key roles in the
development, progression and metastasis of several human cancers
(20–23). However, the precise roles of miR-342
in glioma remain unknown. Therefore, the aim of our study was to
investigate the expression pattern and functions of miR-342 in
glioma. Additionally, we examined the molecular mechanisms involved
in the association of miR-342 with the proliferation, invasion and
apoptosis of glioma cells.
Materials and methods
Clinical samples
Glioma tissues (49 paired) and corresponding normal
adjacent tissues (NATs) were collected from glioma patients
undergoing surgical resection at the Department of Neurosurgery,
The First Affiliated Hospital of Wenzhou Medical University
(Wenzhou, China) from August 2014 to January 2016. None of these
glioma patients received prior radiotherapy or chemotherapy.
Tissues were frozen in liquid nitrogen immediately after collection
and stored at −80°C until further use. This study was approved by
the Ethics Committee of The First Affiliated Hospital of Wenzhou
Medical University. Written informed consent was also obtained from
each patient.
Cell lines, oligonucleotides and cell
transfection
Glioma cell lines U251, U87, A172, and LN229 were
purchased from American Type Culture Collection (Manassas, VA,
USA), and routinely cultured in Dulbecco's modified Eagle's medium
(DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 100 U/ml
penicillin and 100 mg/ml streptomycin (Invitrogen) and 10% fetal
bovine serum (FBS; Invitrogen). Normal human astrocytes (NHAs) were
obtained from ScienCell Research Laboratories (Carlsbad, CA, USA)
and grown in astrocyte medium (ScienCell Research Laboratories).
All cells were maintained at 37°C in a humidified atmosphere
containing 5% CO2.
miR-342 mimics, corresponding miRNA negative control
(miR-NC), PAK4-targeted small interfering RNA (PAK4 siRNA) and
siRNA negative control (NC siRNA) were chemically synthesized by
GenePharma Co., Ltd. (Shanghai, China). PAK4 overexpressed plasmid
(pcDNA3.1-PAK4) and blank plasmid (pcDNA3.1) were purchased from
Chinese Academy of Sciences (Changchun, China). For in vitro
function assays, cells were seeded in 6-well plates at 50–70%
confluence. Transfection and co-transfection was performed through
the use of Opti-MEM and Lipofectamine 2000 (Invitrogen) according
to the manufacturer's protocol. After 6-h transfection, the culture
medium was removed and placed in DMEM containing 10% FBS.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues or cells was extracted using
TRIzol reagent (Invitrogen) according to the manufacturer's
protocol. The quality and concentration of total RNA was examined
using a NanoDrop® ND-1000 spectrophoto-meter.
TaqMan® microRNA assays (Applied Biosystems, Foster
City, CA, USA) was used to detect miR-342 expression, with RUN6B as
an internal control. To quantify PAK4 mRNA, M-MLV Reverse
Transcription system (Promega Corp., Madison, WI, USA) was used to
synthesis single-stranded cDNA and qPCR was conducted using SYBR
Premix Ex Taq (Takara, Dalian, China), with β-actin as an internal
control. The primers used in this study were as follows: miR-342
forward, 5′-GTGCTATCTGTGATTGAGGGA'-3 and reverse,
5′-CGGGTGCGATTTCTGTG'-3; RUN6B forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′. PAK4 forward,
5′-TCCCCCTGAGCCATTGTG-3′ and reverse, 5′-TGACCTGTCTCCCCATCCA-3′;
β-actin forward, 5′-ATGGGTCAGAAGGATTCCTATGTG-3′, and reverse,
5′-CTTCATGAGGTAGTCAGTCAGGTC-3′ Relative expression was determined
using the 2−∆∆Ct method (24).
3-(4, 5-dimethylthiazol-2-yl)-2,
5-diphenyltetra-zolium bromide (MTT) assays
Briefly, cells were harvested and seeded in 96-well
plates at a density of 3×103 cells/well. The following
day, miR-342 mimics, miR-NC, PAK4 siRNA, NC siRNA, pcDNA3.1-PAK4 or
pcDNA3.1 was transfected into cells using Opti-MEM and
Lipofectamine 2000. The culture medium was replaced by DMEM
containing 10% FBS at 6 h post-transfection and then incubated at
37°C in a humidified atmosphere containing 5% CO2 for 0,
24, 48 and 72 h. MTT solution (10 µl) (5 mg/ml; Sigma-Aldrich, St.
Louis, MO, USA) was added into each well and the plates were
incubated at 37°C for another 4 h. Subsequently, the culture medium
was removed carefully and 150 µl dimethyl sulfoxide (Sigma-Aldrich)
was added into each well to solubilize the MTT formazan. The
absorbance was measured at a wavelength of 490 nm with a microplate
spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). All
experiments were performed in triplicate and repeated three
times.
Cell invasion assay
Transfected cells were harvested at 48 h
post-transfection and suspended in FBS-free DMEM. Cells
(5×104) were placed on the top chambers of 24-well
Transwell plates (Corning Inc., Corning, NY, USA) coated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). DMEM
supplemented with 20% FBS was used as a chemoattractant in the
lower chambers. After 48 h of incubation at 37°C with 5%
CO2, the non-invading cells were removed using a cotton
swab. The invasive cells were fixed with 100% methanol for 10 min,
stained with 0.1% crystal violet for 20 min and photographed at
×200 magnification. The number of invasive cells in five random
fields was counted under an inverted microscope (CKX41; Olympus
Corp., Tokyo, Japan). All experiments were performed in triplicate
and repeated three times.
Flow cytometry analysis
Cell apoptosis was assessed 48 h after transfection.
Transfected cells were harvested with trypsinization, washed in
ice-cold PBS and fixed in 80% ice-cold ethanol in PBS.
Subsequently, cells were re-suspended in 1X binding buffer to a
concentration of 1×104 cells. Annexin V-FITC apoptosis
detection kit (Invitrogen Corp.) was utilized to examine cell
apoptosis according to the manufacturer's protocol. Briefly, cells
were stained with FITC-Annexin V and propidium iodide (PI). After
incubation at room temperature in the dark for 15 min, cell
apoptosis was quantified using flow cytometry within 1 h of
staining.
Bioinformatic analysis and luciferase
reporter assay
TargetScan (http://www.Targetscan.org/) and PicTar (http://pictar.mdcberlin.de/) were utilized to
predicate the potential target genes of miR-342. The wild-type
3′-UTR segment of the PAK4 mRNA containing miR-342 binding sites
was amplified and inserted into pMIR Reporter (Ambion, Austin, TX,
USA) and named pMIR-PAK4-3′-UTR-Wt. To mutate the binding site of
miR-342, its complementary sequence in the 3′UTR of PAK4 (GUGUGAU)
was replaced by CACACUC, inserted into pMIR Reporter and named as
pMIR-PAK4-3′-UTR-Mut. For the luciferase reporter assay, miR-342
mimics or miR-NC was transfected into cells, along with
pMIR-PAK4-3′-UTR-Wt or pMIR-PAK4-3′-UTR-Mut, using Lipofectamine
2000 according to the manufacturer's protocol. Luciferase activity
was determined at 48-h post-transfection using a Dual-Luciferase
Reporter assay system (Promega), according to the manufacturer's
instructions. Renilla luciferase activity was used for
normalization.
Western blot analysis
Total proteins were extracted from tissues and cells
using the RIPA lysis buffer with protease inhibitors (Roche
Diagnostics, Basel, Switzerland) and phosphatase inhibitors (Merck
KGaA, Darmstadt, Germany). The concentration of total proteins was
quantified using a BCA Protein assay kit (Pierce Biotechnology,
Rockford, IL, USA). The same amount of protein was separated by 10%
SDS-PAGE gel electrophoresis, transferred to polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA) and
blocked with 5% fat-free milk. The membranes were then incubated
overnight at 4°C with primary antibodies: mouse anti-human
monoclonal PAK4 (sc-390507; 1:1,000 dilution; Santa Cruz
Biotechnology, CA, USA), mouse anti-human monoclonal p-AKT
(sc-271966; 1:1,000 dilution; Santa Cruz Biotechnology), mouse
anti-human monoclonal AKT (sc-81434; 1:1,000 dilution; Santa Cruz
Biotechnology), mouse anti-human monoclonal p-ERK (sc-81492;
1:1,000 dilution; Santa Cruz Biotechnology), mouse anti-human
monoclonal ERK (sc-514302; 1:1,000 dilution; Santa Cruz
Biotechnology), and mouse anti-human monoclonal GAPDH antibody
(sc-47724; 1:1,000 dilution; Santa Cruz Biotechnology). After
washing three times with Tris-buffered saline with 0.5% Tween-20
(TBST; Beyotime Institute of Biotechnology, Haimen, China), the
membranes were probed with goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (sc-2005; 1:5,000
dilution; Santa Cruz Biotechnology) at room temperature for 1 h.
The protein blots were visualized using the ECL Protein Detection
kit (Pierce Biotechnology). GAPDH was used as a loading
control.
Statistical analysis
The data in this study are presented as mean ± SD.
Data were compared using Student's t-test or one-way ANOVA with
SPSS 16.0 software (SPSS, Chicago, IL, USA). The association
between miR-342 and clinicopathologic features of glioma was
evaluated using Pearson's χ2 test. Differences were
considered significant at P<0.05.
Results
miR-342 is downregulated in glioma
tissues and cell lines
To investigate the role of miR-342 in glioma, we
measured miR-342 expression levels in 49 paired glioma tissues and
NATs. The RT-qPCR results showed that miR-342 expression was
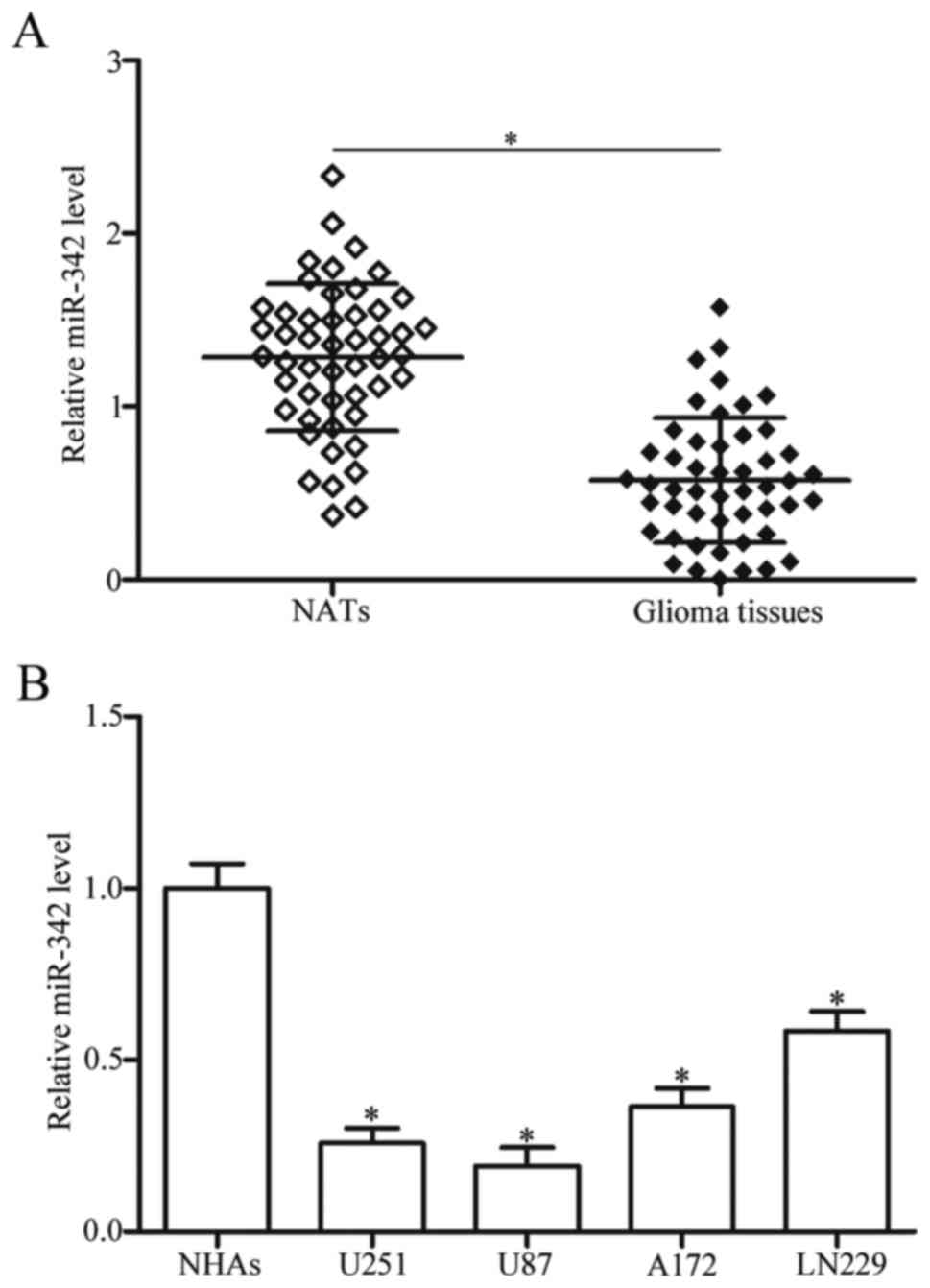
significantly lower in glioma tissues than in NATs (Fig. 1A, P<0.05). We further evaluated
the association between miR-342 expression and clinicopathological
factors of glioma patients. We found that the expression levels of
miR-342 were strongly correlated with advanced WHO grades (P=0.001)
and low KPS scores (P=0.035). No significant association was
observed between miR-342 and each of the following parameters: sex,
age and tumour size (all P>0.05; Table I). In addition, the expression
levels of miR-342 in four glioma cell lines (U251, U87, A172 and
LN229) and normal human astrocytes (NHAs) were examined. As shown
in Fig. 1B, expression level of
miR-342 decreased in the glioma cell lines in comparison with that
in NHAs (P<0.05). These results suggested that low miR-342
expression is correlated with advanced malignancy of gliomas, and
miR-342 may serve as a tumour suppressor in glioma occurrence and
progression.
 | Table I.Association between microRNA-342
expression and clinicopathological factors of glioma patients. |
Table I.
Association between microRNA-342
expression and clinicopathological factors of glioma patients.
|
|
| microRNA-342
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
factors | Cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.409 |
|
Male | 32 | 19 | 13 |
|
|
Female | 17 | 8 | 9 |
|
| Age, years |
|
|
| 0.740 |
|
<55 | 21 | 11 | 10 |
|
|
≥55 | 28 | 16 | 12 |
|
| Tumour size,
cm |
|
|
| 0.647 |
|
<5 | 34 | 18 | 16 |
|
| ≥5 | 15 | 9 | 6 |
|
| KPS score |
|
|
| 0.035 |
|
<80 | 26 | 18 | 8 |
|
|
≥80 | 23 | 9 | 14 |
|
| WHO grade |
|
|
| 0.001 |
|
I–II | 23 | 7 | 16 |
|
|
III–IV | 26 | 20 | 6 |
|
miR-342 overexpression inhibits cell
proliferation and invasion and activates cell apoptosis in
glioma
To investigate miR-342 functions in glioma, we
transfected miR-342 mimics into the U251 and U87 cells, which have
the lowest miR-342 expression level, to increase its endogenous
expression level (Fig. 2A and B,
P<0.05). MTT assay was then used to determine the effect of
miR-342 overexpression on the cell proliferation of glioma. As
shown in Fig. 2C and D, the
proliferation of the U251 and U87 cells was suppressed in the group
transfected with the miR-342 mimics compared with that in the
miR-NC group (P<0.05). We then utilized cell invasion assay to
examine the effect of miR-342 on glioma cell invasion. The results
indicated that the U251 and U87 cells with high miR-342 level
showed lower invasion capacities than the cells with miR-NC
(Fig. 2E, P<0.05). Next, flow
cytometry analysis was performed to explore the role of miR-342 in
glioma cell apoptosis. As shown in Fig.
2F, the upregulation of miR-342 improved apoptosis in the U251
and U87 cells considerably (P<0.05). These results highlighted
that miR-342 plays tumour-suppressive roles in glioma by inhibiting
cell proliferation and invasion and inducing cell apoptosis.
Overall, these results indicated that miR-342 affects the growth,
metastasis and apoptosis of glioma cells.
miR-342 directly targets and inhibits
PAK4 expression in glioma
After observing the tumour-suppressing roles of
miR-342 in glioma, we explored its potential targets using
bioinformatics analysis. The results indicated a large number of
candidate targets of miR-342. Of these targets, P21 activated
kinases 4 (PAK4) was selected for further target identification
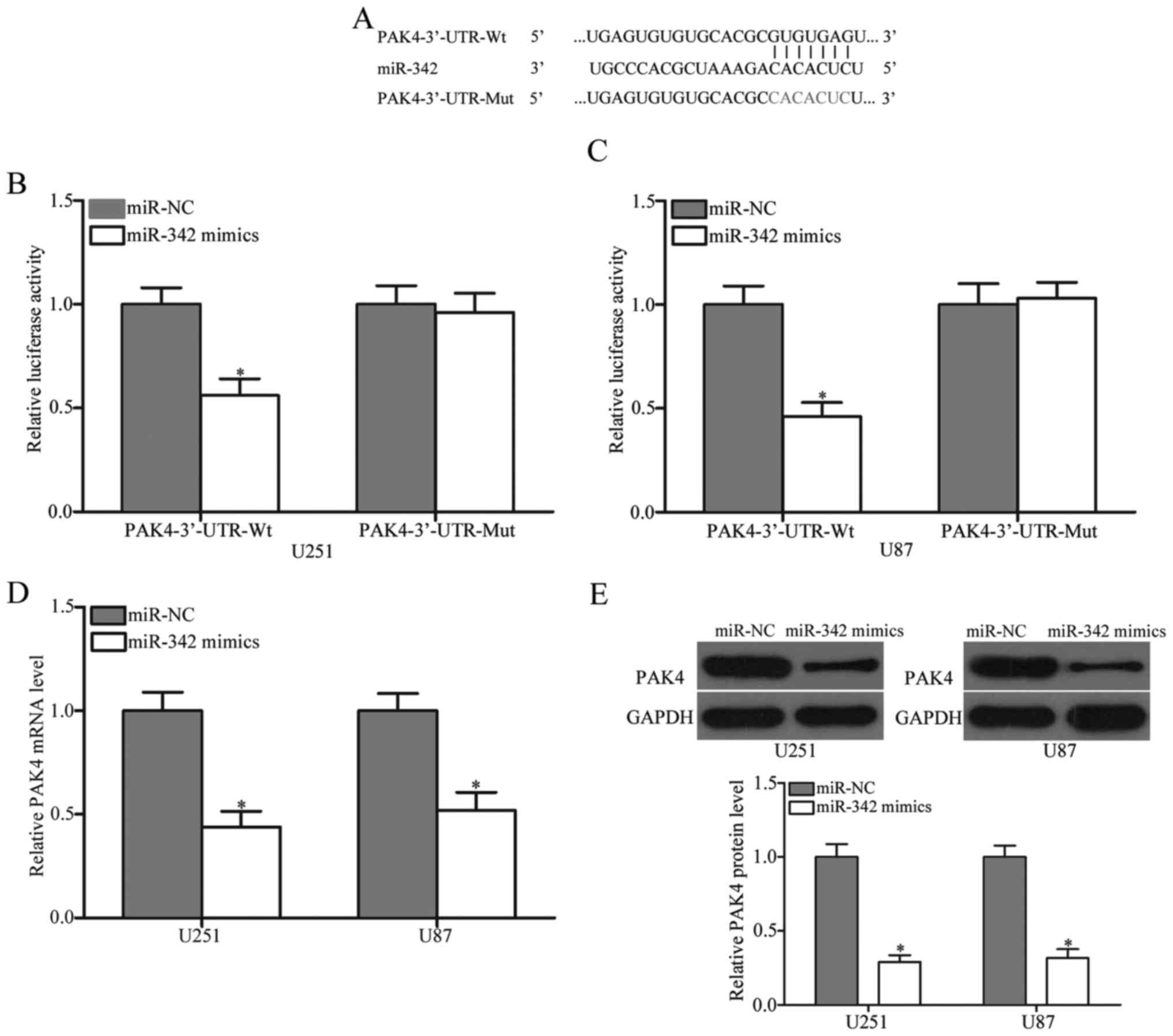
(Fig. 3A), because it was
upregulated in glioma tissues and involved in glioma formation and
progression (25). To test this
hypothesis, we conducted luciferase reporter assay to validate the
interaction between miR-342 and the predicted binding site in the
3′-UTR of the PAK4 gene. As illustrated in Fig. 3B and C, the restoration of miR-342
expression inhibited the luciferase activity of pMIR-PAK4-3′-UTR-Wt
(P<0.05), but not of pMIR-PAK4-3′-UTR-Mut, suggesting that
miR-342 specifically targeted the 3′-UTR of PAK4. RT-qPCR and
western blot analysis were then performed to detect PAK4 mRNA and
protein expression in the U251 and U87 cells transfected with the
miR-342 mimics or miR-NC. Our findings revealed that, at the mRNA
and protein levels, the PAK4 expression levels decreased in the
U251 and U87 cells transfected with the miR-342 mimics compared
with that in the miR-NC group (Fig. 3D
and E, P<0.05). These data demonstrated that miR-342 is a
post-transcriptional regulator of PAK4 and binds directly to the
3′UTR of the PAK4 gene.
PAK4 expression is upregulated and
inversely correlates with miR-342 expression in glioma tissues
To confirm the association between miR-342 and PAK4,
we calculated PAK4 mRNA and protein expression levels in glioma
tissues and NATs. The data obtained from RT-qPCR and western blot
analysis showed that PAK4 mRNA and protein were significantly
upregulated in glioma tissues relative to those with NATs (Fig. 4A and B, P<0.05). Additionally,
Spearman's correlation analysis results confirmed that the negative
association between PAK4 mRNA expression and miR-342 in glioma
tissues (Fig. 4C; r=−0.5261,
P=0.0001).
PAK4 downregulation imitates the roles
of miR-342 in glioma
PAK4 was identified as a direct target of miR-342 in
glioma. We hypothesized that the roles of PAK4 knockdown on glioma
cells are similar with those induced by miR-342 overexpression. To
confirm this hypothesis, we transfected the U251 and U87 cells with
PAK4 siRNA or NC siRNA. As expected, the PAK4 expression decreased
in the U251 and U87 cells after transfection with PAK4 siRNA
(Fig. 5A, P<0.05). The results
of the subsequent functional assays showed that downregulation of
PAK4 repressed the proliferation (Fig.
5B and C, P<0.05) and invasion (Fig. 5D, P<0.05) and induced apoptosis
(Fig. 5E, P<0.05) in the U251
and U87 cells. This result supported the hypothesis that PAK4
underexpression plays the same role as that of miR-342 mimics in
glioma cells. These results further demonstrated that PAK4 may be a
functional target of miR-342 in glioma.
PAK4 reverses the tumour-suppressing
effects of miR-342 on glioma cells
Rescue experiments were performed to evaluate the
contribution of PAK4 to the roles of miR-342 in glioma. After
introducing pcDNA3.1-PAK4 or pcDNA3.1 into the U251 and U87 cells,
we performed western blot analysis 72 h post-transfection. The
results showed that PAK4 was upregulated in the
pcDNA3.1-PAK4-transfected U251 and U87 cells. In addition, PAK4
expression was recovered in the miR-342 mimic-transfected cells
after being transfected with pcDNA3.1-PAK4 (Fig. 6A, P<0.05). The results of the
functional rescue experiments showed that reintroduction of PAK4
effectively rescued the effects of miR-342 overexpression on the
proliferation (Fig. 6B and C,
P<0.05), invasion (Fig. 6D,
P<0.05) and apoptosis (Fig. 6E,
P<0.05) of the U251 and U87 cells. These data showed clearly
that miR-342 exerted tumour-suppressive roles in glioma, at least
in part, by suppressing PAK4.
miR-342 is associated with the AKT and
ERK pathway in glioma
Previous studies reported that PAK4 plays essential
roles in the AKT and ERK pathways (26,27).
Thus, to assess the function of miR-342 on these pathways, we
transfected miR-342 mimics or miR-NC into the U251 and U87 cells.
As shown in Fig. 7, miR-342
overexpression reduced both the p-AKT and p-ERK expression levels
in the U251 and U87 cells (P<0.05). However, it did not affect
the entire AKT and ERK expression. These results illustrated that
miR-342 can inactivate the AKT and ERK pathways in glioma.
Discussion
Glioma has high mortality, high recurrence rate, and
low cure rate, because of its rapid growth and metastasis behaviour
(6,7). miRNA dysregulation has been implicated
in the occurrence and progression of many human cancers, such as
bladder cancer (28), gastric
cancer (29), hepatocellular
carcinoma (30), colorectal cancer
(31) and glioma (32). Thus, some specific miRNAs are
potential therapeutic targets for cancer diagnosis, therapy and
prognosis (33,34). The present study indicated that
miR-342 expression was low in both glioma tissues and cell lines.
Additionally, reduced expression of miR-342 was significantly
correlated with advanced WHO grades and low KPS scores of glioma
patients. According to these results, we supposed that miR-342 may
act as a tumour suppressor in glioma. To confirm this hypothesis,
MTT assay, cell invasion assay and flow cytometry analysis were
performed to examine the effects of miR-342 on cell proliferation
and invasion and apoptosis of glioma. We found that the restoration
of miR-342 expression suppresses the proliferation and invasion and
enhances the apoptotic abilities of glioma cells. Moreover, PAK4
was identified as a direct and functional target of miR-342 in
glioma. The upregulation of miR-342 expression inactivated the AKT
and ERK pathways in glioma.
miR-342 was previously found to be aberrantly
expressed and plays important roles in various human cancers. For
example, miR-342 was observed to be downregulated in ERα-positive
breast cancer tissues and cell lines and significantly correlated
with HER2 and VEGF expression status (20,21).
miR-342 was found to be decreased in tamoxifen-resistant tumour
cells. Enforced expression of miR-342 improved the chemosensitivity
of breast cancer cells to tamoxifen (22). Wang et al reported that
miR-342 overexpression inhibits colorectal cancer cell growth and
metastasis both in vitro and in vivo (23). Zhao and Zhang (35) then revealed that miR-342
upregulation suppresses cell proliferation in hepatocellular
carcinoma by regulating the NF-κB pathway. A study by Xie et
al (36) found that resumption
of miR-342 expression can reduce cell proliferation and invasion
in vitro and cell growth in vivo in non-small cell
lung cancer. Li et al (37)
demonstrated that miR-342 re-expression attenuated cervical cancer
cell proliferation, migration and invasion. These findings
indicated that miR-342 has fundamental roles in carcinogenesis and
progression of malignant tumours and illustrated potential of
miR-342 as a therapeutic target for various cancers.
Exploring the downstream targets of miR-342 can
improve our understanding regarding the underlying molecular
mechanisms involved in the tumour-suppressing roles of miR-342 on
glioma cells. Notably, several tumour suppressors have been
identified as direct targets of miR-342 for example, DNMT1
(23), FOXM1 (38) and FOXQ1 (38) in colorectal cancer; IKK-γ (35), TAB2 (35) and TAB3 (35) in hepatocellular carcinoma; RAP2B
(36) in non-small cell lung
cancer; and FOXM1 (37) in cervical
cancer. Through online bioinformatics analysis, PAK4 was predicted
to contain a miR-342 seed match at position 488–494 of the
PAK4-3′UTR. To test whether miR-342 directly targets 3′UTR of PAK4,
we performed luciferase report assays. By measuring changes in the
luciferase activity, we demonstrated that miR-342 directly targets
3′UTR of PAK4. Additionally, ectopic expression of miR-342 reduced
the PAK4 expression level in glioma cells at the mRNA and protein
levels. The expression level of PAK4 was upregulated and negatively
correlated with miR-342 expression in glioma tissues. PAK4
knockdown have roles similar to those of miR-342 overexpression in
glioma cells, and the upregulation of PAK4 reverses the effects of
miR-342 in glioma cells. Moreover, miR-342 can inactivate the AKT
and ERK pathways in glioma cells. These results suggested that
miR-342 served as a tumour suppressor in glioma, at least in part,
by directly targeting PAK4 and indirectly regulating the AKT and
ERK pathways.
PAKs belong to a family of serine or threonine
kinases, which are best characterized as downstream effectors of
Rac and Cdc42 (39). This family
comprise 6 mammalian isoforms, and can be divided into two groups:
group A, which includes PAKs 1, 2 and 3 and group B, which contains
PAKs 4, 5 and 6 (40,41). PAK4, located at 19q13.2, was
reported to be upregulated in multiple human cancers, such as
pancreatic cancer (42), colorectal
cancer (43), gastric cancer
(44) and renal cell carcinoma
(45). Increasing evidence
indicated that PAK4 plays a significant role in a variety of
cellular functions, such as cell proliferation, migration,
invasion, apoptosis, actin cytoskeletal changes and cytoskeletal
organisation (46). Furthermore,
PAK4 was identified to serve important functions in tumourigenesis
and tumour development (47–49).
In glioma, PAK4 is overexpressed and significantly correlated with
pathological grades. PAK4 downregulation repressed glioma cell
proliferation, motility and adhesion (25). These data indicate a central role of
PAK4 in glioma pathogenesis. The findings of the present study
identified PAK4 as a direct gene target of miR-342 and suggested
that the miR-342/PAK4 pathway is a promising therapeutic target for
the treatment of gliomas.
In conclusion, this study showed a significantly low
expression level of miR-342 in glioma tissues and cell lines. Our
results indicated that miR-342 suppresses tumour in glioma by
directly targeting PAK4 and indirectly regulating the AKT and ERK
pathways. Thus, miR-342 may be a candidate diagnostic marker of
glioma and a potential therapeutic target for the treatment of
patients with this disease.
Acknowledgements
This study was supported by the Zhejiang Provincial
Natural Science Foundation (no. LY16H160051); Medical Scientific
Research Foundation of Zhejiang Province, China (2012RCA042);
Zhejiang Provincial Surgical ‘Top Key’ Discipline (no.
kfjj2011005).
References
|
1
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vigneswaran K, Neill S and Hadjipanayis
CG: Beyond the World Health Organization grading of infiltrating
gliomas: Advances in the molecular genetics of glioma
classification. Ann Transl Med. 3:952015.PubMed/NCBI
|
|
3
|
Lou M and Zhao Y: Satisfactory therapy
results of combining nimustine with nicardipine against glioma at
advanced stage. J Cancer Res Ther. 11:10302015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li BO, Meng C, Zhang X, Cong D, Gao X, Gao
W, Ju D and Hu S: Effect of photodynamic therapy combined with
torasemide on the expression of matrix metalloproteinase 2 and
sodium-potassium-chloride cotransporter 1 in rat peritumoral edema
and glioma. Oncol Lett. 11:2084–2090. 2016.PubMed/NCBI
|
|
5
|
Paw I, Carpenter RC, Watabe K, Debinski W
and Lo HW: Mechanisms regulating glioma invasion. Cancer Lett.
362:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Giese A, Bjerkvig R, Berens ME and
Westphal M: Cost of migration: Invasion of malignant gliomas and
implications for treatment. J Clin Oncol. 21:1624–1636. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laks DR, Visnyei K and Kornblum HI: Brain
tumor stem cells as therapeutic targets in models of glioma. Yonsei
Med J. 51:633–640. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Orang Valinezhad A, Safaralizadeh R and
Kazemzadeh-Bavili M: Mechanisms of miRNA-mediated gene regulation
from common downregulation to mRNA-specific upregulation. Int J
Genomics. 2014:9706072014.PubMed/NCBI
|
|
10
|
Luo W and Sehgal A: Regulation of
circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell.
148:765–779. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rachagani S, Macha MA, Menning MS, Dey P,
Pai P, Smith LM, Mo YY and Batra SK: Changes in microRNA (miRNA)
expression during pancreatic cancer development and progression in
a genetically engineered KrasG12D;Pdx1-Cre mouse (KC) model.
Oncotarget. 6:40295–40309. 2015.PubMed/NCBI
|
|
12
|
Bagnoli M, Canevari S, Califano D, Losito
S, Maio MD, Raspagliesi F, Carcangiu ML, Toffoli G, Cecchin E,
Sorio R, et al: Multicentre Italian Trials in Ovarian cancer (MITO)
translational group: Development and validation of a microRNA-based
signature (MiROvaR) to predict early relapse or progression of
epithelial ovarian cancer: A cohort study. Lancet Oncol.
17:1137–1146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang L, Wang YL, Liu S, Zhang PP, Chen Z,
Liu M and Tang H: miR-181b promotes cell proliferation and reduces
apoptosis by repressing the expression of adenylyl cyclase 9 (AC9)
in cervical cancer cells. FEBS Lett. 588:124–130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Gao DY and Huang L: In vivo
delivery of miRNAs for cancer therapy: Challenges and strategies.
Adv Drug Deliv Rev. 81:128–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao Y, Pang D, Wang C, Zhong S and Wang
S: MicroRNA-134 modulates glioma cell U251 proliferation and
invasion by targeting KRAS and suppressing the ERK pathway. Tumour
Biol. 37:11485–11493. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hong L, Ya-Wei L, Hai W, Qiang Z, Jun-Jie
L, Huang A, Song-Tao Q and Yun-Tao L: MiR-519a functions as a tumor
suppressor in glioma by targeting the oncogenic STAT3 pathway. J
Neurooncol. 128:35–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui R, Guan Y, Sun C, Chen L, Bao Y, Li G,
Qiu B, Meng X, Pang C and Wang Y: A tumor-suppressive microRNA,
miR-504, inhibits cell proliferation and promotes apoptosis by
targeting FOXP1 in human glioma. Cancer Lett. 374:1–11. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Ding H, Han Y, Sun D, Wang H and
Zhai XU: The significance of microRNA-184 on JAK2/STAT3 signaling
pathway in the formation mechanism of glioblastoma. Oncol Lett.
10:3510–3514. 2015.PubMed/NCBI
|
|
19
|
Li Y, Min D, Wang K, Yin S, Zheng H and
Liu L: MicroRNA-152 inhibits cell proliferation, migration and
invasion by directly targeting MAFB in nasopharyngeal carcinoma.
Mol Med Rep. 15:948–956. 2017.PubMed/NCBI
|
|
20
|
He YJ, Wu JZ, Ji MH, Ma T, Qiao EQ, Ma R
and Tang JH: miR-342 is associated with estrogen receptor-α
expression and response to tamoxifen in breast cancer. Exp Ther
Med. 5:813–818. 2013.PubMed/NCBI
|
|
21
|
Savad S, Mehdipour P, Miryounesi M,
Shirkoohi R, Fereidooni F, Mansouri F and Modarressi MH: Expression
analysis of MiR-21, MiR-205, and MiR-342 in breast cancer in Iran.
Asian Pac J Cancer Prev. 13:873–877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cittelly DM, Das PM, Spoelstra NS,
Edgerton SM, Richer JK, Thor AD and Jones FE: Downregulation of
miR-342 is associated with tamoxifen resistant breast tumors. Mol
Cancer. 9:3172010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu
R, Pan Z, Kang T and Huang W: MicroRNA-342 inhibits colorectal
cancer cell proliferation and invasion by directly targeting DNA
methyltransferase 1. Carcinogenesis. 32:1033–1042. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kesanakurti D, Chetty C, Maddirela
Rajasekhar D, Gujrati M and Rao JS: Functional cooperativity by
direct interaction between PAK4 and MMP-2 in the regulation of
anoikis resistance, migration and invasion in glioma. Cell Death
Dis. 3:e4452012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tyagi N, Bhardwaj A, Singh AP, McClellan
S, Carter JE and Singh S: p-21 activated kinase 4 promotes
proliferation and survival of pancreatic cancer cells through AKT-
and ERK-dependent activation of NF-κB pathway. Oncotarget.
5:8778–8789. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu X, Feng J, Zeng D, Ding Y, Yu C and
Yang B: PAK4 confers cisplatin resistance in gastric cancer cells
via PI3K/Akt- and MEK/ERK-dependent pathways. Biosci Rep.
34:342014. View Article : Google Scholar
|
|
28
|
Yu G, Jia Z and Dou Z: miR-24-3p regulates
bladder cancer cell proliferation, migration, invasion and
autophagy by targeting DEDD. Oncol Rep. 37:1123–1131.
2017.PubMed/NCBI
|
|
29
|
Lee SW, Park KC, Kim JG, Moon SJ, Kang SB,
Lee DS, Sul HJ, Ji JS and Jeong HY: Dysregulation of
MicroRNA-196b-5p and MicroRNA-375 in Gastric Cancer. J Gastric
Cancer. 16:221–229. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie
RT, Yang HQ, Chang ZY, Sun R, Chai L, et al: High expression of
miR-105-1 positively correlates with clinical prognosis of
hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget.
8:11896–11905. 2017.PubMed/NCBI
|
|
31
|
Li Z, Li B, Niu L and Ge L: miR-592
functions as a tumor suppressor in human non-small cell lung cancer
by targeting SOX9. Oncol Rep. 37:297–304. 2017.PubMed/NCBI
|
|
32
|
Wei Y, Sun J and Li X: MicroRNA-215
enhances invasion and migration by targeting retinoblastoma tumor
suppressor gene 1 in high-grade glioma. Biotechnol Lett.
39:197–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee KT, Tan JK, Lam AK and Gan SY:
MicroRNAs serving as potential biomarkers and therapeutic targets
in nasopharyngeal carcinoma: A critical review. Crit Rev Oncol
Hematol. 103:1–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li G, Shen Q, Li C, Li D, Chen J and He M:
Identification of circulating MicroRNAs as novel potential
biomarkers for hepatocellular carcinoma detection: A systematic
review and meta-analysis. Clin Transl Oncol. 17:684–693. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao L and Zhang Y: miR-342-3p affects
hepatocellular carcinoma cell proliferation via regulating NF-κB
pathway. Biochem Biophys Res Commun. 457:370–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie X, Liu H, Wang M, Ding F, Xiao H, Hu
F, Hu R and Mei J: miR-342-3p targets RAP2B to suppress
proliferation and invasion of non-small cell lung cancer cells.
Tumour Biol. 36:5031–5038. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li XR, Chu HJ, Lv T, Wang L, Kong SF and
Dai SZ: miR-342-3p suppresses proliferation, migration and invasion
by targeting FOXM1 in human cervical cancer. FEBS Lett.
588:3298–3307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Weng W, Okugawa Y, Toden S, Toiyama Y,
Kusunoki M and Goel A: FOXM1 and FOXQ1 are promising prognostic
biomarkers and novel targets of tumor-suppressive miR-342 in human
colorectal cancer. Clin Cancer Res. 22:4947–4957. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bokoch GM: Biology of the p21-activated
kinases. Annu Rev Biochem. 72:743–781. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rane CK and Minden A: P21 activated
kinases: Structure, regulation, and functions. Small GTPases.
5:52014. View Article : Google Scholar
|
|
41
|
Menges CW, Sementino E, Talarchek J, Xu J,
Chernoff J, Peterson JR and Testa JR: Group I p21-activated kinases
(PAKs) promote tumor cell proliferation and survival through the
AKT1 and Raf-MAPK pathways. Mol Cancer Res. 10:1178–1188. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yeo D, He H, Baldwin GS and Nikfarjam M:
The role of p21-activated kinases in pancreatic cancer. Pancreas.
44:363–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Song B, Wang W, Zheng Y, Yang J and Xu Z:
P21-activated kinase 1 and 4 were associated with colorectal cancer
metastasis and infiltration. J Surg Res. 196:130–135. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kobayashi K, Inokuchi M, Takagi Y, Otsuki
S, Fujimori Y, Sato Y, Yanaka Y, Higuchi K, Aburatani T, Tomii C,
et al: Prognostic significance of PAK4 expression in gastric
cancer. J Clin Pathol. 69:580–585. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu W, Yang Y, Liu Y, Liu H, Zhang W, Xu
L, Zhu Y and Xu J: p21-Activated kinase 4 predicts early recurrence
and poor survival in patients with nonmetastatic clear cell renal
cell carcinoma. Urol Oncol. 33:205.e13–21. 2015. View Article : Google Scholar
|
|
46
|
Gnad F, Young A, Zhou W, Lyle K, Ong CC,
Stokes MP, Silva JC, Belvin M, Friedman LS, Koeppen H, et al:
Systems-wide analysis of K-Ras, Cdc42, and PAK4 signaling by
quantitative phosphoproteomics. Mol Cell Proteomics. 12:2070–2080.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dart AE and Wells CM: P21-activated kinase
4 - not just one of the PAK. Eur J Cell Biol. 92:129–138. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Siu MK, Chan HY, Kong DS, Wong ES, Wong
OG, Ngan HY, Tam KF, Zhang H, Li Z, Chan QK, et al: p21-activated
kinase 4 regulates ovarian cancer cell proliferation, migration,
and invasion and contributes to poor prognosis in patients. Proc
Natl Acad Sci USA. 107:18622–18627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ahmed T, Shea K, Masters JR, Jones GE and
Wells CM: A PAK4-LIMK1 pathway drives prostate cancer cell
migration downstream of HGF. Cell Signal. 20:1320–1328. 2008.
View Article : Google Scholar : PubMed/NCBI
|