Introduction
Human genome sequence data have revealed that more
than 90% of the human genome is transcribed mainly as non-coding
RNAs (ncRNAs) and sparsely as protein-encoding RNAs (<2%). Long
non-coding RNAs (lncRNAs) are ncRNAs with transcripts greater than
200 nucleotides. They participate in different biological
processes, including transcription, alternative splicing,
epigenetic regulation, RNA decay, miRNA silencing, modulation of
protein activity, structural and organizational roles of the cell,
and alteration of protein localization (1). Recently, various lncRNAs have been
proven to be closely related with both normal development and
diseases, especially tumorigenesis (2). However, the mechanisms of action of
most lncRNAs remain unclear (1,3).
As a long intergenic non-coding RNA,
metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1)
was identified in non-small cell lung cancer (NSCLC) (4), with a length of about 8,700 nt, and is
located on human chromosome 11q13.1. MALAT-1 has been documented to
be involved in a number of diseases, especially in the
proliferation, invasion and metastasis of multiple types of cancers
(5). Recent studies have reported
its invasive and metastatic role in lung cancer (4,6),
hepatocellular carcinoma (7),
gastric cancer (8), colorectal
cancer (9) and bladder cancer
(10). It has been generally
accepted as an important marker to predict prognosis in early-stage
lung adenocarcinoma or lung squamous carcinoma (4), pancreatic and renal cell carcinoma
(11). It is also involved in
cisplatin resistance in lung cancer (12). However, little literature is
available to illustrate its role in acute leukemia.
Acute myeloid leukemia (AML) is a malignancy
originating from the hematopoietic system and is characterized by
clonal expansion of myeloid cells and a maturation arrest in bone
marrow, which results in loss of normal hematopoietic function. As
the most common type of acute leukemia in adults, the incidence of
AML increases along with age and mainly affects middle-aged and
elderly populations (13,14). In recent years, complete remission
(CR) and disease-free survival (DFS) of AML patients have been
improved with the optimization of chemotherapy regimens and
supportive treatments (15).
However, 50–70% of AML patients who are able to achieve a CR with
primary induction chemotherapy encounter relapse, with subsequent
survival probability lowered to approximately 10% (16). Thus, most AML patients face a gloomy
prognosis, and the identification of new predictive markers and
therapeutic targets for AML is an urgent quest.
The present study aimed to investigate the potential
role of MALAT-1 in AML. Firstly, we examined the expression levels
of MALAT-1 in bone marrow samples of AML patients and healthy
individuals. Second, we explored the possible mechanisms underlying
of the affects of MALAT-1 on acute myeloid leukemia via cell
experiments. The results showed that upregulation of MALAT-1 was
closely associated with the poor prognosis of M5 patients and its
knockdown prohibited the proliferation of M5 cell lines and
promoted cell apoptosis, suggesting that MALAT-1 may serve as a
potential prognostic marker and therapeutic target.
Materials and methods
Patients and clinical samples
Bone marrow cells were collected from 95 newly
diagnosed AML patients (47 males and 48 females with a median age
of 51 years; range, 14–89 years) admitted to the Affiliated Union
Hospital of Fujian Medical University from December 2012 to
November 2016. The diagnosis was classified according to the
French-American-British (FAB) classification criteria and confirmed
by morphology, immunophenotyping, cytogenetics, molecular cell
biology (MICM classification criteria published by WHO) and
additional examinations. Patients who underwent chemotherapy or
radiotherapy prior to the study were excluded. Thirty-seven healthy
bone marrow donors served as normal controls. Informed consent was
obtained from the patients and healthy individuals before the use
of these clinical samples and the study protocol was approved by
the Ethics Committee of Affiliated Union Hospital. A telephone
follow-up was conducted after obtaining consent from the patients'
families, which was supplemented with the examination of inpatient
and/or outpatient medical records.
After bone marrow puncture, 3–5 ml of the bone
marrow was harvested. Mononuclear cells in bone marrow were
extracted using lymphocyte separation medium (Hao Yang Bio,
Tianjin, China). Cells were processed for extraction of DNA, RNA
and proteins, or were stored at −80°C for later use.
Cell culture
The acute monocytic leukemia cell lines, U-937
(catalog no. TCHu159) and THP-1 (catalog no. TCHu 57), were chosen
according to the results of RT-qPCR. Both cell lines were obtained
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). All cell lines were maintained in RPMI-1640 medium
(Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS;
Hao Yang Bio) and maintained at 37°C in a humidified incubator
containing 5% CO2. The cells were passaged every two
days, and those in logarithmic growth were used for further
study.
Cell transfection
U-937 and THP-1 cell lines were used for further
transfection experiments. Lentiviral vectors of MALAT-1 siRNA
(si-MALAT-1) or scrambled negative control (si-NC) were designed
and synthesized by Genechem (cat no. GIDL80771; Shanghai, China).
Both U-937 and THP-1 cells were transfected with si-MALAT-1 and the
si-NC lentivirus according to the manufacturer's instructions. The
transfection efficiency was determined by fluorescence microscopy
96 h later.
Extraction of total cellular RNA and
cDNA synthesis
Total RNA was extracted with TRIzol reagent
(Invitrogen, Carlsbad, CA, USA). Two microliters of RNA were used
to measure the concentration and purity on a quantitation analyzer
(Thermo Fisher Scientific, Waltham, MA, USA). The purity of the RNA
was within 1.8–2.0 and was confirmed by OD260/280 ratios.
First-strand cDNA was synthesized using the RevertAid First-Strand
cDNA Synthesis kit (Thermo Fisher Scientific). Reverse
transcription was performed to generate complementary DNA in a
final volume of 20 µl, containing 1 µg of RNA, 1 µl of random
primer, 2 µl of dNTP mix (10 mM), 1 µl of RNase inhibiter, 1 µl of
reverse transcriptase, 4 µl of 5X reverse transcriptase buffer and
diethylpyrocarbonate (DEPC)-treated water. The procedure was
performed according to the manufacturer's protocol (Thermo Fisher
Scientific). The cDNA was either immediately used as a template for
RT-qPCR or stored at −20°C for later use.
Real-time quantitative reverse
transcription polymerase chain reaction (RT-qPCR) analysis of
MALAT-1 expression
An ABI7500 real-time RCR module and FastStart
Universal SYBR-Green Master kit (ROX; Roche, Indianapolis, IN, USA)
were employed to investigate reverse-transcription products using
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal
control. The primer sequences of MALAT-1 were as follows: forward,
5′-AAAGCAAGGTCTCCCCACAAG-3′ and reverse,
5′-GGTCTGTGCTAGATCAAAAGGCA-3′. The primer sequences of GAPDH were
as follows: forward, 5′-CCCCTTCATTGACCTCAACTACAT-3′ and reverse,
5′-CGCTCCTGGAAGATGGTGA-3′. The total reaction volume was 25 µl,
including 12.5 µl of SYBR-Green qPCR Mix (Roche), 1 µl of cDNA,
0.75 µl of forward and reverse primers at 10 µmol/l, respectively,
and 10 µl of DEPC water. The reaction conditions were as follows:
50°C for 2 min for 1 cycle (first stage); 95°C for 2 min for 1
cycle (second stage); followed by 95°C for 15 sec and then 60°C for
30 sec for 40 cycles (third stage). Melting curve analysis involved
one cycle of 95°C for 15 sec; 60°C for l min; 95°C for 15 sec; and
then 60°C for 15 sec. Every reaction mixture was triplicated and
RT-qPCR was also tripled for each sample and all cell groups. The
expression levels of MALAT-1 were normalized to the internal
control GAPDH reference to obtain the relative threshold cycle
(ΔCt). The relative levels were calculated by the comparative Ct
(ΔΔCt) method, and the relative expression fold (2−ΔΔCt)
was calculated.
Cell proliferation assay and colony
formation analysis
Cell proliferation rates were measured using Cell
Counting Kit-8 (CCK-8; Dojindo, Japan) according to the
manufacturer's instructions. U-937 cells were seeded into 96-well
plates at a density of 7×103 cells/100 µl and THP-1
cells at a density of 2.5×104 cells/100 µl. In each
plate, there were three groups (3 wells/group): blank control group
(no cells but medium), negative control group (U-937/THP-1 cells
transfected with si-NC), and the experimental group (U-937/THP-1
cells transfected with si-MALAT-1). Cell proliferation was
determined after 24, 48, 72 and 96 h, respectively. An amount of 10
µl of CCK-8 solution was added to each well. Cells were incubated
at 37°C for 2 h, and then the optical density (OD) was measured
with a microplate reader (Biotek, Richmond, VT, USA) at a
wavelength of 450 nm using the following formula:
ODexperiment - ODblank. The experiment was
repeated three times.
To assess the colony formation, U-937 and THP-1
cells transfected with si-NC or si-MALAT-1 lentivirus were seeded
into 24-well plates (500 µl/well, 200 cells/well, three
wells/group), which were pre-coated with methyl cellulose solution
(500 µl/well). The cells were then incubated at 37°C in a
humidified incubator containing 5% CO2. After 10–14
days, the colonies were counted under an inverted microscope
(Nikon, Tokyo, Japan). A colony was defined as an aggregate of
>40 cells. The methyl cellulose solution was prepared for use as
mentioned in our previous study (17).
Analysis of the cell cycle and
apoptosis
Cell cycle and apoptosis analyses were performed on
a BD FACSVerse™ flow cytometer (BD Biosciences, San Jose, CA, USA)
according to the manufacturer's instructions.
Analysis of the cell cycle
Cells (1×106) were collected after a 96-h
transfection and washed with cold phosphate-buffered saline (PBS),
and then fixed in 70% ethanol at 4°C overnight. After fixation, the
cells were washed again and resuspended in PBS, and then stained
with PI/RNase Staining Buffer (BD Biosciences) in the dark at 25°C
for 15 min, and finally analyzed with a flow cytometer (BD
Biosciences) and Modlfit software (Verity Software House, Topsham,
ME, USA).
Analysis of apoptosis
After 96-h transfection, cell apoptosis was analyzed
with the PE Annexin V Apoptosis Detection kit I (BD Biosciences)
according to the manufacturer's protocol. Cells were discriminated
into viable cells, dead cells, early apoptotic and late apoptotic
cells. Ratios of early and late apoptotic cells were compared with
those of the control group. Results were analyzed with FlowJo
software (BD Biosciences). All experiments were performed in
triplicate.
In addition, cells were collected and treated with
bisbenzimide (Hoechst 33258; Beyotime, Shanghai, China), following
the manufacturer's instructions. Apoptotic cells were observed
under a fluorescence microscope (Nikon). Each experiment was
conducted in triplicate.
Western blot analysis
Protein was extracted using lysis solution
containing protease inhibitors (all from Lulong Biotech, Fujian,
China). Equal amounts of total protein (80 µg) from every sample
were separated by 10% SDS-PAGE (Beyotime). The proteins were then
transferred onto PVDF membranes (0.2 µm; Millipore, Bedford, MA,
USA), which were blocked with skim milk (Boster, Hubei, China), and
then incubated with the following primary antibodies at 4°C
overnight: GAPDH (Abcam, Cambridge, MA, USA), caspase-3, caspase-8
and caspase-9 (Cell Signaling Technology, Danvers, MA, USA). After
being washed with TBST, the membranes were incubated with secondary
antibodies (Cell Signaling Technology) at room temperature for 2 h
and the signals of the membranes were detected with West Pico
Chemiluminescent substrate (Thermo Fisher Scientific). Band
intensities were analyzed with ImageJ software (1.45s; National
Institutes of Health Bethesda, MD, USA). The expression levels of
proteins were normalized to the internal control GAPDH. The
experiment was repeated three times.
Statistical analysis
Data are expressed as mean ± SD and were analyzed
with SPSS19.0 Statistical Software (IBM, Armonk, NY, USA) and
GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results of lncRNA MALAT-1 expression in clinical samples were
assessed by nonparametric Mann-Whitney U-test, and multiple groups
were compared using Kruskal-Wallis H test. Three independent
experiments were performed for all measurements. The significance
of mean differences between two groups in the cellular function
experiments was calculated by unpaired two-tailed Student's
t-tests. Rates were compared and analyzed by the Chi-square or
Fisher's exact tests. The overall survival (OS) was estimated by
the Kaplan-Meier method and log-rank test. Variables influencing OS
significantly in univariate analysis were analyzed by using a Cox
proportional hazards model. OS was measured from the beginning of
the study to the death of the patient (by any cause) or to the last
follow-up time for surviving patients. P<0.05 was deemed
statistically significant.
Results
Expression of MALAT-1 in bone marrow
cells derived from AML patients, and analysis of clinical features
and prognosis
The RT-qPCR analysis showed that the expression of
MALAT-1 in M5 patients was markedly increased when compared with
that of the healthy controls (p<0.01) (Fig. 1A) and non-M5 AML patients
(p<0.01) (Fig. 1B), while no
significant difference was observed when other AML subtypes (M0,
M1, M2, M3, M4 or M6) were compared with the controls (p>0.05,
respectively) (Fig. 1A). No
significant difference was observed between the non-M5 AML patients
and the controls (p=0.5398) (Fig.
1B). The clinicopathological characteristics of all AML
patients are summarized in Table
I.
 | Table I.Clinicopathological characteristics
of the AML patients. |
Table I.
Clinicopathological characteristics
of the AML patients.
|
| AML subtypes |
|---|
|
|
|
|---|
|
| M0 | M1 | M2 | M3 | M4 | M5 | M6 |
|---|
| Factors | n | n | n | n | n | n | n |
|---|
| Sex |
|
Male | 1 | 1 | 15 | 1 | 1 | 27 | 0 |
|
Female | 1 | 5 | 6 | 6 | 1 | 29 | 1 |
| Age (years) |
|
<60 | 2 | 3 | 15 | 3 | 2 | 37 | 1 |
|
≥60 | 0 | 3 | 6 | 4 | 0 | 19 | 0 |
| WBC
(x109/l) |
|
<100 | 2 | 3 | 16 | 6 | 2 | 40 | 1 |
|
≥100 | 0 | 3 | 5 | 1 | 0 | 16 | 0 |
| HB (g/l) |
|
<60 | 0 | 1 | 3 | 1 | 2 | 10 | 0 |
|
≥60 | 2 | 5 | 18 | 6 | 0 | 46 | 1 |
| PLT
(x109/l) |
|
<30 | 0 | 2 | 9 | 5 | 2 | 19 | 0 |
|
≥30 | 2 | 4 | 12 | 2 | 0 | 37 | 1 |
| Bone marrow blasts
(%) |
|
<60 | 1 | 0 | 9 | 0 | 1 | 13 | 1 |
|
≥60 | 1 | 6 | 12 | 7 | 1 | 43 | 0 |
| Risk stratification
based on karyotypea |
|
Better | 0 | 0 | 2 | 6 | 0 | 2 | 0 |
|
Intermediate | 2 | 4 | 14 | 0 | 2 | 20 | 0 |
|
Poor | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
|
Unkown | 0 | 2 | 5 | 1 | 0 | 30 | 1 |
The markedly increased expression of MALAT-1 in M5
patients necessitated a probe into its clinical relevance in these
patients. Fifty-six M5 patients (27 males and 29 females with a
median age of 51 years; range, 14–89 years) were included for
further analysis. The correlation between MALAT-1 expression and
clinicopathological parameters was then assessed (Table II). The M5 patients were divided
into two groups using the median MALAT-1 2−ΔCt value of
17.4118 (Fig. 2A). Those with an
MALAT-1 expression level lower than the median value (17.4118) were
assigned to the low-expression group (n=28), and those with
expression above the median value was assigned to the
high-expression group (n=28). As shown in Table II, a high MALAT-1 level was
associated with higher white blood cell count (p<0.01) and
higher platelet count (p<0.05). However, it was not directly
associated with other clinical parameters, including sex, age,
hemoglobin level, bone marrow blast count and stratification based
on karyotype.
 | Table II.MALAT-1 expression and
clinicopathological characteristics of the M5 patients. |
Table II.
MALAT-1 expression and
clinicopathological characteristics of the M5 patients.
| Factors | MALAT-1 high
expression (n=28) n | MALAT-1 low
expression (n=28) n | P-value |
|---|
| Sex |
|
Male | 13 | 14 | 0.789 |
|
Female | 15 | 14 |
|
| Age (years) |
|
<60 | 20 | 17 | 0.397 |
|
≥60 | 8 | 11 |
|
| WBC
(x109/l) |
|
<100 | 15 | 25 | 0.003 |
|
≥100 | 13 | 3 |
|
| HB (g/l) |
|
<60 | 6 | 4 | 0.485 |
|
≥60 | 22 | 24 |
|
| PLT
(x109/l) |
|
<30 | 5 | 14 | 0.011 |
|
≥30 | 23 | 14 |
|
| Bone marrow blasts
(%) |
|
<60 | 4 | 9 | 0.114 |
|
≥60 | 24 | 19 |
|
| Risk stratification
based on karyotypea |
|
Better | 1 | 1 | 0.424 |
|
Intermediate | 12 | 8 |
|
|
Poor | 3 | 1 |
|
|
Unkown | 12 | 18 |
|
The follow-up was completed on November 22, 2016.
The Kaplan-Meier survival curve was used to analyze the association
of MALAT-1 expression with the OS of M5 patients. The results
revealed that, compared with the patients with low MALAT-1
expression, OS of the high-expression group was significantly
decreased (p<0.01) (Fig. 2B).
The mean OS over 5 years (2012–2016) was estimated at 182.24±60.558
days [95% confidence interval (CI), 63.547–300.933] for the
high-expression group and at 449.285±72.462 days (95% CI,
307.259–591.311) for the low-expression group. Clinical prognostic
indicators were also analyzed. Univariate analysis of OS showed
that white blood cell count and MALAT-1 expression level were
prognostic indicators while the remaining clinical parameters were
not (Table III). In the
multivariate analysis, the Cox proportional hazards model revealed
that MALAT-1 overexpression was an independent prognostic factor
for OS [hazard ratio (HR), 2.551; 95% CI, 1.309–4.97; p=0.006]
(Table IV).
 | Table III.Univariate analysis of OS of the
patients with AML-M5 (n=56). |
Table III.
Univariate analysis of OS of the
patients with AML-M5 (n=56).
| Variables | Group | n | P-value (OS) |
|---|
| Sex | Male/female | 27/29 | 0.526 |
| Age (years) | <60/≥60 | 37/19 | 0.218 |
| WBC
(x109/l) | <100/≥100 | 40/16 | 0.006 |
| HB (g/l) | <60/≥60 | 10/46 | 0.866 |
| PLT
(x109/l) | <30/≥30 | 19/37 | 0.754 |
| Bone marrow
blasts | <60%/≥60% | 13/43 | 0.921 |
| MALAT-1 | High/low | 28/28 | 0.001 |
 | Table IV.Multivariate analysis of OS of
patients with AML-M5 (n=56). |
Table IV.
Multivariate analysis of OS of
patients with AML-M5 (n=56).
|
|
|
| OS |
|---|
|
|
|
|
|
|---|
| Variables | Group | n | HR | 95% CI | P-value |
|---|
| WBC
(x109/l) | <100/≥100 | 40/16 | 1.911 | 0.934–3.910 | 0.076 |
| MALAT-1 | High/low | 28/28 | 2.551 | 1.309–4.970 | 0.006 |
Effects of MALAT-1 knockdown on
proliferation, colony formation and cell cycle of U-937 and THP-1
cells
Due to its abnormal expression in M5 clinical
samples, we speculated that MALAT-1 may play a role in M5
progression. To assess the role of MALAT-1 in M5 cellular growth,
we silenced MALAT-1 expression in the U-937 and THP-1 cell lines
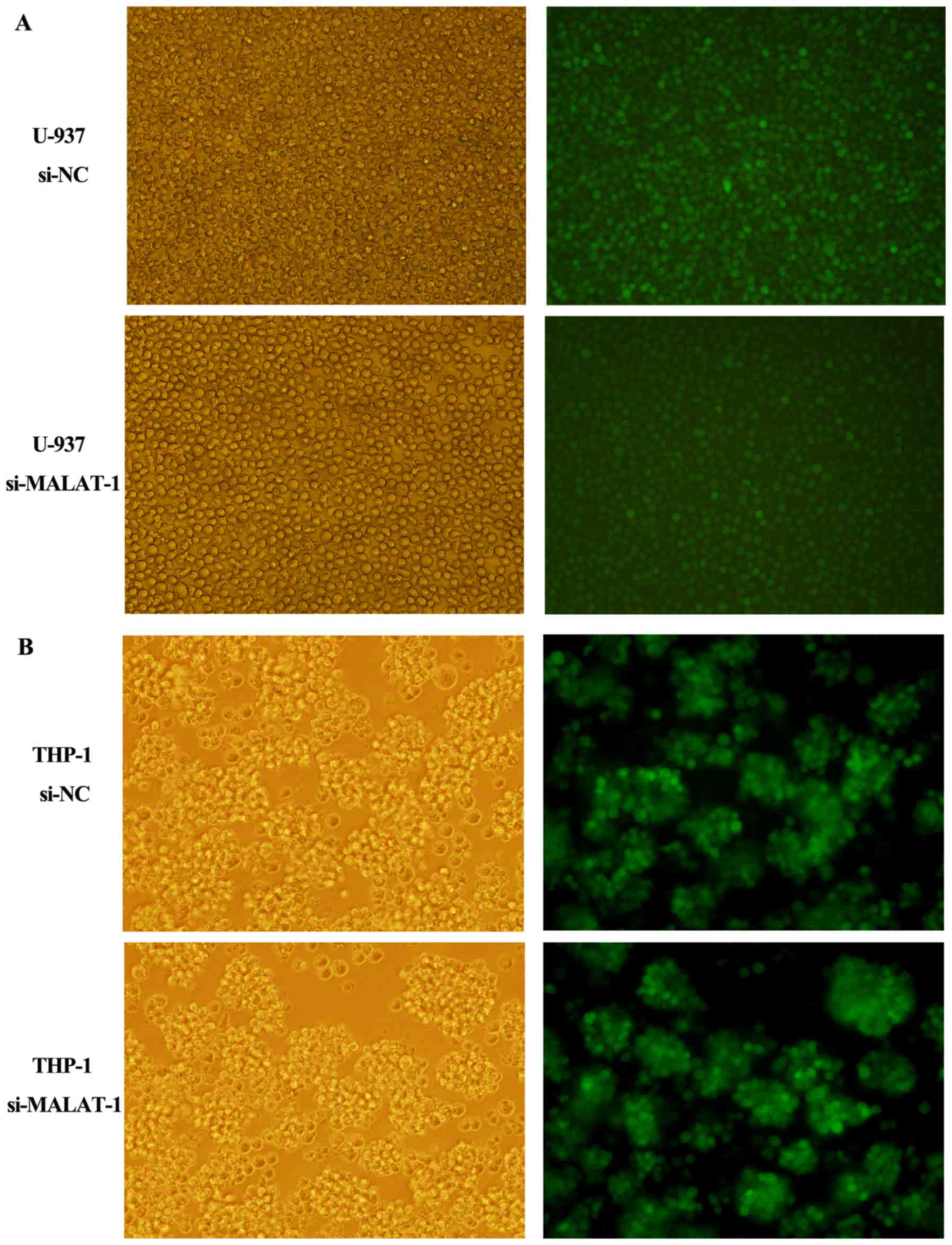
with small interfering RNA (siRNA). The transfection efficiency was
determined by fluorescence microscopy 96 h later as shown in
Fig. 3A (U-937) and B (THP-1). The
siRNA which decreased MALAT-1 expression level by more than 50% was
chosen for further experiments (Fig.
4A). Cells transfected with si-NC were regarded as the
control.
As presented in the results of the CCK8 assays
(Fig. 4B), cell growth was
suppressed in both cell lines transfected with si-MALAT-1 when
compared with cell growth of the si-NC groups. In addition, colony
formation in the U-937 and THP-1 cells was also reduced by MALAT-1
silencing (Fig. 4C). Cell cycle
progression in the U-937 and THP-1 cells was further analyzed by
flow cytometry. In both cell lines, compared with the si-NC groups,
a significantly greater proportion of G0/G1 phase cells was
observed among the MALAT-1 knockdown groups (U-937, p<0.01;
THP-1, p<0.05) (Fig. 5). The
si-NC group had an increased proportion of cells in the S phase in
both cell lines (U-937, p<0.001; THP-1, p<0.01) (Fig. 5).
Effects of MALAT-1 knockdown on U-937
and THP-1 cell apoptosis
We next investigated whether MALAT-1 knockdown
affects cell apoptosis. Flow cytometric analysis revealed that the
percentage of apoptotic U-937 cells was significantly increased in
the MALAT-1-silenced group when compared with that of the si-NC
group (p<0.001) (Fig. 6). A
similar result was observed in the THP-1 cells (p<0.01)
(Fig. 6).
In addition, Hoechst 33258 staining was applied to
confirm cell apoptosis. In the U-937 and THP-1 cell lines, few
apoptotic cells were observed in the si-NC group (Fig. 7A, left panel). However, in the
MALAT-1-silenced cells, the nuclei were dense and stained, and some
nuclei were disintegrated and fragmented into dense granular
particles (apoptotic bodies) (Fig.
7A, arrows, right panel).
Finally, the effects of the silencing of MALAT-1 on
the activation of the apoptosis-associated proteins, caspase-3,
caspase-8, caspase-9, were also determined by western blotting.
Western blot analysis showed that compared with those of the si-NC
groups, protein expression levels of caspase-3, caspase-8 and
caspase-9 were upregulated in the MALAT-1-silenced U-937 and THP-1
cells (Fig. 7B).
Discussion
The present study focused on the roles of lncRNA
MALAT-1 in acute myeloid leukemia. A significantly upregulated
expression of MALAT-1 was found in the M5 patient tissues when
compared with the healthy controls and non-M5 AML patient tissues,
while no statistical difference was found in tissues of other AML
subtypes. Moreover, MALAT-1 silencing inhibited the proliferation
and cycle progression of M5 cells, which also increased the
apoptosis. Compared with other AML subtypes, M5 patients frequently
report a high incidence of hyperleukocytosis, extramedullary
infiltration, disorders of hemostasis, low complete remission (CR)
rate for treatment and poor prognosis (18,19).
Thus, the identification of MALAT-1 in M5 may contribute to
promising strategies for the diagnosis and therapy of AML.
MALAT-1 is broadly expressed in human tissues
including lymphatic tissues, bone marrow and lymphocytes (20,21).
In a recent study of newly diagnosed patients with multiple
myeloma, the expression of MALAT-1 was found to be strongly
correlated with disease status and thus may serve as a predictor of
early progression, suggesting its role in the pathogenesis of
myeloma (22). Another recent study
reported a strong association between expression levels of lncRNAs
(e.g., MALAT-1, TUG1, HOTAIR, lincRNA-p21) and the clinical stages
of chronic lymphocytic leukemia (CLL) (23). In addition, MALAT-1 may play a role
in vincristine resistance in childhood acute lymphoblastic leukemia
(24). These findings on
hematopoietic system cancers lend support to the present study and
hint at the necessity of exploring the role of MALAT-1 in acute
leukemia.
In present study, MALAT-1 expression was upregulated
in M5 patients and was significantly associated with higher white
blood cell count, which is consistent with other research on solid
tumors and myeloma (11) and in
agreement with a recent study regarding the roles of lncRNA HOTAIR
in acute leukemia (12). However,
statistical analysis also revealed a correlation between
upregulated MALAT-1 and a higher platelet level, which was not in
accordance with the previous related evidence on HOTAIR (25,26).
We speculate that this inconsistency may result from the
insufficient number of cases or that MALAT-1 may have a special
role in the hematopoietic system. Previous literature has
documented that MALAT-1 expression is an independent prognostic
parameter for survival in several types of cancers (5). Similarly, univariate and multivariate
analyses in the present research demonstrated that high MALAT-1
expression, instead of other clinical characteristics, was highly
correlated with poor OS. These findings indicate that MALAT-1 may
serve as a candidate indicator for evaluating the prognosis of M5
patients. It should not be ignored that statistical difference was
not found in other AML subtypes when compared with the control,
which may be due to the limited number of cases. Both a larger
cohort and a longer follow-up period would be needed in further
research.
We also speculate that MALAT-1 may function as an
oncogene in the development of M5. To confirm the hypothesis, we
analyzed the effects of MALAT-1 knockdown on the proliferation and
apoptosis of U-937 and THP-1 cell lines. We found that the
downregulation of MALAT-1 in M5 cells inhibited the cell
proliferation, colony formation and cell cycle progression by
inducing G0/G1 stage arrest, which is consistent with some previous
studies (27,28). Silencing of MALAT-1 in gastric
cancer cell line SGC-7901 was found to inhibit cancer progression
by inducing G0/G1 arrest (27), and
a similar result was observed in cervical cancer cell line OVCAR3
(28). MALAT-1 knockdown was found
to induce upregulated protein expression of cyclin E and
cyclin-dependent kinase 2 (CDK2) by silencing tumor suppressor gene
p53 and activating transcription factor E2F, as a consequence of
G1/S phase transition to G2 phase (29). Moreover, a reduction in mRNA levels
of B-MYB and CENPE was observed in MALAT1-depleted cells, thus
influencing mitotic progression and cell cycle (29). However, it is still unclear whether
MALAT-1 influences cell proliferation by interacting with several
cyclins or cyclin-dependent kinases (CDKs) in leukemia, and further
studies are awaited to elucidate the specific molecular
mechanisms.
In addition to reduced cell proliferation, MALAT-1
knockdown increased the apoptosis ratio in both U-937 and THP-1
cells, which was confirmed by Hoechst 33258 staining and flow
cytometric analysis. Furthermore, western blot analysis revealed
that protein expression of caspase-3, caspase-8 and caspase-9 was
upregulated in the MALAT-1-knockdown cells. Apoptosis-associated
proteins play important roles in apoptotic signaling pathways,
which include initiator caspases (caspase-8, and caspase-9) and
executioner caspases (caspase-3, and caspase-7). Caspase-3,
caspase-8 and caspase-9 are key members in the regulation of
programmed cell death (30). Our
finding is consistent with a study of cervical cancer by Guo et
al (31), in which MALAT-1
knockdown upregulated the mRNA expression of pro-apoptotic genes,
caspase-3, caspase-8 and Bax, and suppressed expression of
anti-apoptotic genes, Bcl-2 and Bcl-xl. Our results suggest that
knockdown of MALAT-1 may induce apoptosis by activating
caspase-related apoptotic signaling pathways in leukemia. Further
studies are needed to investigate whether MALAT-1 plays a role in
regulating the expression of upstream genes (Bcl-2 or Bax) in the
caspase pathway, and the underlying mechanism.
In summary, the present study demonstrated that a
high expression of lncRNA MALAT-1 in acute monocytic leukemia is
related to clinicopathologic parameters including patient outcome.
Experiments in vitro showed that knockdown of MALAT-1
inhibited leukemia cell proliferation and induced apoptosis, thus
indicating that MALAT-1 may serve as a promising biomarker of poor
prognosis and a therapeutic target for M5 treatment. However,
several limitations are present in the study: a small number of
cases were assessed in the research; the mechanisms by which
MALAT-1 was upregulated in M5 patients were not assessed; and no
in vivo experiments were conducted to throw light upon the
molecular mechanism involved in acute leukemia progression. Further
studies are warranted to clarify the role of MALAT-1 in various
subtypes of leukemia and its therapeutic potential and prognostic
value in leukemia.
Acknowledgements
The present study was sponsored by the National
Natural Science Foundation of China (no. 81370629), Fujian
Provincial Environmental Protection and Technology Program
(2015R011), and the National and Fujian Provincial Key Clinical
Specialty Discipline Construction Program (China).
Glossary
Abbreviations
Abbreviations:
|
AL
|
acute leukemia
|
|
AML
|
acute myeloid leukemia
|
|
M0
|
acute myeloid leukemia with minimal
differentiation
|
|
M1
|
AML with partial differentiation
|
|
M2
|
AML with maturation
|
|
M3
|
acute promyelocytic leukemia
|
|
M4
|
acute myelomonocytic leukemia with
granules
|
|
M5
|
acute monocytic leukemia
|
|
M6
|
acute erythroleukemia
|
|
NC
|
negative control
|
|
ncRNA
|
non-coding RNA
|
|
lncRNA
|
long non-coding RNA
|
|
MALAT-1
|
metastasis-associated lung
adenocarcinoma transcript 1
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
RT-qPCR
|
real-time quantitative polymerase
chain reaction
|
References
|
1
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qiu MT, Hu JW, Yin R and Xu L: Long
noncoding RNA: An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen L, Chen L, Wang Y, Jiang X, Xia H and
Zhuang Z: Long noncoding RNA MALAT1 promotes brain metastasis by
inducing epithelial-mesenchymal transition in lung cancer. J
Neurooncol. 121:101–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, et
al: Metastasis-associated long non-coding RNA drives gastric cancer
development and promotes peritoneal metastasis. Carcinogenesis.
35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han Y, Liu Y, Nie L, Gui Y and Cai Z:
Inducing cell proliferation inhibition, apoptosis, and motility
reduction by silencing long noncoding ribonucleic acid
metastasis-associated lung adenocarcinoma transcript 1 in
urothelial carcinoma of the bladder. Urology. 81:209.e1–209.e7.
2013. View Article : Google Scholar
|
|
11
|
Tian X and Xu G: Clinical value of lncRNA
MALAT1 as a prognostic marker in human cancer: Systematic review
and meta-analysis. BMJ Open. 5:e0086532015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lopez-Ayllon BD, Moncho-Amor V, Abarrategi
A, de Ibañez Cáceres I, Castro-Carpeño J, Belda-Iniesta C, Perona R
and Sastre L: Cancer stem cells and cisplatin-resistant cells
isolated from non-small-lung cancer cell lines constitute related
cell populations. Cancer Med. 3:1099–1111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dores GM, Devesa SS, Curtis RE, Linet MS
and Morton LM: Acute leukemia incidence and patient survival among
children and adults in the United States, 2001–2007. Blood.
119:34–43. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Löwenberg B, Downing JR and Burnett A:
Acute myeloid leukemia. N Engl J Med. 341:1051–1062. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farag SS, Ruppert AS, Mrózek K, Mayer RJ,
Stone RM, Carroll AJ, Powell BL, Moore JO, Pettenati MJ, Koduru PR,
et al: Outcome of induction and postremission therapy in younger
adults with acute myeloid leukemia with normal karyotype: A cancer
and leukemia group B study. J Clin Oncol. 23:482–493. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mims A and Stuart RK: Developmental
therapeutics in acute myelogenous leukemia: Are there any new
effective cytotoxic chemotherapeutic agents out there? Curr Hematol
Malig Rep. 8:156–162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen JZ, Zhang YY, Fu HY, Wu DS and Zhou
HR: Overexpression of microRNA-143 inhibits growth and induces
apoptosis in human leukemia cells. Oncol Rep. 31:2035–2042.
2014.PubMed/NCBI
|
|
18
|
Azoulay E, Fieux F, Moreau D, Thiery G,
Rousselot P, Parrot A, Le Gall JR, Dombret H and Schlemmer B: Acute
monocytic leukemia presenting as acute respiratory failure. Am J
Respir Crit Care Med. 167:1329–1333. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Porcu P, Cripe LD, Ng EW, Bhatia S,
Danielson CM, Orazi A and McCarthy LJ: Hyperleukocytic leukemias
and leukostasis: A review of pathophysiology, clinical presentation
and management. Leuk Lymphoma. 39:1–18. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lane L, Argoud-Puy G, Britan A, Cusin I,
Duek PD, Evalet O, Gateau A, Gaudet P, Gleizes A, Masselot A, et
al: neXtProt: A knowledge platform for human proteins. Nucleic
Acids Res. 40:(D1). D76–D83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu C, Macleod I and Su AI: BioGPS and
MyGene.info: Organizing online, gene-centric information. Nucleic
Acids Res. 41:(D1). D561–D565. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cho SF, Chang YC, Chang CS, Lin SF, Liu
YC, Hsiao HH, Chang JG and Liu TC: MALAT1 long non-coding RNA is
overexpressed in multiple myeloma and may serve as a marker to
predict disease progression. BMC Cancer. 14:8092014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Isin M, Ozgur E, Cetin G, Erten N, Aktan
M, Gezer U and Dalay N: Investigation of circulating lncRNAs in
B-cell neoplasms. Clin Chim Acta. 431:255–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moqadam F Akbari, Lange-Turenhout EA,
Ariës IM, Pieters R and den Boer ML: miR-125b, miR-100 and miR-99a
co-regulate vincristine resistance in childhood acute lymphoblastic
leukemia. Leuk Res. 37:1315–1321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B
and Chen Y: Overexpression of long non-coding RNA HOTAIR predicts a
poor prognosis in patients with acute myeloid leukemia. Oncol Lett.
10:2410–2414. 2015.PubMed/NCBI
|
|
26
|
Zhang YY, Huang SH, Zhou HR, Chen CJ, Tian
LH and Shen JZ: Role of HOTAIR in the diagnosis and prognosis of
acute leukemia. Oncol Rep. 36:3113–3122. 2016.PubMed/NCBI
|
|
27
|
Wang J, Su L, Chen X, Li P, Cai Q, Yu B,
Liu B, Wu W and Zhu Z: MALAT1 promotes cell proliferation in
gastric cancer by recruiting SF2/ASF. Biomed Pharmacother.
68:557–564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, Li
J and Sheng X: The long noncoding RNA MALAT-1 is highly expressed
in ovarian cancer and induces cell growth and migration. PLoS One.
11:e01552502016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al:
Long noncoding RNA MALAT1 controls cell cycle progression by
regulating the expression of oncogenic transcription factor B-MYB.
PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miura M: Apoptotic and nonapoptotic
caspase functions in animal development. Cold Spring Harb Perspect
Biol. 4:42012. View Article : Google Scholar
|
|
31
|
Guo F, Li Y, Liu Y, Wang J, Li Y and Li G:
Inhibition of metastasis-associated lung adenocarcinoma transcript
1 in CaSki human cervical cancer cells suppresses cell
proliferation and invasion. Acta Biochim Biophys Sin (Shanghai).
42:224–229. 2010. View Article : Google Scholar : PubMed/NCBI
|