Introduction
Lung cancer is one of the most common malignancies
and the leading cause of cancer-related mortality worldwide. Among
all cases of lung cancer, more than 80% of patients present with
non-small cell lung cancer (NSCLC) with a dismal 5-year survival
(1–3). Recently, the high rate of cigarette
smoking and environmental pollution have resulted in an increase in
the incidence and mortality of lung cancer (4,5).
Importantly, despite great improvements in comprehensive diagnosis
and medical treatment, the long-term survival of NSCLC patients
remains unsatisfactory due to the high rates of recurrence and
distant metastasis (6,7). Therefore, it is urgent to elucidate
the underlying mechanism responsible for the development and
progression of NSCLC and identify novel therapeutic targets
involved in NSCLC (8).
MicroRNAs (miRNAs) are a family of endogenous, short
single-stranded and non-coding RNAs that negatively modulate gene
expression by binding to the 3′-untranslated region (3′-UTR) of
target mRNAs to cause degradation or inhibition of translation
(9,10). Increasing evidence has confirmed
that miRNAs play crucial roles in the physiological and
pathological processes in NSCLC including cell differentiation,
proliferation, apoptosis, migration and metastasis (11,12).
Recently, miR-212, which is located on chromosome 17p13.3, was
found to play a critical role in the progression of cancers,
including NSCLC (13–16). Previous studies show that miR-212
functions as a regulator of cell invasion, metastasis,
proliferation and drug sensitivity. miR-212 performs antimetastatic
properties through suppression of SOX4 in breast cancer (17). miR-212 functions as an
epigenetic-silenced tumor-suppressor involving in tumor metastasis
and invasion of gastric cancer by downregulating PXN expression
(18). Moreover, miR-212 inhibits
glioblastoma cell proliferation by targeting SGK3 (19). However, the expression of miR-212 is
upregulated in pancreatic, esophageal and prostate cancer. miR-212
promotes pancreatic cancer cell growth and invasion by targeting
the Hedgehog signaling pathway receptor patched-1 (20). Overexpression of miR-212 predicts a
poor prognosis of esophageal cancer patients (21). Thus, the functional significance of
miR-212 in cancer initiation, development and process seems to be
cancer-type specific. In regards to NSCLC, Li et al reported
that miR-212 displays tumor-promoting properties in NSCLC cells and
targets the Hedgehog pathway receptor PTCH1 (22); however, Lu et al demonstrated
that miR-212 functions as a tumor suppressor in NSCLC by targeting
synaptic acetylcholinesterase (23). The expression level of miR-212 and
its function in NSCLC are contradictory. Therefore, it is important
to identify the roles of miR-212 in NSCLC.
In the present study, we investigated the expression
level and biological function of miR-212 in NSCLC progression. Our
data showed that miR-212 was downregulated in NSCLC and the reduced
miR-212 was correlated with poor prognostic characteristics and
worse 5-year survival of NSCLC patients. We demonstrated that
miR-212 regulated the migration and invasion of NSCLC by targeting
SOX4 in vitro. Furthermore, miR-212 also suppressed EMT
phenotype progression. These data suggest that miR-212 inhibited
cell migration and invasion of NSCLC by targeting SOX4. Thus,
miR-212 is a novel prognostic biomarker for NSCLC patients.
Materials and methods
Clinical tissues and cell culture
Clinical specimens were collected from 115 NSCLC
patients who received surgical resection in our hospital during
2002 and 2011. All the clinical tissues from the patients were
pathologically confirmed as NSCLC before being used for further
experiments in the present study. Informed consent was obtained
from every patient involved in the present study. Approval for
experiments involving patient samples was obtained from the
Institutional Research Ethics Committee of The Affiliated Hospital
of Hangzhou Normal University.
Cell lines including H292, H1299, A549, SPC-A1 and
BEAS-2B were purchased from the Cell Bank of Chinese Academy of
Sciences (Shanghai, China) and the American Type Culture Collection
(ATCC; Rockville, MD, USA). All cells were cultured in Dulbeccos
modified Eagles medium (DMEM) supplemented with 10% fetal bovine
serum (FBS) (both from Gibco Co., New York, NY, USA). Cell cultures
were kept in cell incubators with humidified atmosphere and 5%
CO2 at 37°C.
Quantitative real-time reverse
transcription-PCR (qRT-PCR)
RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA)
was used to extract RNA from the NSCLC tissues and cells.
Transcriptional First Strand cDNA Synthesis kit (Roche,
Indianapolis, IN, USA) and SYBR-Green PCR Master Mix (Applied
Biosystems, Foster City, CA, USA) were used for reverse
transcription reactions and real-time PCR. Primers for miR-212, U6,
SOX4 and GAPDH were purchased from GeneCopoeia (Guangzhou, China).
U6 and GAPDH were the internal controls for miR-212 and SOX4,
respectively.
Western blotting
Total protein was extracted in RIPA buffer
(Beyotime, Shanghai, China) containing a protease and phosphatase
inhibitor, and then the protein concentration was determined using
the BCA Protein Assay kit (both from Thermo Scientific, Rockford,
IL, USA). Equal protein 30 µg was loaded and separated by 10%
SDS-PAGE and transferred to polyvinyllidene diflouride (PVDF)
membranes (Millipore, Billerica, MA, USA). The membranes were
incubated with respective primary antibodies: SOX4 and GAPDH
(1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C
overnight after blocking with 5% non-fat milk in TBS. Then, the
membranes were washed three times with Tris-buffered saline with
Tween-20 (TBST) and incubated with appropriate
peroxidase-conjugated secondary antibody for 2 h at room
temperature (ZSGB-BIO, Beijing, China). Protein bands were
visualized using an enhanced chemiluminescence kit (Amersham,
Little Chalfont, UK).
Immunohistochemical analysis
The tissues were fixed in formalin, embedded with
paraffin and sliced into 4-µm sections for immunohistochemical
staining. After deparaffinizing, dehydrating and antigen retrieval,
the slides were incubated in 3% H2O2 for
blocking endogenous peroxidase activity. Then, after incubating
with 10% goat serum for 30 min, E-cadherin and vimentin (1:300;
Cell Signaling Technology, Inc.) antibodies were applied as the
primary antibodies by a streptavidin peroxidase-conjugated (SP-IHC)
method. The percentage of positive cells was expressed as: 0 for
<10%; 1 for 10–30%; 2 for 31–50%; 3 for >50%.
Cell transfection
miR-212 mimic and miR-212 inhibitor were obtained
from GeneCopoeia Inc. SOX4 expression vector and SOX4-specific
siRNA were purchased from Ruibo Biotechnology Corp. (Guangzhou,
China). The tranfection of these vectors into NSCLC cells was
performed in 6-well plates using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) based on the instructions from the
manufacturer.
Transwell assays
The migration and invasion abilities of the NSCLC
cells were investigated by Transwell assays without or with
Matrigel. Generally, NSCLC suspended in basal DMEM was seeded into
the upper chamber and the lower chambers contained 600 µl DMEM with
20% FBS. Twenty-four to 48 h later, NSCLC cells that had migrated
or invaded through the membranes and stayed on the lower surface
were stained with crystal violet. Cell number for the migrated or
invaded cells was counted under a microscope.
Luciferase reporter assay
Wild-type SOX4 3′-UTR sequence and the mutated SOX4
3′-UTR sequence were constructed into the pGL3 control vector
(Promega, Madison, WI, USA) to obtain wt SOX4-3′-UTR and mt
SOX4-3′-UTR vectors, respectively. For the luciferase reporter
assay, NSCLC cells were co-transfected with the wild-type construct
or mutant construct, and, miR-212 mimics or inhibitor or control or
negative control vector. Forty-eight hours after transfection,
cells were harvested and lysed. The Dual-Luciferase Reporter Assay
System (Promega, Shanghai, China) was used to determine firefly and
Renilla luciferase activities.
Statistical analysis
Data are presented as the mean ± SD and at least
three independent replicates were performed. SPSS software 16.0
(SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad
Software, Inc., La Jolla, CA, USA) were used with a two-tailed
Students t-test, Pearson's correlation analysis, Kaplan-Meier
method and the log-rank test to evaluate the statistical
significance. Significant differences were defined as
P<0.05.
Results
miR-212 expression is downregulated in
NSCLC tissues and cell lines
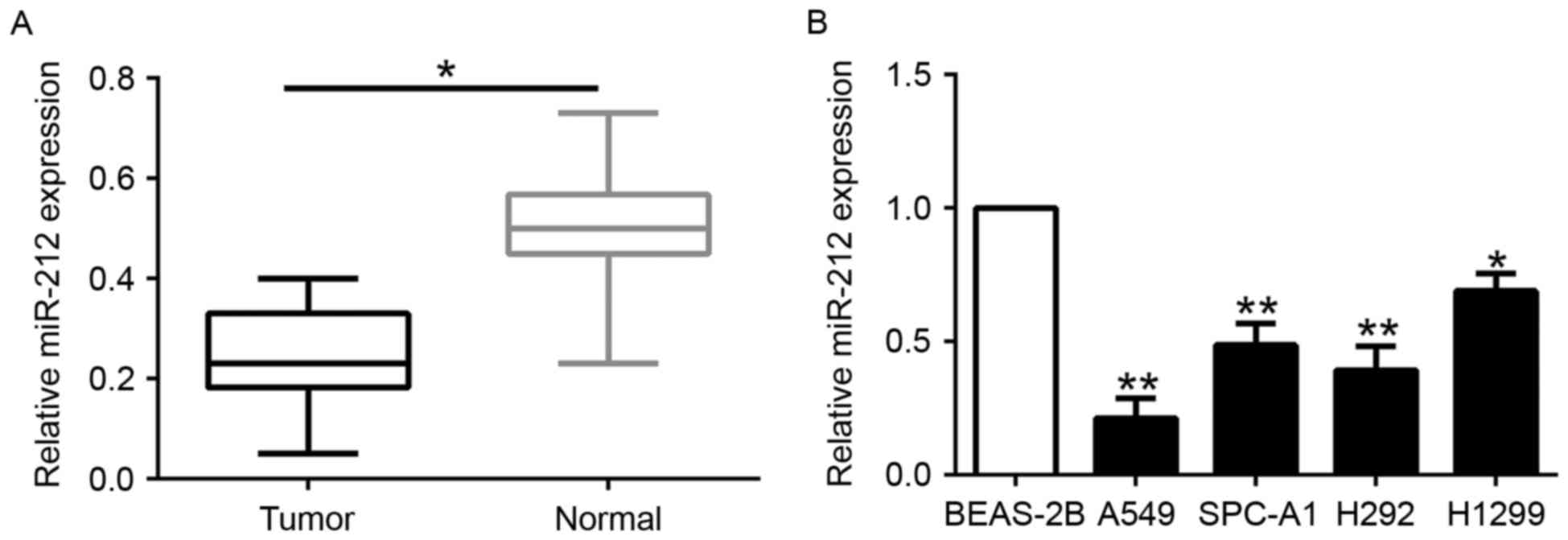
To determine the expression level of miR-212 in
NSCLC, we first evaluated the expression of miR-212 in 115 paired
NSCLC and adjacent normal lung tissues. miR-212 was significantly
decreased in the NSCLC tissues compared to that noted in the
matched tumor-adjacent tissues (P<0.05; Fig. 1A). Similarly, miR-212 was markedly
downregulated in a panel of NSCLC cells (A549, SPC-A1, H292 and
H1299) as compared to that observed in the normal lung epithelial
cell line BEAS-2B (P<0.05; Fig.
1B). These data identified miR-212 as a tumor-suppressor of the
progrssion of NSCLC.
Correlations between miR-212
expression and clinical characteristics of the NSCLC cases
To investigate the clinical significance of reduced
miR-212 in NSCLC, we divided the patients into two different
miR-212 groups according to the median expression level. As shown
in Table I, decreased miR-212
expression was significantly correlated with advanced
tumor-node-metastasis (TNM) stage (P=0.003) and positive lymph node
metastasis (P=0.001). These results suggest that the reduced
miR-212 was associated with poor prognostic features of NSCLC.
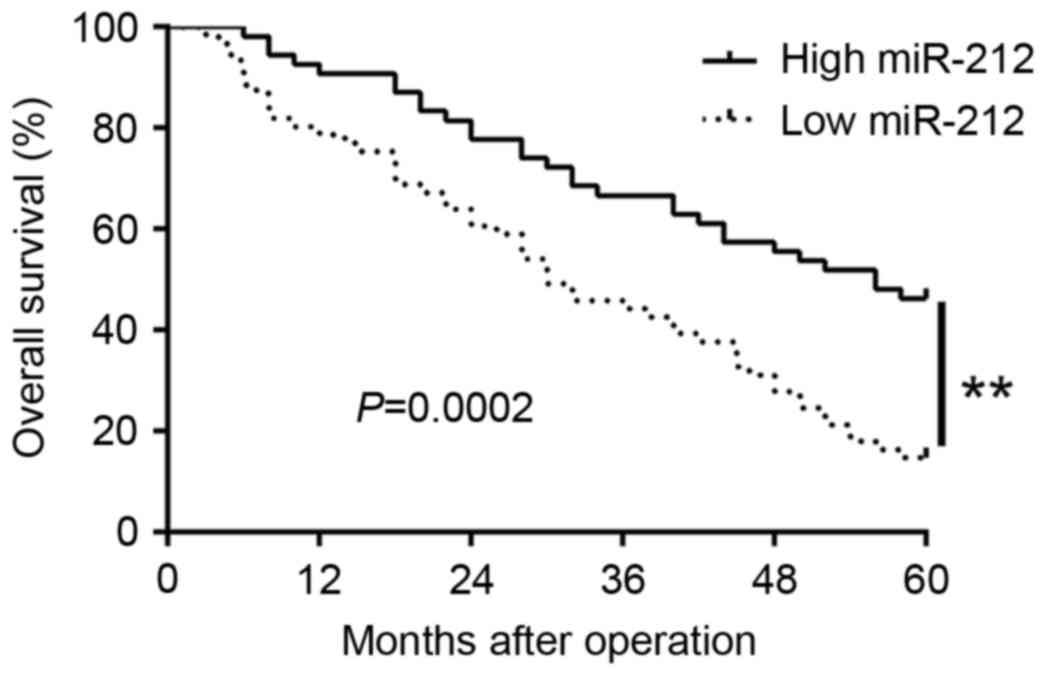
Furthermore, Kaplan-Meier analysis revealed that the patients with
low miR-212 expression had obviously shorter overall survival than
those with high miR-212 expression (P=0.0002; Fig. 2). In addition, miR-212 expression
was an independent factor for predicting 5-year overall survival in
NSCLC patients (P=0.001, 0.001, respectively; Table II). Taken together, these data
indicate that miR-212 is a potential biomarker for predicting the
outcome of NSCLC patients.
 | Table I.Correlation between miR-212 expression
and clinicopathological features of the NSCLC cases (n=115). |
Table I.
Correlation between miR-212 expression
and clinicopathological features of the NSCLC cases (n=115).
|
|
| Expression level |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Cases (n) |
miR-212high (n=54) | miR-212low
(n=61) | P-value |
|---|
| Age (years) |
|
|
| 0.716 |
|
<60 | 36 | 16 | 20 |
|
| ≥60 | 79 | 38 | 41 |
|
| Sex |
|
|
| 0.589 |
| Male | 89 | 43 | 46 |
|
|
Female | 26 | 11 | 15 |
|
| Histologic type |
|
|
| 0.724 |
|
Squamous | 68 | 31 | 37 |
|
|
Adenocarcinoma | 47 | 23 | 24 |
|
| Lymph node
metastasis |
|
|
| 0.001a |
|
Negative | 83 | 47 | 36 |
|
|
Positive | 32 | 7 | 25 |
|
| TNM stage |
|
|
| 0.003a |
|
I+II | 78 | 44 | 34 |
|
|
III+IV | 37 | 10 | 27 |
|
 | Table II.Univariate and multivariate analyses
of prognostic factors in NSCLC patients. |
Table II.
Univariate and multivariate analyses
of prognostic factors in NSCLC patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| TNM stage | 2.013 | 1.409–2.872 | 0.002a | 1.862 | 1.283–2.708 | 0.003a |
| Lymph node
metastasis | 2.798 | 1.879–5.682 | 0.001a | 2.214 | 1.387–3.263 | 0.002a |
| miR-212 | 3.967 | 1.583–7.382 | 0.001a | 3.223 | 1.237–6.962 | 0.001a |
miR-212 inhibits NSCLC cell migration
and invasion in vitro
To investigate the potential role of miR-212 in
NSCLC, we performed gain- and loss-of-function experiments based on
miR-212 mimics and inhibitors, which were transfected into A549
(P<0.05; Fig. 3A) and H1299
cells (P<0.05; Fig. 3C),
respectively. As measured by Matrigel-uncoated (for migration
assay) and Matrigel-coated (for invasion assay) Transwell assays,
ectopic expression of miR-212 in A549 cells resulted in a
significant reduction of cell migration and invasion capacity
(P<0.05; Fig. 3B). In contrast,
transfection of H1299 cells with miR-212 inhibitor obviously
increased cell migration and invasion (P<0.05; Fig. 3D). These data demonstrated that
miR-212 suppressed the migration and invasion of NSCLC cells in
vitro.
miR-212 suppresses
epithelial-to-mesenchymal transition in NSCLC cells
EMT has been identified as having a crucial role in
the induction of metastatic progression of cancer (24). To investigate the potential role of
miR-212 in modulating NSCLC metastasis, EMT markers were assessed.
We found that miR-212 overexpression increased the epithelial
marker E-cadherin and inhibited N-cadherin and vimentin expression
(P<0.05; Fig. 4A). In contrast,
miR-212 knockdown decreased E-cadherin expression and increased
N-cadherin and vimentin expression (P<0.05; Fig. 4B). In addition, we further explored
the correlation between miR-212 expression and EMT markers in NSCLC
tissues. We found that the E-cadherin expression in the high
miR-212 group was higher than that in the low miR-212 group.
Conversely, the expression level of vimentin in the high miR-212
group was markedly lower than that in the low miR-212 group
(P<0.05; Fig. 4C). Taken
together, these results suggest that miR-212 functions as a
suppressor of EMT in NSCLC cells.
SOX4 is a direct target of
miR-212
To explore the molecular mechanism by which miR-212
suppressed NSCLC migration and invasion, we used public
bioinformatic algorithms (TargetScan 6.2 and MiRanda) to search for
candidate target genes. We selected sex-determining region Y-box 4
(SOX4) for proceeding with experimental confirmation, which was
confirmed in other cancers. The 3′-UTR of SOX4 mRNA contains a
putative complementary site of miR-212 (Fig. 5A). To confirm the prediction, we
constructed the wild-type (WT) and mutant (MUT) 3′-UTR of SOX4
plasmids. Luciferase reporter assays revealed that ectopic miR-212
expression significantly inhibited the luciferase activity of SOX4
containing a WT 3′-UTR, but did not suppress the activity of SOX4
with a MUT 3′-UTR; in contrary, miR-212 inhibition increased the
luciferase activity of wild-type of 3′-UTR of SOX4 while had no
effect on the mutant SOX4 3′-UTR (P<0.01; Fig. 5B). Moreover, we demonstrated that
miR-212 overexpression significantly inhibited SOX4 mRNA and
protein expression in A549 cells, whereas the downregulation of
miR-212 prominently increased the mRNA and protein levels of SOX4
in H1299 cells (P<0.05, respectively; Fig. 5C and D).
Subsequently, we examined the correlation between
miR-212 expression and SOX4 in NSCLC tissues. We found that the
expression of SOX4 mRNA and protein in the miR-212 high-expressing
tumors were significantly lower than those in the miR-212
low-expressing tumors (P<0.05, respectively; Fig. 5E and F). Taken together, these
results suggest that miR-212 inhibits SOX4 expression by direct
binding to the 3′-UTR of SOX4.
Alteration in SOX4 expression reverses the
functional effects of miR-212 in NSCLC cells. To clarify that SOX4
is a functional target of miR-212, SOX4 was restored by a plasmid
vector in miR-212-overexpressing A549 cells (P<0.05; Fig. 6A). Furthermore, SOX4 overexpression
increased cell migration, invasion (P<0.05, respectively;
Fig. 6B) and promoted EMT
(P<0.05; Fig. 6E). Similarly,
SOX4 knockdown by a specific siRNA in miR-212-suppressive H1299
cells (P<0.05; Fig. 6C)
significantly inhibited cell migration, invasion (P<0.05,
respectively; Fig. 6D) and EMT
(P<0.05; Fig. 6F). These data
demonstrated that SOX4 is a downstream mediator in the function of
miR-212 in NSCLC.
Discussion
Increasing evidence has confirmed that aberrant
miRNAs play a critical role in NSCLC development and progression.
Previous studies indicated that miR-212 is involved in the
pathogenic process of diverse human cancers. miR-212 downregulates
SMAD2 expression to suppress the G1/S phase transition of the cell
cycle and EMT in cervical cancer cells (25). miR-212 is downregulated and
suppresses methyl-CpG-binding protein MeCP2 in gastric cancer
(26). Moreover, regulation of
heparin-binding EGF-like growth factor by miR-212 was found to be a
potential mechanism for the acquired cetuximab-resistance in head
and neck squamous cell carcinoma (27). miR-212 was found to inhibit
hepatocellular carcinoma cell proliferation and induce apoptosis by
targeting FOXA1 (28). In contrast,
overexpression of miR-212 was found to be associated with poor
prognosis of patients with pancreatic ductal adenocarcinoma
(29). In the present study, we
demonstrated that miR-212 expression was significantly
downregulated in NSCLC tissues and cell lines. These data indicate
that miR-212 may be involved in the pathogenesis of NSCLC as a
tumor suppressor. Following clinical analysis of miR-212 with
pathological characteristics, we found that reduced miR-212 was
obviously associated with advanced TNM stage and positive lymph
node metastasis. Moreover, our data indicated that high expression
of miR-212 predicated a significant better 5-year survival for
NSCLC patients. Multivariate Cox repression analysis indicated that
miR-212 was an independent prognostic factor for predicting the
survival of NSCLC patients. Taken together, these results indicate
that miR-212 expression is pivotal for the prognostic outcome in
NSCLC patients. Functionally, we found that miR-212 overexpression
reduced while miR-212 knockdown increased SOX4 expression in NSCLC
cell lines by directly interacting with the 3′-UTR of SOX4 mRNA. In
addition, we found the SOX4 mRNA and protein expression was
inversely correlated with miR-212 expression in NSCLC tissues. In
addition, alteration of SOX4 expression partially reversed
miR-212-induced function in NSCLC cells. Taken together, SOX4 was
identified as a functional target gene of miR-212 in NSCLC.
Sex-determining region Y-related high-mobility group
box 4 (SOX4) has been reported to be upregulated in multiple
cancers, including NSCLC (30).
SOX4 is involved in early embryogenesis and cell phenotype
decisions. Previous studies confirmed that SOX4 is a critical
component of the PTEN/PI3K/AKT pathway in prostate cancer (31). SOX4 is a frequent target of
retroviral insertional mutagenesis and contributes to
transformation in murine hematopoietic cells. In the present study,
we found that miR-212 inhibited tumor migration and invasion and
the EMT phenotype by targeting SOX4 in vitro. Furthermore,
we found that miR-212 expression was inversely correlated with EMT
marker (E-cadherin and vimentin) expression, which reinforce the
biological function of miR-212 on EMT. These data confirmed that
the functional effect of miR-212 in NSCLC in vitro was
dependent on SOX4.
In summary, we demonstrated that miR-212 was
downregulated in NSCLC tissues and cell lines, and its expression
was correlated with malignant clinicopathological features.
Furthermore, we confirmed that overexpression of miR-212 inhibited
cell migration and invasion in vitro by inhibiting
SOX4-mediated EMT signaling pathway. These results suggest that
miR-212 is a potential invasion-associated tumor suppressor in
NSCLC. In the future, therapeutic interventions concentrating on
miR-212-SOX4 may help suppress the development and metastasis of
NSCLC.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zornosa C, Vandergrift JL, Kalemkerian GP,
Ettinger DS, Rabin MS, Reid M, Otterson GA, Koczywas M, D'Amico TA,
Niland JC, et al: First-line systemic therapy practice patterns and
concordance with NCCN guidelines for patients diagnosed with
metastatic NSCLC treated at NCCN institutions. J Natl Compr Canc
Netw. 10:847–856. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Blais N and Kassouf E: Maintenance
therapies for non-small cell lung cancer. Front Oncol. 4:2132014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
8
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shivdasani RA: MicroRNAs: Regulators of
gene expression and cell differentiation. Blood. 108:3646–3653.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao Q, Mao ZD, Shi YJ, Chen Y, Sun Y,
Zhang Q, Song L and Peng LP: MicroRNA-7 inhibits cell
proliferation, migration and invasion in human non-small cell lung
cancer cells by targeting FAK through ERK/MAPK signaling pathway.
Oncotarget. 7:77468–77481. 2016.PubMed/NCBI
|
|
12
|
Zhu X, Li Y, Shen H, Li H, Long L, Hui L
and Xu W: miR-137 inhibits the proliferation of lung cancer cells
by targeting Cdc42 and Cdk6. FEBS Lett. 587:73–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dou C, Wang Y, Li C, Liu Z, Jia Y, Li Q,
Yang W, Yao Y, Liu Q and Tu K: MicroRNA-212 suppresses tumor growth
of human hepatocellular carcinoma by targeting FOXA1. Oncotarget.
6:13216–13228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ding G, Zhou L, Qian Y, Fu M, Chen J, Chen
J, Xiang J, Wu Z, Jiang G and Cao L: Pancreatic cancer-derived
exosomes transfer miRNAs to dendritic cells and inhibit RFXAP
expression via miR-212-3p. Oncotarget. 6:29877–29888. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ramalinga M, Roy A, Srivastava A,
Bhattarai A, Harish V, Suy S, Collins S and Kumar D: MicroRNA-212
negatively regulates starvation induced autophagy in prostate
cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis
and cellular senescence. Oncotarget. 6:34446–34457. 2015.PubMed/NCBI
|
|
16
|
Luo XJ, Tang DG, Gao TL, Zhang YL, Wang M,
Quan ZX and Chen J: MicroRNA-212 inhibits osteosarcoma cells
proliferation and invasion by down-regulation of Sox4. Cell Physiol
Biochem. 34:2180–2188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hanieh H: Aryl hydrocarbon
receptor-microRNA-212/132 axis in human breast cancer suppresses
metastasis by targeting SOX4. Mol Cancer. 14:1722015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, Li Z, Xiong J, Gong B, Zhang G, Cao
C, Jie Z, Liu Y, Cao Y, Yan Y, et al: MicroRNA-212 functions as an
epigenetic-silenced tumor suppressor involving in tumor metastasis
and invasion of gastric cancer through down-regulating PXN
expression. Am J Cancer Res. 5:2980–2997. 2015.PubMed/NCBI
|
|
19
|
Liu H, Li C, Shen C, Yin F, Wang K, Liu Y,
Zheng B, Zhang W, Hou X, Chen X, et al: MiR-212-3p inhibits
glioblastoma cell proliferation by targeting SGK3. J Neurooncol.
122:431–439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H,
Wang W, Huang X, Yuan Z and Ai K: miR-212 promotes pancreatic
cancer cell growth and invasion by targeting the hedgehog signaling
pathway receptor patched-1. J Exp Clin Cancer Res. 33:542014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qi B, Liu SG, Qin XG, Yao WJ, Lu JG, Guo
L, Wang TY, Li HC and Zhao BS: Overregulation of microRNA-212 in
the poor prognosis of esophageal cancer patients. Genet Mol Res.
13:7800–7807. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Zhang D, Chen C, Ruan Z, Li Y and
Huang Y: MicroRNA-212 displays tumor-promoting properties in
non-small cell lung cancer cells and targets the hedgehog pathway
receptor PTCH1. Mol Biol Cell. 23:1423–1434. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu L and Zhang X, Zhang B, Wu J and Zhang
X: Synaptic acetylcholinesterase targeted by microRNA-212 functions
as a tumor suppressor in non-small cell lung cancer. Int J Biochem
Cell Biol. 45:2530–2540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao JL, Zhang L, Guo X, Wang JH, Zhou W,
Liu M, Li X and Tang H: miR-212/132 downregulates SMAD2 expression
to suppress the G1/S phase transition of the cell cycle and the
epithelial to mesenchymal transition in cervical cancer cells.
IUBMB Life. 67:380–394. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wada R, Akiyama Y, Hashimoto Y, Fukamachi
H and Yuasa Y: miR-212 is downregulated and suppresses
methyl-CpG-binding protein MeCP2 in human gastric cancer. Int J
Cancer. 127:1106–1114. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hatakeyama H, Cheng H, Wirth P, Counsell
A, Marcrom SR, Wood CB, Pohlmann PR, Gilbert J, Murphy B, Yarbrough
WG, et al: Regulation of heparin-binding EGF-like growth factor by
miR-212 and acquired cetuximab-resistance in head and neck squamous
cell carcinoma. PLoS One. 5:e127022010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tu H, Wei G, Cai Q, Chen X, Sun Z, Cheng
C, Zhang L, Feng Y, Zhou H, Zhou B, et al: MicroRNA-212 inhibits
hepatocellular carcinoma cell proliferation and induces apoptosis
by targeting FOXA1. Onco Targets Ther. 8:2227–2235. 2015.PubMed/NCBI
|
|
29
|
Wu Z, Zhou L, Ding G and Cao L:
Overexpressions of miR-212 are associated with poor prognosis of
patients with pancreatic ductal adenocarcinoma. Cancer Biomark.
18:35–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Castillo SD, Matheu A, Mariani N,
Carretero J, Lopez-Rios F, Lovell-Badge R and Sanchez-Cespedes M:
Novel transcriptional targets of the SRY-HMG box transcription
factor SOX4 link its expression to the development of small cell
lung cancer. Cancer Res. 72:176–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bilir B, Osunkoya AO, Wiles WG IV,
Sannigrahi S, Lefebvre V, Metzger D, Spyropoulos DD, Martin WD and
Moreno CS: SOX4 is essential for prostate tumorigenesis initiated
by PTEN ablation. Cancer Res. 76:1112–1121. 2016. View Article : Google Scholar : PubMed/NCBI
|