Introduction
Glioblastoma (GBM) is the most malignant and
prevalent type of primary brain malignancies (1,2). These
tumors are not strictly focal lesions, and disseminate along the
myelinated axons, blood vessels, or through the subarachnoid space
(3), which makes complete resection
difficult. Despite current multimodal treatments, the median
survival time of GBM patients is merely 15 months from the date of
diagnosis and the 5-year survival rate is <10% (4). Clinically, the first-line therapy for
GBM comprises neurosurgery with maximum feasible resection,
followed by alkylating agent based-chemotherapy concomitant with
and after radiotherapy (1,4,5).
Temozolomide, as the most frequently used alkylating agent,
exhibits potent activity against malignant glioma with minimal
additional toxicity. TMZ most often alkylates DNA at the position
of N7 or O6 guanine residues, which induces
DNA double-strand breakage and subsequent apoptosis (6,7).
However, most GBM patients are resistant to this alkylating agent
eventually, greatly compromising the long-term tumor control.
O6 methylguanine-DNA methyltransferase (MGMT), a
cellular DNA repair protein, could rapidly reverse alkylation at
the O6 position of guanine (7,8).
Although a growing number of research indicates that MGMT
expression is responsible for chemoresistance to TMZ in GMB cells,
TMZ-resistant (TR) GBM tissue analysis showed reduced MGMT
expression in >50% of GBM cases (9,10).
Therefore, it is of importance to identify the diverse molecular
mechanisms involved in chemoresistance for finding ways to
sensitize GBM cells to TMZ to ameliorate the therapeutic
efficacy.
MicroRNAs (miRNAs or miRs) are a class of endogenous
small (19–25 nucleotides) non-coding single-stranded RNAs that are
involved in post-transcriptional regulation either by degrading a
specific mRNA(s) or inhibiting translation (11). Through binding to the
3′-untranslated region of mRNAs, miRNAs have been proved to play
crucial roles in a wide range of physiological and pathological
processes, including cancer development and progression (12,13).
Increasing evidence showed that miRNAs are aberrantly expressed in
human cancer tissues, contributing to regulating DNA damage
response, interfering with response to chemotherapy as consequence
(7,14). For example, some studies have found
that many miRNAs, such as miR-20a, miR-195, miR-181b, regulated the
chemosensitivity of glioblastoma cells to TMZ (15–17).
miR-146b-5p, located within 10q24-26, is a frequently missing
chromosomal region in GBM (18–20).
Overexpression of miR-146b-5p could suppress GBM cell invasion and
migration by targeting MMP16, while increasing apoptosis of GBM
cells (18). Furthermore,
upregulation of miR-146b-5p also attenuates cell viability, stem
cell marker expression and neurosphere formation (21). However, little is known about
whether miR-146b-5p is involved in the regulation of the
chemosensitivity of glioblastoma cells to TMZ.
TRAF6, a key member of tumor necrosis factor
receptor-associated factors, plays an important role in the
physiological and pathological processes (22). TRAF6 is characterized to be a signal
transducer of TNFR superfamily and Toll/interleukin-1 (IL-1) family
(23). Furthermore, TRAF6 also
functions as an E3 ubiquitin ligase, catalyzing K63
polyubiquitination of various proteins, such as TAK1 and legumain
(24,25). Elevated expression of TRAF6 has been
found in colon cancer, breast cancer and lung cancer, and TRAF6
promotes proliferation of these cancer cells (25–27).
Liu et al (28) previously
reported that TRAF6 as a direct functional target gene regulated by
miR-146b-5p promoted glioma cell invasion and suppressed apoptosis.
As TMZ-resistant cells usually are characterized by increased
invasiveness and declined apoptosis, the function of TRAF6 in TMZ
resistance deserves to be understood.
Thus, in the present study, we investigated the
expression and function of miR-146b-5p and TRAF6 in GBM cancerous
tissues and TMZ-resistant cell lines. Our results indicated that
miR-146b-5p was down regulated in TMZ-resistant cell lines, and
TRAF6 was a direct functional target of miR-146b-5p. By
upregulation of miR-146b-5p or silencing TRAF6 can sensitize GBM
cells to TMZ. Our studies indicate that miR-146b-5p and TRAF6 may
be potential therapeutic targets for GBM treatment.
Materials and methods
Cell culture
The human GBM cell lines U87-MG and U251-MG
(purchased from the Cell Bank of the Shanghai Branch of the Chinese
Academy of Sciences, 2014) were cultured in Dulbecco's modified
Eagle's medium (Invitrogen, USA) supplemented with 10% fetal bovine
serum (Invitrogen) and maintained in a humidified atmosphere with
5% carbon dioxide-humidified atmosphere at 37°C.
Patients and tissue samples
The study was approved by the Ethics Committee of
the Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong
University. Written consent was obtained from patients or guardians
on behalf of the minors enrolled in the study. Six GBM patients
receiving standard chemoradiotherapy at Ren Ji Hospital, Shanghai
Jiao-Tong University between January 2012 and December 2013 were
recruited and followed for this study. The initial survival time of
patients with primary GBM after standard chemoradiotherapy of less
than six months was designated as resistant to TMZ treatment and
the initial survival time greater than ten months was designated as
sensitive to TMZ.
Generation of temozolomide-resistant
glioblastoma cells
The parental U87-MG and U251-MG cells were treated
with temozolomide at 400 µM or DMSO solvent control at a final
concentration of 0.1% for 4 weeks. In brief, to generate
TMZ-resistant GBM cells, cells were treated with fresh TMZ every
day for 5 consecutive days and then exposed to the fresh TMZ every
3 days to a total of 3 weeks. Survival cells were continued its
culture for 4 weeks until deriving stable resistant cell lines
(TMZ-resistant U251 cells (U251-TR) and U87 cells (U87-TR) for
subsequent experiments.
Western blot analysis
To analyze the expression of proteins, western blot
assays were performed using the following primary antibodies:
rabbit anti-human TRAF6 (1:1,000 dilution, ab33915; Abcam, USA),
AKT (1:1,000 dilution, 9272; Cell Signaling Technology), p-AKT
(1:1,000 dilution, 4060; Cell Signaling Technology), P65 (1:1,000
dilution, 8242; Cell Signaling Technology), p-p65 (1:1,000
dilution, 3033; Cell Signaling Technology) and mouse anti-actin
(1:10,000; Millipore). Briefly, tissues were lysed with RIPA buffer
[50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.5%
Na-deoxycholate] containing protease inhibitors (Complete, Mini;
Roche); 20–30 µg samples of the lysates were separated on 8–12%
SDS-PAGE gels and transferred to PVDF membranes. The membranes were
incubated with primary antibodies overnight at 4°C. The primary
antibody incubation was followed by incubation with an
HRP-conjugated secondary antibody. The bound antibodies were
detected using an ECL kit (PI32209; Pierce).
cDNA synthesis and real-time PCR
reaction
Total RNA was extracted from cells using TRIzol
reagent (Life Technology, USA). After that, mRNA was reverse
transcribed using SuperScript First-Strand Synthesis-System for
RT-PCR (Invitrogen) according to the manufacturer's instructions.
Real-time PCR was performed using SYBR Green reagents (Takara,
Japan) containing 100 µM final concentration of primers (primer
sequences were listed in Table I).
PCR conditions were: 95°C for 15 sec, followed by 40 cycles of 95°C
for 5 sec, 60°C for 30 sec. PCRs were performed in triplicates
using the Roche LightCycler 480® RT-PCR system
(Roche).
 | Table I.Sequences of qPCR primers. |
Table I.
Sequences of qPCR primers.
| Primer name | Sequence |
|---|
| Human-TRAF6-F |
5′-TGATGTAGAGTTTGACCCACCC-3′ |
| Human-TRAF6-R |
5′-GTCAACTGGACATTTGTGACCTG-3′ |
| hsa-miR146b-5p |
TGAGAACTGAATTCCATAGGCT |
Lentivirus-mediated gene knockdown and
overexpression
The target sequences for TRAF6 siRNAs were
5′-GCTCTTGGTGGATCATCAA-3′ (TRAF6-KD1) and
5′-GCATGTTCAATGGGAGCTTGGA-3′ (TRAF6-KD2). After 48 h, the
efficiency of TRAF6 knockdown was confirmed via quantitative
real-time polymerase chain reaction (qRT-PCR) and western blot
analysis.
Lentiviral vectors for human TRAF6-shRNA carrying a
green fluorescent protein (GFP) sequence were constructed by Hanyin
Co. (Shanghai, China). The recombinant TRAF6 knockdown lentivirus
and the negative control (NC) lentivirus (GFP-lentivirus; Hanyin
Co. Shanghai, China) were prepared and titered to 109
TU/ml (transfection unit). To obtain the stable TRAF6-knockdown
cell line, cells were seeded in 6-well dishes at a density of
2×105 cells per well. The cells were then infected with
the same titer virus with 8 µg/ml polybrene the following day.
Approximately 72 h after viral infection, GFP expression was
confirmed under a fluorescence microscope, and the culture medium
was replaced with selection medium containing 4 µg/ml puromycin.
The cells were then cultured for ≥14 days. The puromycin-resistant
cell clones were isolated, amplified in medium containing 2 µg/ml
puromycin for 7–9 days, and transferred to a medium without
puromycin. The clones were designated as TRAF6-KD or NC cells.
TRAF6 was cloned into a pMSCV–IRES-GFP vector. For overexpression
of TRAF6, GBM cells were transfected by viral supernatant from 293T
cells transfected with TRAF6 or control vector. The clones were
designated as TRAF6-OE or NC cells.
CCK8
A CCK8 assay was conducted according to the kit
instructions (DH343-2, Beijing Dongsheng Biotechnology, China).
Briefly, cells were plated at equal cell density (2,000 cell/100 µl
per well) in 96-well plates with cetuximab (300 µg/ml) for
continuous detection over a 5-day period. At the beginning of the
second day, the culture was terminated by adding 10 µl CCK8 (5
mg/ml) to the original culture medium. After 2 h, the plates were
measured using a microplate reader (Elx800; Biotek, USA). Cell
proliferation was measured using OD490 values.
Construction of luciferase reporter
vectors
The TRAF6 gene 3′-UTR was generated by inserting the
3′ flanking region of the TRAF6 gene into pGL3-basic vector
(Promega, Fitchburg, WI, USA). To identify the critical miRNA
binding region of the TRAF6, subsequently, mutated luciferase
reporter constructs were generated by PCR. All the constructs were
verified by sequencing to rule out the possibility of any PCR
error.
Transient transfections and luciferase
assays
Cells were seeded in 96-well plates and grown to a
density of 80% confluence before transfection. Each luciferase
reporter construct (100 ng) and 0.3 µl Lipofectamine™ 2000
(Invitrogen) reagent in Opti-MEM were transfected according to the
manufacturer's instructions. In order to correct transfection
efficiency, additional 4 ng of the pRL-TK vector which contained
the Renilla luciferase gene (hRluc) under the control of the
Herpes simplex virus thymidine kinase promoter was co-transfected
in each well. The cells were harvested after 48-h transfection, and
then lysed with 20 µl passive lysis buffer for each well (Promega).
Luciferase activity was measured using Dual-Luciferase Reporter
assay system (Promega) on a luminescence reader (Acuu FLEX Lumi
400; Aloka, Tokyo, Japan).
The nude mouse tumor xenograft
model
For brain xenografts, briefly, 1×106
U87-TR cells with or without miR-146b-5p were stereotactically
injected into the brains of the 6-week-old athymic nude mouse. A
1-mm drill was then used to make a burr hole 1 mm anterior of the
bregma and 2 mm to the right of the skull. A 21-gauge Hamilton
syringe was advanced 3.5 mm deep, and then retracted 3 mm for
implanting 5 µl of tumor cell suspension. After 1 week, half of the
mice in each group were treated by TMZ by tail vein injection. Mice
were monitored for the development of tumors by magnetic resonance
imaging (MRI). The animal study was approved by the Institutional
Animal Care and Use Committee of Renji Hospital, Shanghai Jiao Tong
University.
Statistical analysis
All statistical analyses were performed using SPSS
for Windows v.17.0 (SPSS, Chicago, IL, USA). All results were
considered significant at two-sided p<0.05 value.
Results
TRAF6 was upregulated in TMZ-resistant
GBM cells and tissues
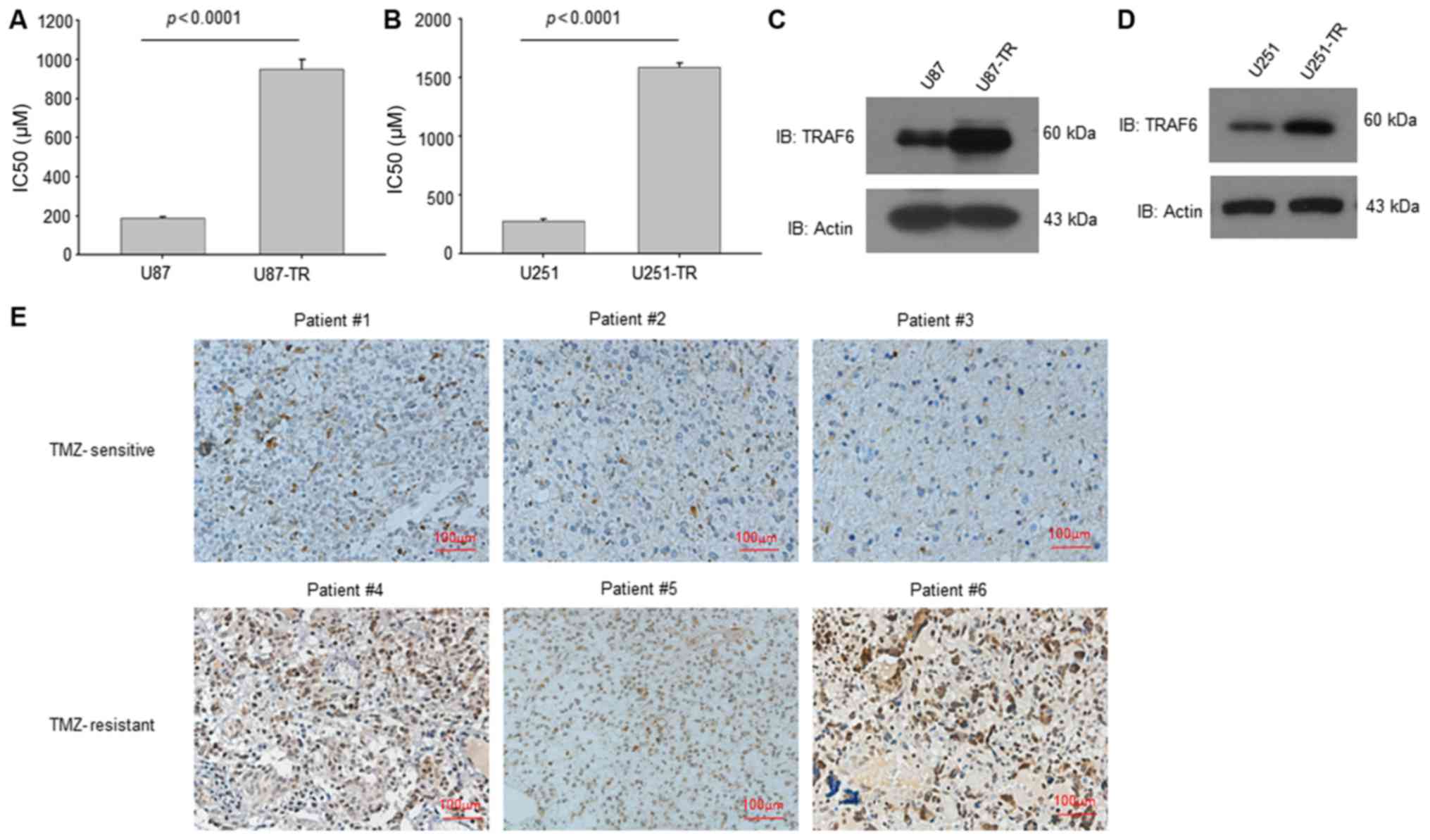
We established TMZ-resistant GBM cells (U87-TR and
U251-TR) with gradually elevated TMZ concentration in wild-type GBM
cells (U87 and U251). The IC50 value of resistant cells
increased nearly 5-fold compared with parental wild-type cells
(Fig. 1A and B). The western
blotting results showed that TRAF6 expression was upregulated in
both TMZ-resistant GBM cells (Fig. 1C
and D). Further we collected specimens from GBM patients with
different response to TMZ treatment. The immunohistochemistry
staining results showed that the expression of TRAF6 was higher in
TMZ-resistant GBM tissues than that of TMZ sensitive GBM tissues
(Fig. 1E). Taken together, these
results suggest that TRAF6 was upregulated in TMZ-resistant GBM
cells and cancerous tissues.
Knockdown of TRAF6 in TMZ-resistant
GBM cells reduced the TMZ resistance
We then knocked down TRAF6 in U87-TR and U251-TR
cells. Both the RT-PCR and western blotting results indicated that
TRAF6 were efficiently suppressed by shRNAs (Fig. 2A-D). The CCK8 analysis revealed that
knockdown of TRAF6 significantly reduced cell proliferation ability
(Fig. 2E and F). CCK8 assay of
IC50 of these cells showed that knockdown of TRAF6 in
TMZ-resistant GBM cells reduced the TMZ resistance (Fig. 2G and H).
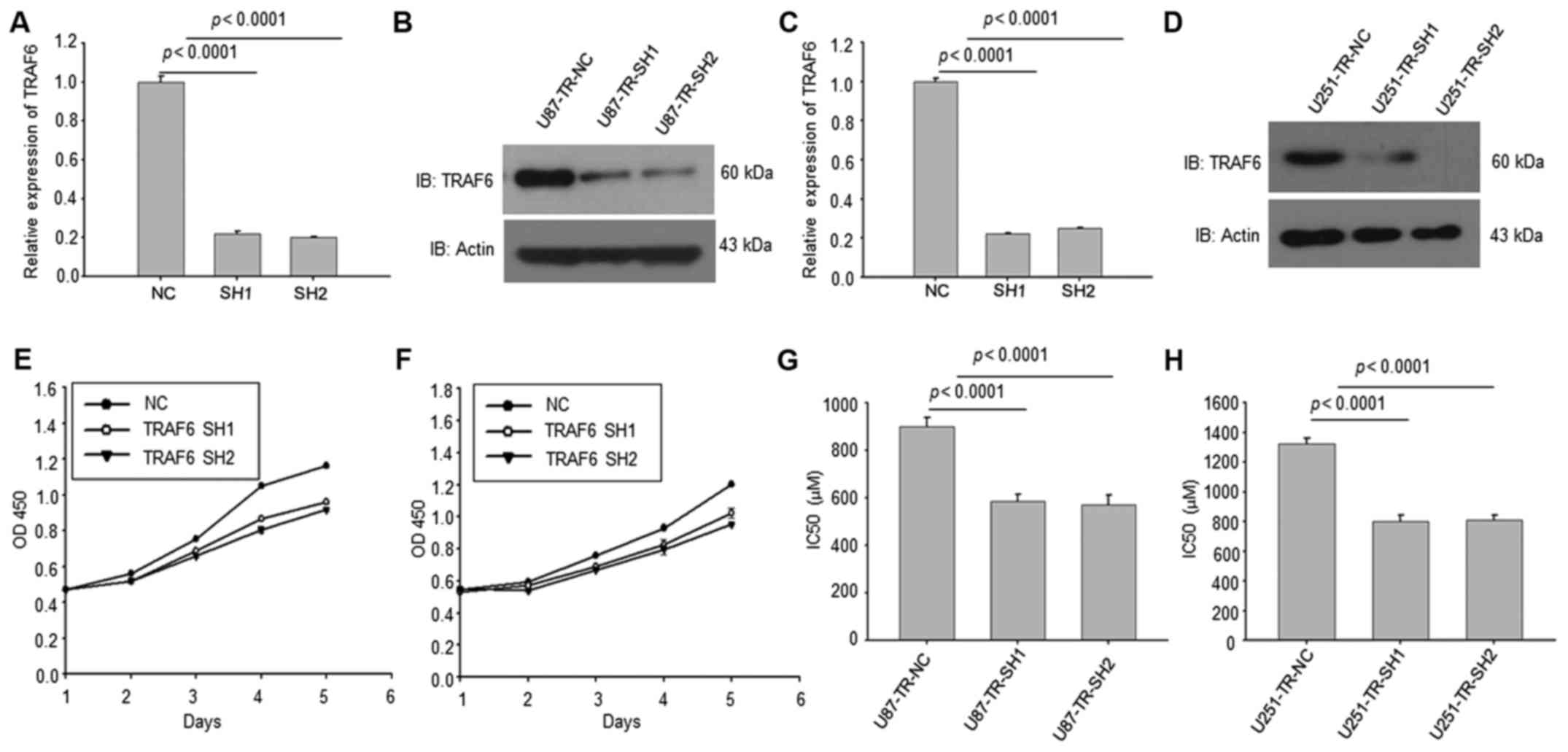 | Figure 2.TRAF6 knockdown reverses sensitivity
of TR cells to TMZ. (A-D) By transfecting shRNAs into TR cells,
western blot analysis and RT-PCR results showed mRNA and protein
level of TRAF6 decreased efficiently compared to control group,
p<0.0001. (E and F) The CCK8 analysis revealed that knockdown of
TRAF6 reduced cell proliferation ability. (G and H) With the
knockdown of TRAF6 in TR cells, the IC50 value decreased
obviously compared to control group. All experiments were performed
in triplicate and the data in (A), (C) and (E-H) are presented as
mean ± SD, p<0.0001. TRAF6, tumor necrosis factor receptor
associated factor 6; TR, temozolomide-resistant; TMZ, temozolomide;
shRNAs, short hairpin RNA; RT-PCR, real-time polymerase chain
reaction; CCK8, cell counting kit-8; IC50, half maximal
inhibitory concentration. |
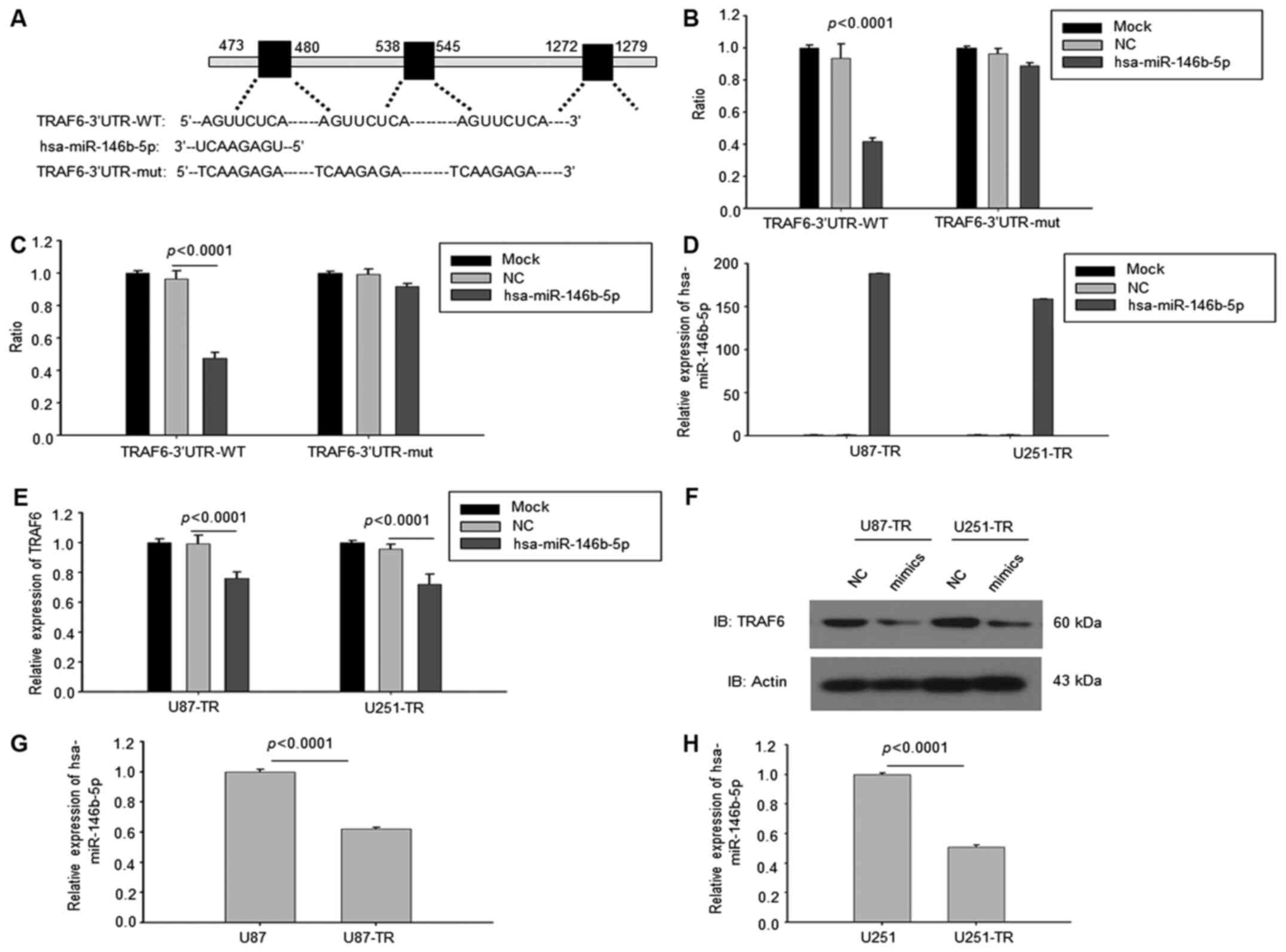
Downregulated miR-146b-5p targeted
TRAF6 in temozolomide-resistant GBM cells
We further searched for the upstream regulator of
TRAF6 in TMZ-resistant GBM cells. Analyzing the potential miRNA
binding to the 3′-UTR of TRAF6 by TargetScan found that there were
three binding motifs to miR-146b-5p, suggesting that miR-146b-5p
might be the regulator of TRAF6 (Fig.
3A). According to the results of luciferase reporter assay,
overexpression of miR-146b-5p inhibited the luciferase activity
while mutated the conserved binding sequences of miR-146b-5p,
miR-146b-5p failed to inhibit the luciferase activity in U87-TR and
U251-TR cells (Fig. 3B and C). The
overexpression of miR-146b-5p was validated by RT-PCR in two cell
lines (Fig. 3D). Consistent with
these results, the mRNAs of TRAF6 were downregulated in miR-146b-5p
overexpressed U87-TR and U251-TR cells (Fig. 3E). The western blotting results also
showed that TRAF6 was reduced in miR-146b-5p overexpressed U87-TR
and U251-TR cells (Fig. 3F).
Finally the expression of miR-146b-5p was examined in GBM wild-type
cells and TMZ-resistant cells. The RT-PCR results showed that the
expression of miR-146b-5p was downregulated in U87-TR and U251-TR
cells (Fig. 3G and H). Thus, all of
these results demonstrated that downregulated miR-146b-5p targeted
TRAF6 in temozolomide-resistant GBM cells.
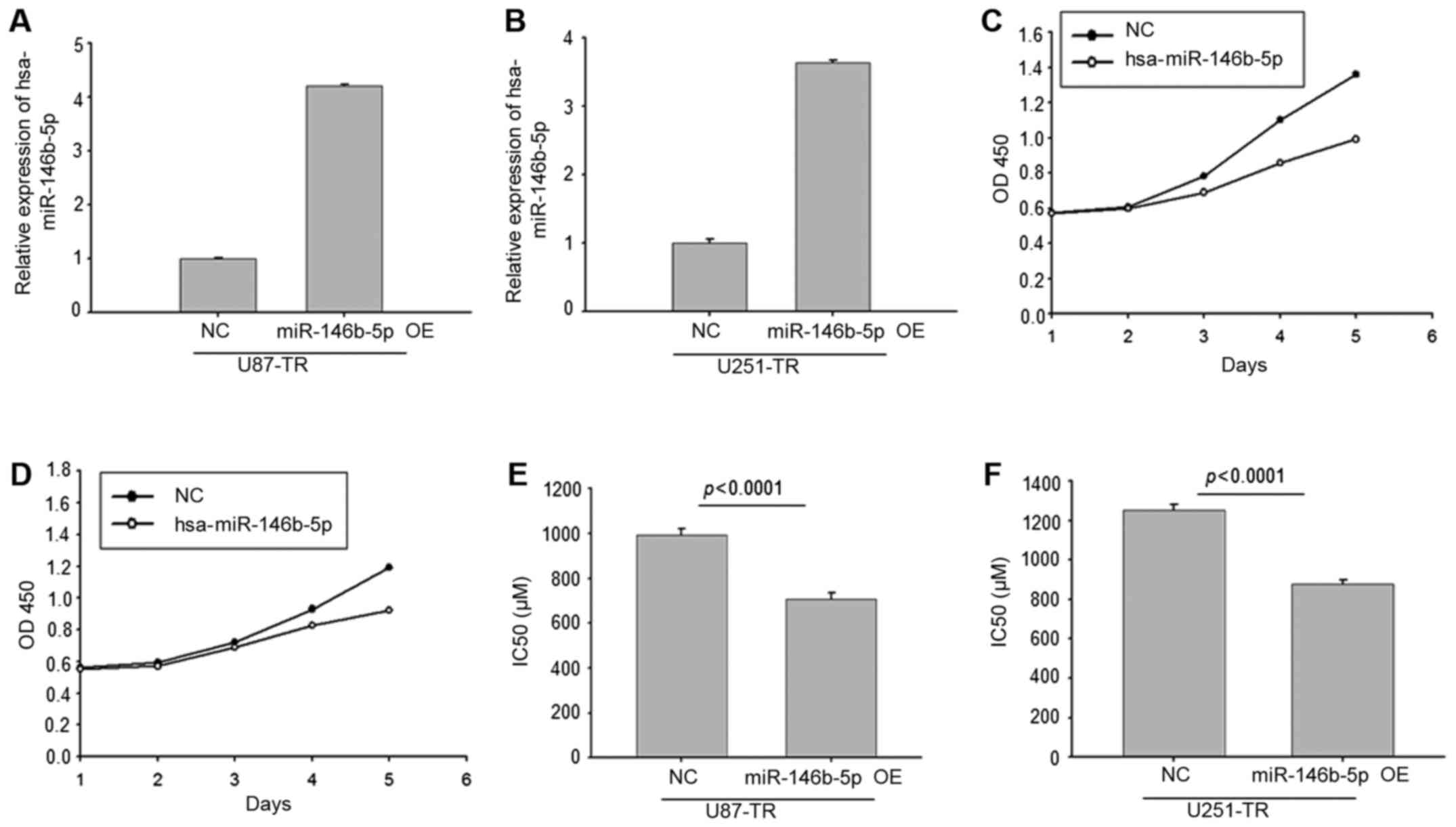
Overexpression of miR-146b-5p in
TMZ-resistant GBM cells reduced the resistance to TMZ
We then constructed miR-146b-5p overexpressing
TMZ-resistant GBM cells by lentivirus. The RT-PCR results validated
that miR-146b-5p was overexpressed in U87-TR and U251-TR cells
(Fig. 4A and B). The CCK8 analysis
revealed that overexpression of miR-146b-5p significantly reduced
cell proliferation ability (Fig. 4C and
D). Moreover, the comparison of IC50 showed that
overexpression of miR-146b-5p in TMZ-resistant GBM cells reduced
the TMZ resistance (Fig. 4E and F).
These results revealed that miR-146b-5p was a negative regulator of
TMZ resistance in GBM cells.
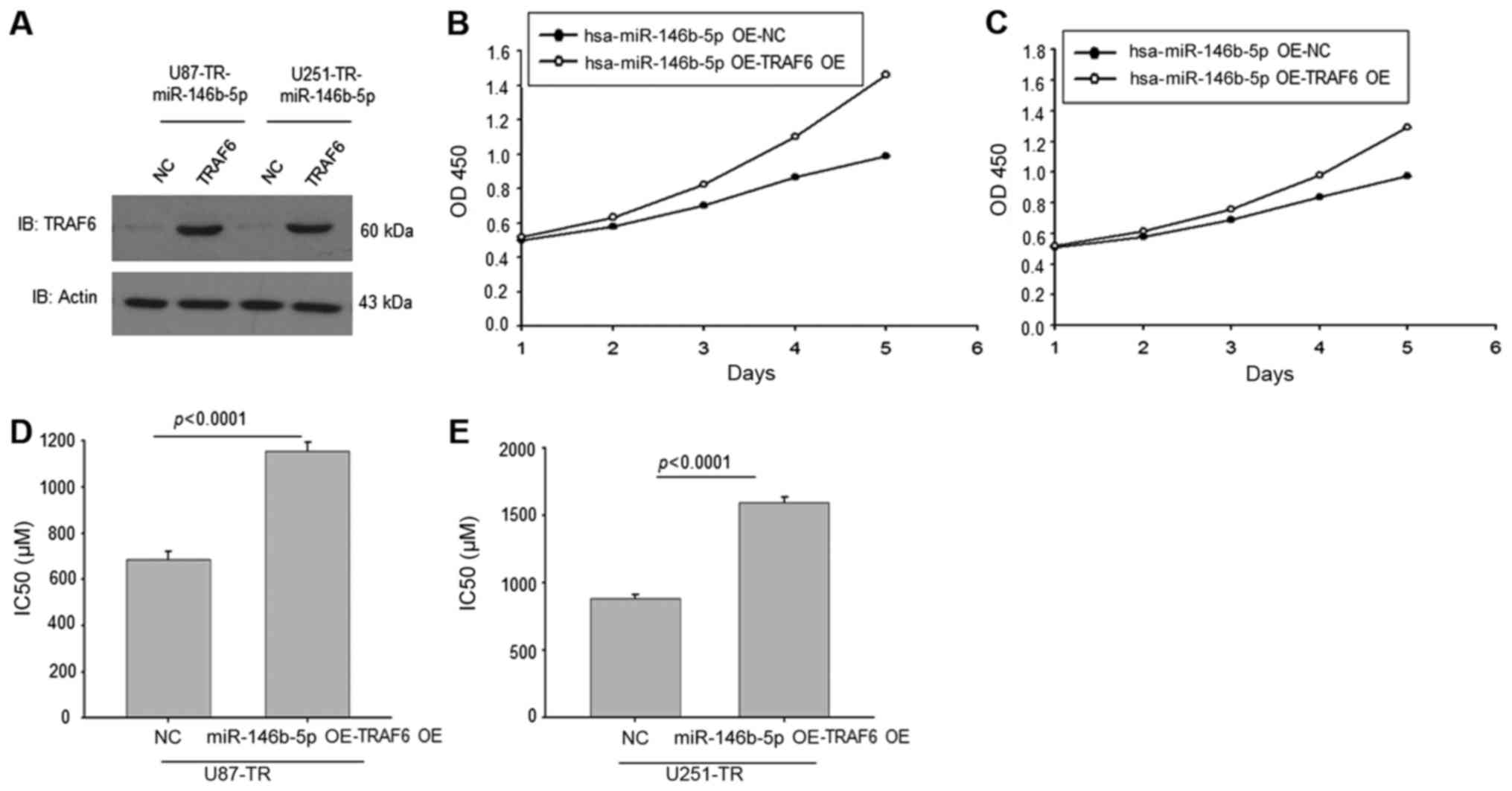
Overexpression of TRAF6 in
TMZ-resistant miR-146b-OE GBM cells elevated the IC50 of
TMZ
As we have found that miR-146b-5p targeted TRAF6 in
TMZ-resistant GBM cells, we overexpressed TRAF6 in TMZ-resistant
miR-146b-OE GBM cells. The western blotting result showed that
TRAF6 expression was elevated in these cells compared to the NC
control (Fig. 5A). The CCK8
analysis revealed that overexpression of TRAF6 recued the cell
proliferation ability in miR-146b-5p-OE cells (Fig. 5B and C). Calculation of the
IC50 of these cells showed that overexpression of TRAF6
in miR-146b-5p-OE GBM cells regained the TMZ resistance (Fig. 5D and E). Thus, these data revealed
that downregulated miR-146b-5p increased TMZ-resistance might
partially be through targeting TRAF6.
miR-146b-5p enhances the
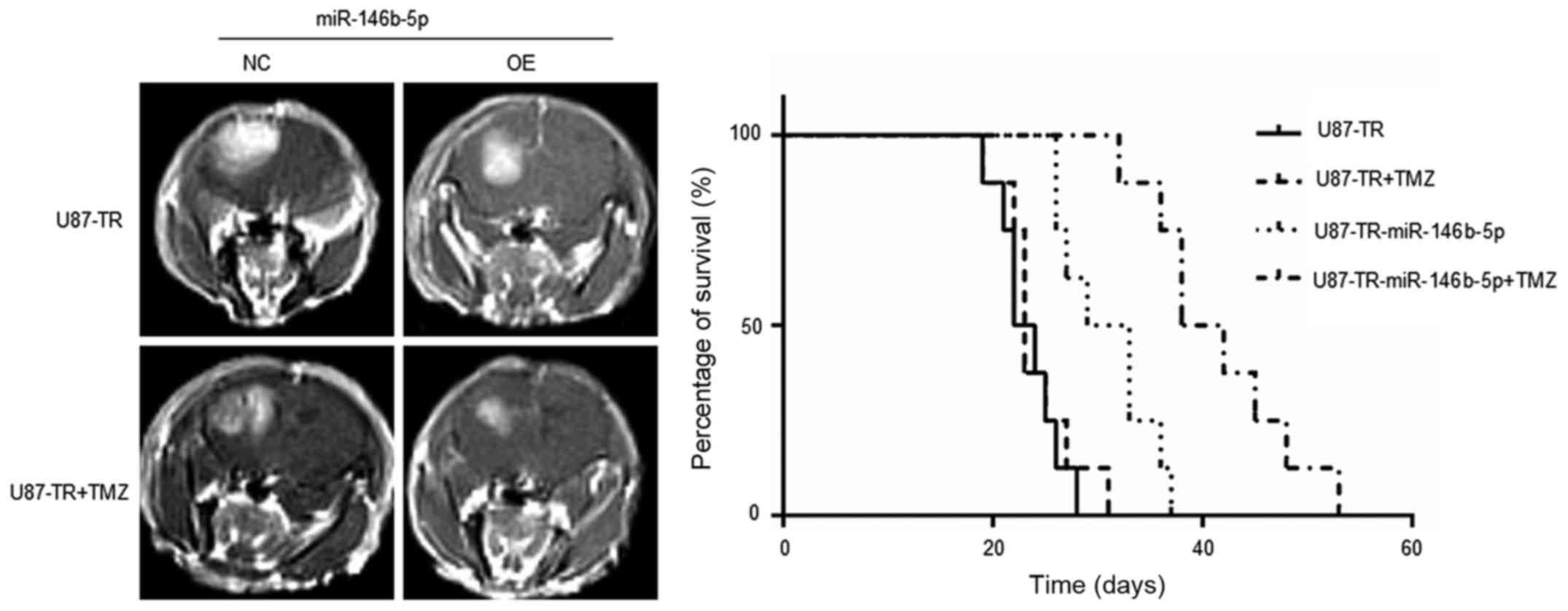
chemo-sensitivity of TMZ in vivo
To assess the functions of miR-146b-5p in GBM
chemo-sensitivity in vivo, an orthotopic xenotransplanted
GBM model was employed. U87-TR cells overexpressing miR-NC or
miR-146b-5p were stereotactically injected into the brain of the
athymic nude mouse, respectively. After 1 week, the miR-NC group
was randomly divided into two groups: miR-NC and miR-NC+TMZ. TMZ
was delivered to the miR-NC+TMZ group by tail vein injection. The
same procedure was also applied to the miR-146b-5p overexpression
group. When a few mice started to show signs of morbidity, mice in
each experimental group were assessed by MRI to confirm
intracranial tumor formation. The survival times of every mouse of
each group were recorded and analyzed by the Kaplan-Meier method.
The results demonstrated that with TMZ treatment, mice in the
miR-146b-5p-overexpressing group survived longer than the control
group (Fig. 6). These data
demonstrated that miR-146b-5p enhances the sensitivity of TMZ
chemotherapy of GBM cells in vivo.
miR-146b-5p/TRAF6 regulates AKT and
NF-κB pathway activation in TMZ-resistant GBM cells
TRAF6 has been found regulating the AKT and NF-κB
pathway in cancer. We analyzed both pathway in TMZ-resistant GBM
cells. Compared to the wild-type, phosphorylated Akt and p65 were
increased in U87-TR and U251-TR cells (Fig. 7A and B). Knockdown of TRAF6 in
U87-TR and U251-TR cells reduced the activation of Akt and p65
(Fig. 7C and D). Moreover,
overexpression of miR-146b-5p in U87-TR and U251-TR cells
consistently reduced the activation of Akt and p65 (Fig. 7E), and overexpression of TRAF6 in
miR-146b-5p-OE TR cells would reverse this effect (Fig. 7F). Altogether, miR-146b-5p/TRAF6
regulates AKT and NF-κB (nuclear factor-κB) pathway activation in
TMZ-resistant GBM cells.
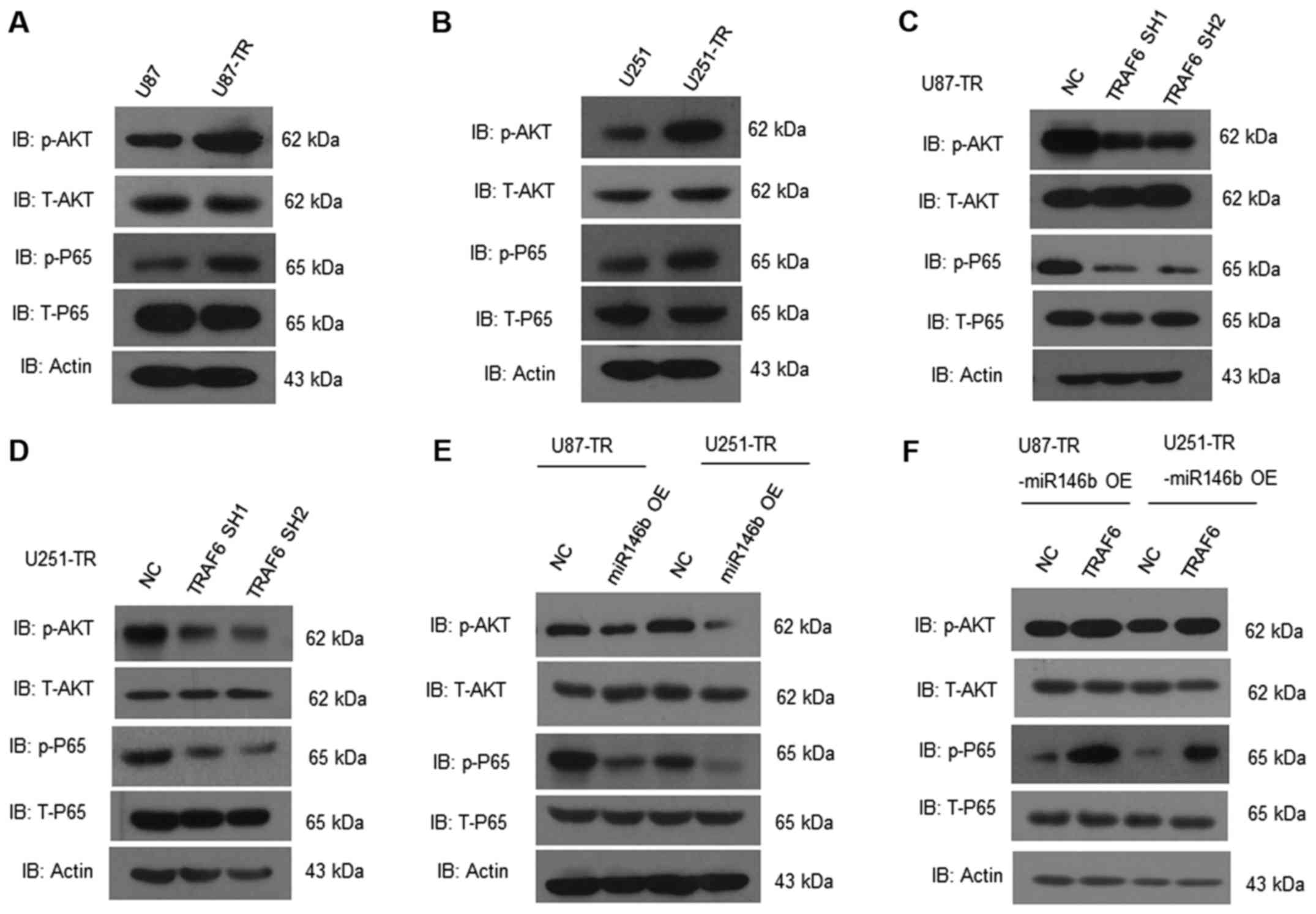 | Figure 7.miR-146b-5p and TRAF6 knockdown
suppress the activation of AKT/NF-κB pathway. (A and B) Western
blot analyses of p-AKT, T-AKT, p-P65, T-P65 in U87, U87-TR, U251,
U251-TR cells. (C and D) Expression of p-AKT, T-AKT, p-P65, T-P65
in U87/U251-TR cells and TRAF6 knockdown U87/U251-TR cell were
analyzed by western blot analysis. (E) Above proteins were analyzed
in miR-146b-5p-OE U87/U251-TR cells and control group by western
blot analysis. (F) With overexpression of TRAF6 in miR-146b-5p-OE
U87/U251-TR cells, relative proteins of AKT/NF-κB pathway were
activated. Data are representative of three independent
experiments. AKT, protein kinase B; NF-κB, nuclear factor-κB; TR,
temozolomide-resistant; OE, overexpression. |
Discussion
The regimen of postoperative usage of temozolomide
combined with radiotherapy has become the standard therapy for GBM,
which significantly improves the overall survival for patients with
glioma (1). However, TMZ resistance
rapidly progresses among most patients, becoming the main reason
why treatment fails and tumor recurs. Besides the critical role of
MGMT in enhancing TMZ resistance, other mechanisms mediating
intrinsic or acquired resistance to TMZ have also been recognized.
As previous studies reported, epithelial-to-mesenchymal transition,
re-organization of the cytoskeleton, activation of pro-survival
pathway genes, disturbance of the mismatch repair system,
autophagy, glioma stem cells, and abnormal expression of microRNAs
were closely associated with temozolomide resistance (10,15–17,29–34).
In the present study, we acquired a stable TMZ-resistant cell line,
in which miR-146b-5p was significantly downregulated and
upregulated expression can sensitize GBM cells to TMZ.
MicroRNAs have been found to play critical oncogenic
roles in tumor development and drug resistance. Some previous
studies have reported the relationship between miRNAs and TMZ
resistance in GBM. Overexpression of miR-17, miR-20a, and miR-181b
sensitizes the glioma cells to TMZ-induced cytotoxicity, while
upregulation of miR-195, miR-29c, miR-221/222 enhances TMZ
resistance (7,15–17,35,36).
However, the function of miR-146b-5p in TMZ resistance has not been
recognized. Previous studies demonstrated that miR-146b-5p was
downregulated in recurrent GBM samples compared to that of primary
ones, indicating its function in treatment resistance (37). Moreover, by binding to
3′-untranslated region of EGFR mRNA, miR-146b-5p suppresses the
stemness and induces differentiation of GSCs, which also has a good
part in TMZ resistance (21). In
this study, we found that miR-146b-5p was downregulated in
TMZ-resistant cells, and overexpression of miR-146b-5p conferred
sensitivity to TMZ on GBM cells by targeting TRAF6. Although a
previous study has reported that low expression of miR-146b-5p
would enhance the glioma cell invasive capacity and correlate with
poor prognosis for glioma patients (28), its role in TMZ resistance was first
clarified. Since the miR-146b-5p also involves in modulating other
target genes including MMP16 and EGFR (18,38),
its mechanisms of TMZ resistance need to be further investigated in
the future.
TRAF6, as the E3 ubiquitin ligase, takes part in
promoting oncogenesis. Overexpression of TRAF6 in mouse marrow
cells leads to a myelodysplastic syndrome, which develops into a
fatal acute myeloid leukemia eventually (39). It promotes proliferation and
regulates apoptosis in breast cancer, osteosarcoma and lung
adenocarcinoma cells (25,26,40).
Furthermore, overexpression of TRAF6 enhances the resistance of
colon cancer cells to 5-fluorouracil and acute leukemia cells to
bortezomib (27). In our study, we
found that TRAF6, as a target gene of miR-146b-5p, not only played
a substantial role in tumor proliferation ability, but also
contributed TMZ resistance to GBM cells. Silencing of TRAF6 could
mimic the antitumor effect of miR-146b-5p through reversing TMZ
resistance and decreasing proliferation in GBM cells. Activation of
some signaling pathways including AKT and NF-κB has been found to
be involved in TRAF6-mediated oncogenesis (26,28,41).
Constitutive activation of AKT/NF-κB pathway is frequently observed
in different types of cancer and contributes to tumor progression,
and chemoresistance (42–44). A previous study in metastatic
melanoma cell line demonstrated that management of TMZ could
activate the AKT/NF-κB, while inhibiting the activity of AKT and
NF-κB could sensitize cancer cells to TMZ (42). In order to confirm whether
miR-146b-5p targeted TRAF6 overexpression contributes to TMZ
resistance in glioblastoma by activating the AKT/NF-κB pathway,
signaling array experiments were conducted. Western blot analysis
revealed that TR cells had a higher level of p-AKT and p-p65 which
is an indicator of NF-κB activation compared to control group, and
overexpression of miR-146b-5p or TRAF6 knockdown caused a marked
reduction p-AKT and p-p65. It is well known that AKT/NF-κB
signaling pathway plays an essential role in regulation of
autophagy which also contributes to TMZ resistance (33,45).
However, whether autophagy is regulated by miR-146b-5p targeted
TRAF6 through AKT/NF-κB pathway in TMZ resistance is under
investigation in our lab.
In conclusion, the results of the present study
showed that TRAF6, the target gene of miR-146b-5p, was highly
expressed in TR cells and associated with TMZ resistance. The
integrated analyses depending on miRNAs and their related
regulatory networks provide new insights into addressing TMZ
resistance in GBM, and miR-146b-5p may serve as a novel and
potential therapeutic agent for overcoming TMZ resistance in
patients with gliomas.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81402042), Shanghai Science and
Technology (14140903400, 14YF1402600), the State Key Laboratory of
Oncogenes and Related Genes (no. 90-14-01), the Shanghai Municipal
Population and Family Planning Commission (2013SY024), the Key
Specialty Construction Project and Science Technology Development
Project of the Pudong Health and Family Commission of Shanghai
(nos. PWZz2013-18 and PW2013A-19), and the Training Plan for
Scientific Research of Renji Hospital (RJZZ13-021).
References
|
1
|
Stupp R, Mason WP, Van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups, ; National Cancer Institute of
Canada Clinical Trials Group, : Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Walecki J, Tarasów E, Kubas B, Czemicki Z,
Lewko J, Podgórski J, Sokól M and Grieb P: Hydrogen-1 MR
spectroscopy of the peritumoral zone in patients with cerebral
glioma: Assessment of the value of the method. Acad Radiol.
10:145–153. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Hegi ME, Mason WP, Van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al European Organisation for Research and Treatment
of Cancer Brain Tumour and Radiation Oncology Groups, ; National
Cancer Institute of Canada Clinical Trials Group, : Effects of
radiotherapy with concomitant and adjuvant temozolomide versus
radiotherapy alone on survival in glioblastoma in a randomised
phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet
Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stupp R and Hegi ME: Brain cancer in 2012:
Molecular characterization leads the way. Nat Rev Clin Oncol.
10:69–70. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Knizhnik AV, Roos WP, Nikolova T, Quiros
S, Tomaszowski KH, Christmann M and Kaina B: Survival and death
strategies in glioma cells: Autophagy, senescence and apoptosis
triggered by a single type of temozolomide-induced DNA damage. PLoS
One. 8:e556652013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao S, Yang Z, Qiu X, Lv R, Liu J, Wu M,
Liao Y and Liu Q: miR-29c contribute to glioma cells temozolomide
sensitivity by targeting O6-methylguanine-DNA
methyltransferases indirectely. Oncotarget. 7:50229–50238; Epub
ahead of print. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hermisson M, Klumpp A, Wick W, Wischhusen
J, Nagel G, Roos W, Kaina B and Weller M:
O6-methylguanine DNA methyltransferase and p53 status
predict temozolomide sensitivity in human malignant glioma cells. J
Neurochem. 96:766–776. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang K, Wang XQ, Zhou B and Zhang L: The
prognostic value of MGMT promoter methylation in glioblastoma
multiforme: A meta-analysis. Fam Cancer. 12:449–458. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Feng W, Lu Y, Li H, Xiang W, Chen
Z, He M, Zhao L, Sun X, Lei B, et al: Expression of dynein,
cytoplasmic 2, heavy chain 1 (DHC2) associated with glioblastoma
cell resistance to temozolomide. Sci Rep. 6:289482016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
14
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: Predictors and modifiers of chemo- and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei J, Qi X, Zhan Q, Zhou D, Yan Q, Wang
Y, Mo L, Wan Y, Xie D, Xie J, et al: miR-20a mediates
temozolomide-resistance in glioblastoma cells via negatively
regulating LRIG1 expression. Biomed Pharmacother. 71:112–118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ujifuku K, Mitsutake N, Takakura S,
Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K,
Nagata I, et al: miR-195, miR-455-3p and miR-10a(*) are implicated
in acquired temozolomide resistance in glioblastoma multiforme
cells. Cancer Lett. 296:241–248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Sai K, Chen FR and Chen ZP:
miR-181b modulates glioma cell sensitivity to temozolomide by
targeting MEK1. Cancer Chemother Pharmacol. 72:147–158. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Wang Y, Yu L, Sun C, Cheng D, Yu S,
Wang Q, Yan Y, Kang C, Jin S, et al: miR-146b-5p inhibits glioma
migration and invasion by targeting MMP16. Cancer Lett.
339:260–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Thuijl HF, Scheinin I, Sie D, Alentorn
A, van Essen HF, Cordes M, Fleischeuer R, Gijtenbeek AM, Beute G,
Van den Brink WA, et al: Spatial and temporal evolution of distal
10q deletion, a prognostically unfavorable event in diffuse
low-grade gliomas. Genome Biol. 15:4712014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weller M, Weber RG, Willscher E, Riehmer
V, Hentschel B, Kreuz M, Felsberg J, Beyer U, Löffler-Wirth H,
Kaulich K, et al: Molecular classification of diffuse cerebral WHO
grade II/III gliomas using genome- and transcriptome-wide profiling
improves stratification of prognostically distinct patient groups.
Acta Neuropathol. 129:679–693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang W, Yu H, Shen Y, Liu Y, Yang Z and
Sun T: MiR-146b-5p overexpression attenuates stemness and
radioresistance of glioma stem cells by targeting
HuR/lincRNA-p21/β-catenin pathway. Oncotarget. 7:41505–41526. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu H, Tamashiro S, Baritaki S, Penichet
M, Yu Y, Chen H, Berenson J and Bonavida B: TRAF6 activation in
multiple myeloma: A potential therapeutic target. Clin Lymphoma
Myeloma Leuk. 12:155–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Inoue J, Gohda J and Akiyama T:
Characteristics and biological functions of TRAF6. Adv Exp Med
Biol. 597:72–79. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen ZJ: Ubiquitination in signaling to
and activation of IKK. Immunol Rev. 246:95–106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin Y, Qiu Y, Xu C, Liu Q, Peng B,
Kaufmann GF, Chen X, Lan B, Wei C, Lu D, et al: Functional role of
asparaginyl endopeptidase ubiquitination by TRAF6 in tumor invasion
and metastasis. J Natl Cancer Inst. 106:dju0122014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Starczynowski DT, Lockwood WW, Deléhouzée
S, Chari R, Wegrzyn J, Fuller M, Tsao MS, Lam S, Gazdar AF, Lam WL,
et al: TRAF6 is an amplified oncogene bridging the RAS and NF-κB
pathways in human lung cancer. J Clin Invest. 121:4095–4105. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun H, Li X, Fan L, Wu G, Li M and Fang J:
TRAF6 is upregulated in colon cancer and promotes proliferation of
colon cancer cells. Int J Biochem Cell Biol. 53:195–201. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu J, Xu J, Li H, Sun C, Yu L, Li Y, Shi
C, Zhou X, Bian X, Ping Y, et al: miR-146b-5p functions as a tumor
suppressor by targeting TRAF6 and predicts the prognosis of human
gliomas. Oncotarget. 6:29129–29142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yi GZ, Liu YW, Xiang W, Wang H, Chen ZY,
Xie SD and Qi ST: Akt and β-catenin contribute to TMZ resistance
and EMT of MGMT negative malignant glioma cell line. J Neurol Sci.
367:101–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim SH, Joshi K, Ezhilarasan R, Myers TR,
Siu J, Gu C, Nakano-Okuno M, Taylor D, Minata M, Sulman EP, et al:
EZH2 protects glioma stem cells from radiation-induced cell death
in a MELK/FOXM1-dependent manner. Stem Cell Rep. 4:226–238. 2015.
View Article : Google Scholar
|
|
31
|
Shi L, Fei X, Wang Z and You Y: PI3K
inhibitor combined with miR-125b inhibitor sensitize TMZ-induced
anti-glioma stem cancer effects through inactivation of
Wnt/β-catenin signaling pathway. In Vitro Cell Dev Biol Anim.
51:1047–1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fukushima T, Takeshima H and Kataoka H:
Anti-glioma therapy with temozolomide and status of the DNA-repair
gene MGMT. Anticancer Res. 29:4845–4854. 2009.PubMed/NCBI
|
|
33
|
Yan Y, Xu Z, Dai S, Qian L, Sun L and Gong
Z: Targeting autophagy to sensitive glioma to temozolomide
treatment. J Exp Clin Cancer Res. 35:232016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun YC, Wang J, Guo CC, Sai K, Wang J,
Chen FR, Yang QY, Chen YS, Wang J, To TS, et al: MiR-181b
sensitizes glioma cells to teniposide by targeting MDM2. BMC
Cancer. 14:6112014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Comincini S, Allavena G, Palumbo S, Morini
M, Durando F, Angeletti F, Pirtoli L and Miracco C: microRNA-17
regulates the expression of ATG7 and modulates the autophagy
process, improving the sensitivity to temozolomide and low-dose
ionizing radiation treatments in human glioblastoma cells. Cancer
Biol Ther. 14:574–586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen L, Zhang J, Han L, Zhang A, Zhang C,
Zheng Y, Jiang T, Pu P, Jiang C and Kang C: Downregulation of
miR-221/222 sensitizes glioma cells to temozolomide by regulating
apoptosis independently of p53 status. Oncol Rep. 27:854–860.
2012.PubMed/NCBI
|
|
37
|
Bo LJ, Wei B, Li ZH, Wang ZF, Gao Z and
Miao Z: Bioinformatics analysis of miRNA expression profile between
primary and recurrent glioblastoma. Eur Rev Med Pharmacol Sci.
19:3579–3586. 2015.PubMed/NCBI
|
|
38
|
Katakowski M, Zheng X, Jiang F, Rogers T,
Szalad A and Chopp M: MiR-146b-5p suppresses EGFR expression and
reduces in vitro migration and invasion of glioma. Cancer Invest.
28:1024–1030. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Starczynowski DT, Kuchenbauer F,
Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra
M, Wells RA, et al: Identification of miR-145 and miR-146a as
mediators of the 5q- syndrome phenotype. Nat Med. 16:49–58. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meng Q, Zheng M, Liu H, Song C, Zhang W,
Yan J, Qin L and Liu X: TRAF6 regulates proliferation, apoptosis,
and invasion of osteosarcoma cell. Mol Cell Biochem. 371:177–186.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han F, Zhang L, Qiu W and Yi X: TRAF6
promotes the invasion and metastasis and predicts a poor prognosis
in gastric cancer. Pathol Res Pract. 212:31–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Caporali S, Levati L, Graziani G, Muzi A,
Atzori MG, Bonmassar E, Palmieri G, Ascierto PA and D'Atri S: NF-κB
is activated in response to temozolomide in an AKT-dependent manner
and confers protection against the growth suppressive effect of the
drug. J Transl Med. 10:2522012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu JF, Tsao YT and Hou CH:
Fractalkine/CX3CL1 induced intercellular adhesion
molecule-1-dependent tumor metastasis through the
CX3CR1/PI3K/Akt/NF-κB pathway in human osteosarcoma. Oncotarget.
Aug 12–2016.(Epub ahead of print). doi:
10.18632/oncotarget.11250.
|
|
44
|
Zou W, Ma X, Hua W, Chen B and Cai G:
Caveolin-1 mediates chemoresistance in cisplatin-resistant ovarian
cancer cells by targeting apoptosis through the Notch-1/Akt/NF-κB
pathway. Oncol Rep. 34:3256–3263. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Barré B and Perkins ND: The Skp2 promoter
integrates signaling through the NF-kappaB, p53, and Akt/GSK3beta
pathways to regulate autophagy and apoptosis. Mol Cell. 38:524–538.
2010. View Article : Google Scholar : PubMed/NCBI
|