Introduction
Osteosarcoma is one of the most common primary bone
tumours, affecting patients of all ages, however there are two
incidence peaks. The first peak is in childhood and adolescense and
the second peak is at 75–79 years of age (1,2). The
primary tumour originates in the distal femur, proximal tibia or
humerus and its rapid cell division may contribute to the
oncogenesis of osteosarcoma during childhood (3). Adjuvant chemotherapy following
surgery, has improved the long-term survival rates from <20 to
70% in localized osteosarcoma. However, this combined therapy has
shown limited progress in the long-term survival rates of
metastatic osteosarcoma cases, which account for one-quarter of all
patients at the time of initial diagnosis. The higher incidence of
osteosarcoma in children and adolescents, its tendency to
metastasize and the poor prognosis in metastatic cases render
osteosarcoma the leading cause of cancer-related deaths in
childhood, although it accounts only for 5% of childhood cancers
(4). A better understanding of
tumour initiation, metastasis and recurrence in osteosarcoma is
urgently needed to improve patient prognosis.
Mature microRNAs (miRNAs) are endogenic regulatory,
single-stranded, non-coding RNAs that are ~22 nt in length. miRNAs
regulate one-third of all genes by complete or partial base-pairing
at the 3′-untranslated region of their target mRNAs, resulting in
translation inhibition and/or cleavage (5,6). Thus,
miRNAs can serve as either oncogenes or tumour suppressors
depending on their target genes (7,8).
Accumulating evidence indicates that some miRNAs are differentially
expressed in osteosarcoma tissues, cell lines and patient sera and
that these miRNAs play important roles in tumorigenesis, metastasis
and angiogenesis (9,10). These findings have provided new
insights into the genetic mechanisms of osteosarcoma tumorigenesis
and have contributed to the identification of novel osteosarcoma
biomarkers and the development of miRNA-targeted treatments
(11).
miR-504 has rarely been studied and not much is
known about its roles in oncogenesis and cancer metastasis.
Computational prediction for potential targets of miR-504 indicated
that this miRNA may directly interact with the mRNAs of FOXP1,
NRF1, CDK6, P53 and tumour protein p53-inducible nuclear protein 1
(TP53INP1) genes. Recent studies demonstrated that miR-504 directly
targets the FOXP1, NRF1, CDK6 and P53 genes (12–15).
TP53INP1 was demonstrated to regulate the transcriptional activity
of P53 or P73 through direct interaction. Furthermore, TP53INP1
participates in autophagy by binding LC3, PINK1 and Parkin
(16). Notably, TP53INP1 and P53
are functional partners because TP53INP1 is a target gene of the
transcription factor P53. In return, TP53INP1 can activate the
transcription activity of P53 (17). To avoid interference among miR-504,
P53 and TP53INP1, the P53-null osteosarcoma cell line 143B (IARC
TP53 database, http://p53.iarc.fr/Manual.aspx) was used to explore
the influence of miR-504/TP53INP1 modulation on tumour growth and
metastasis in vitro and in vivo.
Materials and methods
Statement of ethics
All procedures involving human participants were
performed in accordance to the 1964 Declaration of Helsinki and its
later amendments. The animal research protocol was approved by the
Animal Experiment Centre Committee of Xi'an Jiaotong University.
All animal experimental procedures were performed according to the
policy of Xi'an Jiaotong University Animal Experiment Centre.
Clinical samples and cell line
A total of 20 pairs of primary osteosarcoma and
matched adjacent non-cancerous paraffin-embedded specimens were
obtained at the authors' institution between 2012 and 2015. The
osteosarcoma diagnosis and the confirmation that the adjacent
tissues were normal were performed by two independent pathologists
based on the pathological changes associated with osteosarcoma and
the microscopic morphology of the adjacent normal tissues. The
tumour stage was assessed according to the Enneking-Musculoskeletal
Tumour Staging System (18). The
human osteosarcoma cell line 143B was purchased from the Type
Culture Collection of the Chinese Academy of Science, (Wuhan,
China) and was cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% foetal
bovine serum (FBS; Gibco) and 0.015 mg/ml 5-bromo-2′-deoxyuridine
(Sigma-Aldrich, St. Louis, MO, USA) in a humidified incubator at
37°C with 5% CO2.
Modulation of miR-504 in vitro
miR-504 mimics or inhibitors (RiboBio, Guangzhou,
China) were transfected into 143B cells with the Ribo FECT™ CP
Transfection kit (all reagents from RiboBio) according to the
manufacturer's instructions to achieve the overexpression or
knockdown of miR-504, respectively. Using the Ribo FECT™ CP
Transfection kit, a negative control mimic (normal control)
(RiboBio) and siR-Ribo™ Transfection Control (Cy3; RiboBio) were
transfected into 143B cells as the normal and transfection
controls, respectively.
RNA preparation and quantitative
real-time PCR
The microRNAs were extracted from the clinical
samples and cell lines using the miRNAprep Pure FFPE kit (Tiangen
Biotech, Beijing, China) or the TRIzol reagent (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's instructions.
The miRcute miRNA First-Strand cDNA Synthesis kit and miRcute miRNA
qPCR Detection kit (SYBR Green; Tiangen Biotech) were used for cDNA
synthesis and RT-qPCR according to the manufacturer's instructions.
The RT-qPCR conditions were 95°C for 5 min followed by 40 cycles of
95°C for 15 sec and 60°C for 20 sec and a final dissociation stage.
The PCR was performed using the Roche LightCycler® 96
fluorescent quantitative PCR instrument. The relative miR-504
expression level was determined using the 2−ΔCt method
with the nuclear U6 RNA as the endogenous control (19). The forward primers were designed as
follows: miR-504, 5′-GACCCTGGTCTGCACTCTATCA-3′; and U6,
5′-ACGCAAATTCGTGAAGCGTTC-3′. The reverse primers were provided with
the kit.
Luciferase reporter system
The potential targets of miR-504 were predicted by
miRanda (http://www.microrna.org). The predicted
miR-504 binding site at the TP53INP1 3′-UTR and corresponding
mutant 3′-UTR sequence were cloned in the psiCHECK™-2 Vector
(Promega, Madison, WI, USA). The psiCHECK-2-TP53INP1-WT and
psiCHECK-2-TP53INP1-MT were co-transfected with miR-504 NC or
miR-504 mimic into 293T cells. The relative luciferase activity was
analysed using a Promega Dual-Luciferase system (Promega) after 48
h of transfection.
Colony formation assay
Four hundred cells were seeded in 60-mm dishes and
cultured for nine days. Subsequently, the medium was removed. The
colonies were fixed with 4% paraformaldehyde for 10 min, stained
with 1% crystal violet for 15 min and then counted.
Cell proliferation assay
The cells were transfected and seeded in 96-well
plates at a density of 1,000 cells/well. The medium was changed
every three days. The cells were harvested each day from the third
to the seventh day and the cell viability was detected with Cell
Counting Kit-8 (Beyotime Institute of Biotechnology, Shanghai,
China) according to the manufacturer's instructions.
Cell cycle and apoptosis assay
The transfected cells were stained with 50 µg/ml
propidium iodide (Sigma-Aldrich) and 250 µg/ml RNAase
(Sigma-Aldrich). After 30 min of incubation in the dark, the cell
cycle was analysed using a FACSCalibur flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA). Cell apoptosis was analysed
using an Annexin V-FITC/PI Apoptosis Detection kit (Wanleibio,
Shenyang, China) by flow cytometry according to the manufacturer's
instructions.
Cell migration and invasion assay
To determine the cell migration, 3×104
cells were reseeded in the top of a Transwell chamber 48 h after
transfection. The cells that remained in the upper chamber after 24
h were carefully removed using a cotton swab. The migrated cells
were fixed and stained with 1% crystal violet for 20 min. The
procedure for the cell invasion was similar to the migration assay,
except for the following two differences: the upper Transwell
chamber was precoated with 60 µl of diluted Matrigel (BD
Biosciences) and incubated at 37°C overnight with 6×104
transfected cells seeded in the upper chamber.
Western blot analysis
Seventy-two hours after transfection, the total
cellular protein was extracted using RIPA lysis buffer and
Phosphatase Inhibitor II Cocktail (50X) (Heart, Xi'an, China)
according to the manufacturer's instructions. The total protein was
separated and transferred onto polyvinylidene membranes (Merck
Millipore, Billerica, MA, USA). Subsequently, the membranes were
blocked and incubated with primary antibodies overnight at 4°C.
Following incubation with an HRP-conjugated goat anti-rabbit IgG
secondary antibody (dilution, 1:5,000; Boster Biological
Technology, Wuhan, China) at room temperature for 1 h, the protein
signals were detected using the Immobilon™ Western Chemiluminescent
HRP Substrate (Merck Millipore). The following primary antibodies
were used: rabbit anti-TP53INP1 (dilution, 1:2,000; Abcam,
Cambridge, MA, USA), rabbit anti-P53 (phospho S46; dilution,
1:1,000; Abcam), rabbit anti-GAPDH (dilution, 1:1000; Wanleibio),
rabbit anti-P53 (dilution, 1:800; Wanleibio), rabbit anti-P73
(dilution, 1:500; Wanleibio), rabbit anti-P21 (dilution, 1:500;
Wanleibio), rabbit anti-Bax (dilution, 1:500; Wanleibio), rabbit
anti-caspase-3 (dilution, 1:500; Wanleibio), rabbit
anti-cleaved-caspase-3 (dilution, 1:300; Wanleibio) and rabbit
anti-SPARC (dilution, 1:1000; Sanying Biotechnology, Wuhan,
China).
Xenografts in nude mice and associated
histopathology
Ten five-week-old nude mice were randomly and
equally divided into the miR-504 agomir and the miR-504 NC groups.
Subsequently, 3×106 cells were subcutaneously injected
in the flanks of the nude mice. The cells had been pre-transfected
with agomir-hsa-miR-504 (miR-504 agomir; GenePharma, Shanghai,
China) or agomir negative control (miR-504 NC; GenePharma) based on
the grouping. One week after the tumour-cell implantation, 1 nmol
of miR-504 agomir or 1 nmol of agomir miR-504 NC was injected into
the tumours twice a week; the tumour volume was concurrently
assessed according to the formula: tumour volume = length ×
width2/2. All mice were sacrificed four weeks after
inoculation and orthotopic tumours and lungs were harvested. After
weighing the orthotopic tumours, both tumours and lungs were
stained with haematoxylin and eosin (H&E).
Immunohistochemistry
The slides containing formalin-fixed,
paraffin-embedded, 5-µm thick tissue sections were subjected to
microwave antigen retrieval and then incubated with the primary
antibody overnight at 4°C. Subsequently, the slides were incubated
with an HRP-conjugated secondary antibody for 30 min at room
temperature and the proteins were chemically visualised using a DAB
kit (ZSGB-Bio, Shanghai, China).
Statistical analysis
Statistical comparisons of the results were
performed using the SPSS 15.0 software (SPSS Inc., Chicago, IL,
USA) with Student's t-test, independent-samples t-test or Fisher's
exact probability test, depending on the particular data type.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-504 is highly expressed in the
osteosarcoma tissues
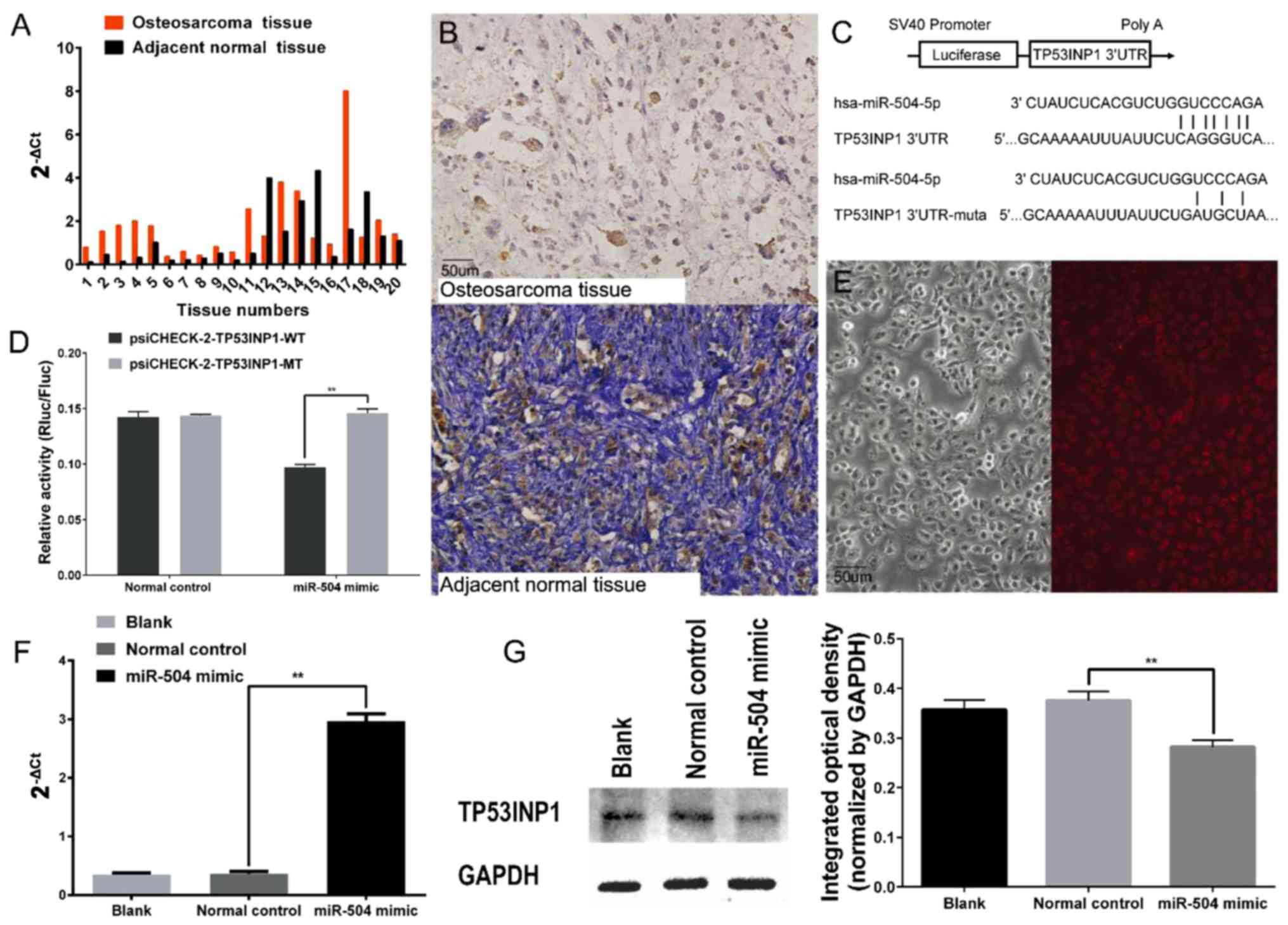
Expression of miR-504 was higher in 17 osteosarcoma
specimens compared to the matched adjacent non-cancerous tissues
and the remaining tumour specimens had a relatively lower
expression of miR-504 based on RT-qPCR assay (Fig. 1A). As shown in Table I, miR-504 was more highly expressed
in patients with distant metastasis, advanced clinical stage and
larger tumours. Additionally, TP53INP1-positive specimens had a
statistically lower expression of miR-504. There was no significant
difference between miR-504 expression and age or sex.
 | Table I.Relationship between the expression
of miR-504, TP53INP1 and the clinical characteristics in 20
osteosarcoma patients. |
Table I.
Relationship between the expression
of miR-504, TP53INP1 and the clinical characteristics in 20
osteosarcoma patients.
|
|
|
|
| TP53INP1
expression |
|
|---|
|
|
|
|
|
|
|
|---|
| Clinical
characteristics | N | miR-504
expression | P-value | Positive | Negative | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
Female | 8 | 2.069±0.896 | 0.630 | 4 | 4 | 0.535 |
|
Male | 12 | 1.676±0.284 |
| 5 | 7 |
|
| Age (years) |
|
|
|
|
|
|
|
<18 | 9 | 2.254±0.766 | 0.336 | 3 | 6 | 0.311 |
|
≥18 | 11 | 1.488±0.319 |
| 6 | 5 |
|
| Tumour size
(cm) |
|
|
|
|
|
|
|
<10 | 13 | 1.251±0.235 | <0.05 | 7 | 6 | 0.272 |
|
≥10 | 7 | 2.914±0.914 |
| 2 | 5 |
|
| Clinical stage |
|
|
|
|
|
|
|
IA-IIA | 7 | 0.793±0.138 | <0.05 | 6 | 1 | <0.05 |
|
IIB-III | 13 | 2.393±0.530 |
| 3 | 10 |
|
| Distal
metastasis |
|
|
|
|
|
|
|
Yes | 7 | 3.125±0.912 | <0.05 | 1 | 6 | 0.058 |
| No | 13 | 1.137±0.149 |
| 8 | 5 |
|
| Tumour site |
|
|
|
|
|
|
| Femur
or tibia | 14 | 1.984±0.509 | 0.563 | 5 | 9 | 0.217 |
|
Other | 6 | 1.481±0.519 |
| 4 | 2 |
|
| TP53INP1 |
|
|
|
|
|
|
|
Positive | 9 | 0.778±0.106 | <0.05 |
|
|
|
|
Negative | 11 | 2.696±0.582 |
|
|
|
|
TP53INP1 is not frequently expressed
in osteosarcoma tissues
TP53INP1 was detected in nine osteosarcoma specimens
(45%) using immunohistochemical staining, which was tan-coloured
and located in the nucleus. Seventeen adjacent non-cancerous
specimens (85%) exhibited TP53INP1 expression (Fig. 1B). Furthermore, the detectable
expression of TP53INP1 was infrequent in patients with advanced
clinical stage according to Fisher's exact probability test
(Table I).
miR-504 directly targets TP53INP1
We searched for potential targets of miR-504 using
the miRanda databases and found a potential miR-504-binding site in
the 3′UTR of the TP53INP1 mRNA. To validate the direct binding
between miR-504 and the predicted seed sequence, the predicted
potential binding sequence and paired mutated sequence were
inserted into the psiCHECK™-2 vector (Fig. 1C). Luciferase activity was
significantly decreased after co-transfection of
psiCHECK-2-TP53INP1-WT and miR-504 mimic, whereas miR-504 had no
effect on luciferase activity when co-transfected with
psiCHECK-2-TP53INP1-MT (Fig. 1D).
The feasibility of the transfection was verified using Cy3-labelled
normal control siRNA and RT-qPCR in 143B cells (Fig. 1E and F). Furthermore, the results of
western blot analysis confirmed that the expression of TP53INP1 was
downregulated in the 143B cells after the miR-504 mimic
transfection (Fig. 1G).
miR-504 promotes 143B cell growth,
migration and invasion in vitro
The CCK-8 assay was performed to investigate the
effects of miR-504 on the proliferation of 143B cells.
Proliferation was significantly enhanced in 143B cells transfected
with miR-504 mimic and decreased with miR-504 inhibition compared
with the normal control and blank groups (Fig. 2A). Consistently, the cells
transfected with the miR-504 mimic had more colonies in the colony
formation assay, whereas fewer colonies survived in the normal
control and blank groups. The lowest number of colonies was found
in the miR-504 inhibitor group (Fig.
2B).
Based on a cell cycle assay, the G1 subpopulation
proportion of cells was decreased in the miR-504 mimic-transfected
cells compared with other groups and the proportion of cells in the
G2 subpopulation was increased. In contrast, G1 arrest was obvious
in the cells transfected with the miR-504 inhibitor, which blocked
the G1 to S-phase transition, compared with the blank and normal
control groups (Fig. 2C). The
transfection of miR-504 mimic mitigated apoptosis in the 143B cells
compared with the normal control and blank groups and the cells
transfected with the miR-504 inhibitor had the highest proportion
of apoptosis (Fig. 2D).
To investigate the effects of miR-504 on migration
and invasion in 143B cells, a Transwell assay was performed. Cells
in the miR-504 mimic group exhibited a strengthened migration and
invasion capacity compared with the normal control and blank
groups, while minimal numbers of cells traversed the polycarbonate
membrane of the Transwell chamber in the miR-504 inhibitor group
for both the migration and invasion assays (Fig. 2E and F).
Finally, we evaluated TP53INP1, phosphorylated P53,
P73, P21, Bax, cleaved-caspase-3 and SPARC expression and found
that TP53INP1, P73, P21, Bax and cleaved-caspase-3 expression was
decreased 72 h after the miR-504 mimic transfection, but
upregulated in the miR-504 inhibitor group. SPARC is correlated
with pancreatic cancer cell migration and was upregulated in 143B
cells after the miR-504 mimic transfection compared with the normal
control and blank groups. However, phosphorylated P53 was
undetectable in all four groups (Fig.
3).
miR-504 promotes xenograft growth and
distant metastasis in vivo
The tumour growth was faster in the miR-504 agomir
group than in the miR-504 NC group (Fig. 4A). The tumours were harvested and
weighed at the fourth week and the tumour weights ascertained
faster growth in the miR-504 agomir group (1.29 vs. 0.95 g)
(Fig. 4B). The microscopical
morphology of the tumour was also visualised using H&E staining
(Fig. 4C).
All lungs were harvested and stained with H&E to
detect distal metastases. Only one mouse developed pulmonary
metastasis in the miR-504 NC group and three mice developed
pulmonary metastasis in the miR-504 agomir group. Mice in the
latter group developed larger metastatic tumours (Fig. 4D).
Although all tumours had detectable TP53INP1
expression, the expression level was lower in the miR-504
agomir-treated tumours (Fig.
4E).
Discussion
Many studies have revealed that miRNAs serve as
oncogenes or tumour suppressors in various types of cancer
depending on the particular miRNA (20). miR-504 has been demonstrated to
participate in tumour development and metastasis by targeting
FOXP1, NRF1 and P53 in several tumours, acting as an oncogene
(13,15,21,22).
Clinical samples demonstrated that higher expression levels of
miR-504 are correlated with worse radio-therapeutic effects in
nasopharyngeal carcinoma and with a poor prognosis for pancreatic
ductal adenocarcinoma patients (13,23).
In the present study, the expression of miR-504 was higher in most
clinical osteosarcoma samples compared to adjacent non-cancerous
tissues, which statistically correlated with tumour size,
metastasis and clinical stage. Similar to the expression of
miR-504, TP53INP1 was negatively correlated with the clinical stage
and the expression of TP53INP1 was almost non-existent in patients
with distant metastases.
One miRNA can regulate hundreds of proteins and one
protein can be regulated by multiple miRNAs. These complex
interactions between miRNAs and target proteins constitute a
regulatory network (24,25). The miRNA target prediction database
provides thousands of potential target proteins for one particular
miRNA. With the assistance of the luciferase reporter system,
RT-qPCR and western blotting, only a few proteins have been
demonstrated to be directly regulated by a particular miRNA.
TP53INP1 is a computationally analysed target of miR-504. The
luciferase reporter assay system confirmed a direct interaction of
the miR-504 mimic with the predicted seed region in the TP53INP1
3′-UTR. Moreover, the western blot analysis results demonstrated
lower TP53INP1 expression in the osteosarcoma 143B cells
transfected with the miR-504 mimic than in the control cells, which
further ascertained the direct regulation between miR-504 and
TP53INP1 in 143B cells.
Several studies have revealed that TP53INP1
functions as a tumour suppressor through P53 phosphorylation at
serine-46 by forming a protein complex with the protein kinase
homeodomain-interacting protein kinase-2 (HIPK2) or protein kinase
Cδ (PKCδ) to induce G1 arrest and apoptosis (17,26,27).
Because of the high degree of structural similarity with P53, P73
activates the transcription activity of the majority of
P53-responsive genes and serves as a tumour suppressor. These
responsive genes include P21 and Bax, as reported by Tomasini et
al (28). Importantly, P73 is
more stable in tumours than P53, which is often mutated (29,30).
Due to a missense mutation at exon 5, TP53 is inactive in the
osteosarcoma 143B cell line and serine-46-phosphorylated P53 was
undetectable in all four groups, confirming the loss-of-function
mutation of P53 in 143B cells. In the present study, miR-504
promoted proliferation and mitigated apoptosis in 143B cells in
vitro. Additionally, larger xenograft tumours were detected in
nude mice with increased expression of miR-504. Furthermore,
protein detection revealed that TP53INP1, P73, P21 and Bax were
suppressed by miR-504. Both P73α and P73β, which promote tumour
apoptosis, were downregulated after the miR-504 transfection
(31). Phosphorylation,
acetylation, ubiquitylation and sumoylation have been demonstrated
to occur and to determine the stabilization and transactivation
ability of P73. TP53INP1 has been shown to stimulate the cell cycle
arrest and pro-apoptotic functions of P73, however the specific
underlying mechanism remains unknown (28,32–35).
The variation in protein expression further confirms the results of
the cell cycle and apoptosis analyses. In conclusion, miR-504
promotes tumour growth through TP53INP1, which partly depends on
suppressing P73 and downstream proteins and this effect occurs
independently of P53 in 143B cells. The present study also reveals
that miR-504 inhibition further inhibits tumour growth.
The suppression of TP53INP1 promotes tumour
metastasis in endometrial carcinoma and non-small cell lung cancer.
TP53INP1 also participates in decreasing the epithelial-mesenchymal
transition in liver cancer cells. Additionally, the TP53INP1
expression level is inversely correlated with positive lymph node
metastasis in clinical breast carcinoma tissue specimens (36–39).
TP53INP1 suppresses pancreatic cancer cell migration by regulating
the expression of SPARC, which is a matrix cellular protein that
has a pivotal role in regulating cell-matrix interaction and
migration (40). The expression of
SPARC was increased and osteosarcoma cells were more aggressive
after the miR-504 mimic transfection in vitro and the lung
metastasis model confirmed the strengthened invasion ability of the
miR-504 expressing cells in the present study.
Concerning limitations to the present study the use
of miR-504 as a biomarker to predict the prognosis of early-stage
osteosarcoma requires further validation with a larger sample size.
The physical interaction between P73 and the miR-504/TP53INP1 axis
warrants further exploration using rescue experiments with
miR-504-resistant TP53INP1-plasmid in 143B cells, analysis using a
yeast two-hybrid system and co-immunoprecipitation.
In conclusion, miR-504 directly targets TP53INP1 in
osteosarcoma 143B cells. miR-504 promotes the proliferation and
invasion of 143B cells and mitigates their apoptosis by targeting
TP53INP1. As a result, P73 plays a role in tumour suppression
independently of P53. The results of this study indicate that
miR-504 and TP53INP1 expression are correlated with the tumour size
and metastatic burden.
Acknowledgements
This study is supported by the Shaanxi Provincial
Health and Family Planning Commission Project (grant no. 2016D067)
and the Foundation of the First Affiliated Hospital of Medical
College, Xi'an Jiaotong University (grant no. 2013YK19;
2016MS-08).
References
|
1
|
Whelan J, McTiernan A, Cooper N, Wong YK,
Francis M, Vernon S and Strauss SJ: Incidence and survival of
malignant bone sarcomas in England 1979–2007. Int J Cancer.
131:E508–E517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Savage SA and Mirabello L: Using
epidemiology and genomics to understand osteosarcoma etiology.
Sarcoma. 2011:5481512011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gill J, Ahluwalia MK, Geller D and Gorlick
R: New targets and approaches in osteosarcoma. Pharmacol Ther.
137:89–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pasquinelli AE, Hunter S and Bracht J:
MicroRNAs: A developing story. Curr Opin Genet Dev. 15:200–205.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou X and Yang PC: MicroRNA: A small
molecule with a big biological impact. Microrna. 1:12012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sassen S, Miska EA and Caldas C: MicroRNA
- implications for cancer. Virchows Arch. 452:1–10. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang L, Shrestha S, LaChaud G, Scott MA
and James AW: Review of microRNA in osteosarcoma and
chondrosarcoma. Med Oncol. 32:6132015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sampson VB, Yoo S, Kumar A, Vetter NS and
Kolb EA: MicroRNAs and potential targets in osteosarcoma: Review.
Front Pediatr. 3:692015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Yan YG, Wang C, Zhang SJ, Yu XH
and Wang WJ: MicroRNAs in osteosarcoma. Clinica Chimica Acta.
444:9–17. 2015. View Article : Google Scholar
|
|
12
|
Cui R, Guan Y, Sun C, Chen L, Bao Y, Li G,
Qiu B, Meng X, Pang C and Wang Y: A tumor-suppressive microRNA,
miR-504, inhibits cell proliferation and promotes apoptosis by
targeting FOXP1 in human glioma. Cancer Lett. 374:1–11. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao L, Tang M, Hu Z, Yan B, Pi W, Li Z,
Zhang J, Zhang L, Jiang W, Li G, et al: miR-504 mediated
down-regulation of nuclear respiratory factor 1 leads to
radio-resistance in nasopharyngeal carcinoma. Oncotarget.
6:15995–16018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kikkawa N, Kinoshita T, Nohata N, Hanazawa
T, Yamamoto N, Fukumoto I, Chiyomaru T, Enokida H, Nakagawa M,
Okamoto Y, et al: microRNA-504 inhibits cancer cell proliferation
via targeting CDK6 in hypopharyngeal squamous cell carcinoma. Int J
Oncol. 44:2085–2092. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song
JS, Tang LH, Levine AJ and Feng Z: Negative regulation of tumor
suppressor p53 by microRNA miR-504. Mol Cell. 38:689–699. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saadi H, Seillier M and Carrier A: The
stress protein TP53INP1 plays a tumor suppressive role by
regulating metabolic homeostasis. Biochimie. 118:44–50. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tomasini R, Samir AA, Carrier A, Isnardon
D, Cecchinelli B, Soddu S, Malissen B, Dagorn JC, Iovanna JL and
Dusetti NJ: TP53INP1s and homeodomain-interacting protein kinase-2
(HIPK2) are partners in regulating p53 activity. J Biol Chem.
278:37722–37729. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Enneking WF, Spanier SS and Goodman MA: A
system for the surgical staging of musculoskeletal sarcoma. 1980.
Clin Orthop Relat Res. 415:4–18. 2003. View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Inukai S and Slack F: MicroRNAs and the
genetic network in aging. J Mol Biol. 425:3601–3608. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang MH, Lin BR, Chang CH, Chen ST, Lin
SK, Kuo MY, Jeng YM, Kuo ML and Chang CC: Connective tissue growth
factor modulates oral squamous cell carcinoma invasion by
activating a miR-504/FOXP1 signalling. Oncogene. 31:2401–2411.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Soutto M, Chen Z, Saleh MA, Katsha A, Zhu
S, Zaika A, Belkhiri A and El-Rifai W: TFF1 activates p53 through
down-regulation of miR-504 in gastric cancer. Oncotarget.
5:5663–5673. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang B, Gu Y and Chen Y: Identification
of novel predictive markers for the prognosis of pancreatic ductal
adenocarcinoma. Cancer Invest. 32:218–225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT,
Xu LD, Wang YY, Du L, Zhang YP, et al: MiRNA-miRNA synergistic
network: Construction via co-regulating functional modules and
disease miRNA topological features. Nucleic Acids Res. 39:825–836.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
D'Orazi G, Cecchinelli B, Bruno T, Manni
I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal
G, et al: Homeodomain-interacting protein kinase-2 phosphorylates
p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 4:11–19. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hofmann TG, Möller A, Sirma H, Zentgraf H,
Taya Y, Dröge W, Will H and Schmitz ML: Regulation of p53 activity
by its interaction with homeodomain-interacting protein kinase-2.
Nat Cell Biol. 4:1–10. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tomasini R, Seux M, Nowak J, Bontemps C,
Carrier A, Dagorn JC, Pébusque MJ, Iovanna JL and Dusetti NJ:
TP53INP1 is a novel p73 target gene that induces cell cycle arrest
and cell death by modulating p73 transcriptional activity.
Oncogene. 24:8093–8104. 2005.PubMed/NCBI
|
|
29
|
Pflaum J, Schlosser S and Müller M: p53
family and cellular stress responses in cancer. Front Oncol.
4:2852014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dötsch V, Bernassola F, Coutandin D, Candi
E and Melino G: p63 and p73, the ancestors of p53. Cold Spring Harb
Perspect Biol. 2:a0048872010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yoon MK, Ha JH, Lee MS and Chi SW:
Structure and apoptotic function of p73. BMB Rep. 48:81–90. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Satija YK and Das S: Tyr99 phosphorylation
determines the regulatory milieu of tumor suppressor p73. Oncogene.
35:513–527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gonzalez S, Prives C and Cordon-Cardo C:
p73alpha regulation by Chk1 in response to DNA damage. Mol Cell
Biol. 23:8161–8171. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ferraris VA, Brown JR, Despotis GJ, Hammon
JW, Reece TB, Saha SP, Song HK, Clough ER, Shore-Lesserson LJ,
Goodnough LT, et al: Society of Thoracic Surgeons Blood
Conservation Guideline Task Force; Society of Cardiovascular
Anesthesiologists Special Task Force on Blood Transfusion;
International Consortium for Evidence Based Perfusion: 2011 update
to the Society of Thoracic Surgeons and the Society of
Cardiovascular Anesthesiologists blood conservation clinical
practice guidelines. Ann Thorac Surg. 91:944–982. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mantovani F, Piazza S, Gostissa M, Strano
S, Zacchi P, Mantovani R, Blandino G and Del Sal G: Pin1 links the
activities of c-Abl and p300 in regulating p73 function. Mol Cell.
14:625–636. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang F, Liu T, He Y, Yan Q, Chen X, Wang
H and Wan X: MiR-125b promotes proliferation and migration of type
II endometrial carcinoma cells through targeting TP53INP1 tumor
suppressor in vitro and in vivo. BMC Cancer. 11:4252011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Q, Han Y, Wang C, Shan S, Wang Y, Zhang
J and Ren T: MicroRNA-125b promotes tumor metastasis through
targeting tumor protein 53-induced nuclear protein 1 in patients
with non-small-cell lung cancer. Cancer Cell Int. 15:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu F, Kong X, Lv L and Gao J: TGF-β1 acts
through miR-155 to down-regulate TP53INP1 in promoting
epithelial-mesenchymal transition and cancer stem cell phenotypes.
Cancer Lett. 359:288–298. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ito Y, Motoo Y, Yoshida H, Iovanna JL,
Takamura Y, Miya A, Kuma K and Miyauchi A: Decreased expression of
tumor protein p53-induced nuclear protein 1 (TP53INP1) in breast
carcinoma. Anticancer Res. 26(6B): 1–4395. 2006.PubMed/NCBI
|
|
40
|
Seux M, Peuget S, Montero MP, Siret C,
Rigot V, Clerc P, Gigoux V, Pellegrino E, Pouyet L, N'Guessan P, et
al: TP53INP1 decreases pancreatic cancer cell migration by
regulating SPARC expression. Oncogene. 30:3049–3061. 2011.
View Article : Google Scholar : PubMed/NCBI
|