Introduction
Lung cancer is the leading cause of cancer-related
deaths worldwide, particularly in China, and non-small cell lung
cancer (NSCLC; including squamous, adenocarcinoma and large cell)
accounts for 80% of all cases (1,2).
Despite advances in surgery and radiochemotherapy, the 5-year
survival rate of NSCLC patients remains unsatisfactory (1).
La-related protein 1 (LARP1), a member of the LARP
family, regulates both mRNA translation and stability (3). In cervical cancer, the knockdown of
LARP1 suppressed cell migration, while the overexpression of LARP1
promoted migration through post-transcriptionally regulated genes
such as mTOR (4,5). The protein and mRNA levels of LARP1
were upregulated in hepatocellular carcinoma (HCC), and
overexpressed LARP1 indicated poor survival in patients (6). Loss of tumour-suppressive miR-26a/b
increased the invasive ability of prostate cancer cells via
targeting of LARP1 (7). However,
the biological effect of LARP1 on NSCLC cells is not well
understood.
MicroRNAs (miRNAs) are a class of small, non-coding
single-stranded RNAs that participate in various biological
processes (8). During
carcinogenesis, miRNAs have been functionally classified as
proto-oncogenes or tumour-suppressor genes and contribute to tumour
progression by modulating target genes (9,10).
Among these miRNAs, miR-374a functioned as an oncogene in gastric
and breast cancer by promoting cell proliferation and invasion
(11,12). However, low expression levels of
miR-374a in early-stage NSCLC were associated with poor survival in
patients (13). miR-374a was
significantly decreased in lung adenocarcinoma tissues, and the
overexpression of miR-374a resulted in the inhibition of cell
proliferation, migration and invasion (14). Thus, the exact function and
mechanism of miR-374a in the development of NSCLC warrants further
investigation.
It has been widely accepted that the dysregulation
of a single miRNA may affect a multitude of mRNAs involved in
tumour-related pathways. For example, the activation of STAT3 led
to cell proliferation (15), and
knockdown of phosphorylated-STAT3 (p-STAT3) inhibited the invasive
capacity of pancreatic cancer cells (16). Positive p-STAT3 expression in
cervical cancer was associated with lymph node metastasis (17). Both miR-375 and miR-133b suppressed
cell growth through the regulation of STAT3 signalling (18,19).
All of these findings suggest that the biological behaviour of
tumour cells may be regulated by STAT3 signalling activity.
In the present study, we found that silencing of
LARP1 inhibited cell proliferation, migration and invasion, as well
as the downstream STAT3 pathway and epithelial-mesenchymal
transition (EMT). Knockdown of LARP1 inhibited tumour growth in
vivo. LARP1 was a direct and functional target of miR-374a,
which was affected by lncRNA XIST. We revealed that the
XIST/miR-374a/LARP1 axis was involved in pulmonary
carcinogenesis.
Materials and methods
Clinical samples and cells
This protocol was approved by the Ethics Committee
of the Institutional Review Board at Zhejiang University School of
Medicine, and written informed consent was collected before
surgery. In total, 84 pairs of primary NSCLC (including
adenocarcinoma and squamous cell carcinoma) and adjacent non-tumour
tissues were surgically obtained from patients who underwent
surgery at the hospital from 2010 to 2012. None of the patients
received radiotherapy or chemotherapy prior to surgery. The
adjacent non-cancerous tissues were obtained at least 3 cm away
from the tumour margin. All of the samples were snap-frozen in
liquid nitrogen and stored at −80°C.
NSCLC cell lines (H520, A549, H1299 and H1975) and
the lung/bronchial normal epithelial cell line BEAS-2B were
obtained from the American Type Culture Collection (ATCC; Manassas,
VA, USA), and maintained in RPMI-1640 medium (Gibco, Karlsruhe,
Germany) supplemented with 10% fetal bovine serum (FBS)
(Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere of 5%
CO2 at 37°C.
RNA isolation and quantitative
real-time PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen) and NanoDrop 2000 (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used to assess the quality of RNA.
Complementary DNA (cDNA) was synthesized using High Capacity cDNA
Reverse Transcription kit (ABI, Carlsbad, CA, USA). qRT-PCR was
carried out using 10 ng of cDNA with SYBR-Green Master Mix (ABI) on
an LightCycler 480 (Roche, Basel, Switzerland). Primers were as
follows: LARP1 forward, 5′-CATTAGCTAAGCACATGAAGTG-3′ and reverse,
5′-GTACGTACGAATCTTGCAA-3′. For the miRNA assay, the cDNA was
synthesized using the One Step PrimeScript miRNA cDNA Synthesis kit
(Qiagen, Hilden, Germany), and miRNA was quantified using
TaqMan® MicroRNA Assays under the ABI 7900 system
(Thermo Fisher Scientific, Inc.). All samples were assessed in
triplicates. β-actin and U6 were used as endogenous controls. Data
were analysed using the 2−ΔΔCt method.
Plasmid construction, oligonucleotides
and transfection
To knock down LARP1, we used a lentivirus infection
strategy. Lentivirus containing shRNA targeting the LARP1 gene or
non-silencing control shRNA were constructed by GeneChem (Shanghai,
China). Cells were plated at a density of 30% and were in good
condition before the day of infection. The aforementioned
lentivirus was used to treat the H520 and A549 cells according to
the manufacturer's protocol.
LARP1 cDNA (XM_005268404) without its 3-untranslated
region (3′-UTR) was amplified by PCR using the primers containing
the MluI and KpnI restriction sites and was then
inserted into the pcDNA 3.1(+) vector (Invitrogen) to generate the
recombinant vector pcLARP1. The primers used were:
5′-GGATTTCCAAGACGCGTCCCATACCTAGCTGCCC-3′ (forward) and
5′-CGAGTGGTACCAGAGTGATGGGGCTGGTG-3′ (reverse). The empty vector
served as a negative control. The miR-374a mimics, inhibitor and
corresponding negative controls were purchased from GenePharma, and
transfections were performed using Lipofectamine 2000
(Invitrogen).
For the knockdown of XIST, shRNA against lncRNA XIST
or scrambled oligonucleotides was ligated into the LV-3 (pGLVH1/GFP
+ Puro) vector (GenePharma). The 293 cells were co-transfected with
Lenti-Pac HIV Expression Packaging Mix and the lentiviral vectors
(or the control lentivirus) using Lipofectamine 2000. Lentiviral
particles in the supernatant were harvested. The H520 cells were
transfected with lentivirus or control virus. The cells were
treated with puromycin (2 µg/ml) for 14 days, and GFP-positive
cells were selected as sh-XIST and sh-ctrl (control).
Cell proliferation assay
Cell proliferation was determined using the MTT
method. Cells (2×103) were seeded into 96-well plates
and were maintained at 37°C. MTT (100 µl; 0.5 mg/ml; Sigma, St.
Louis, MO, USA) was added to each well at the indicated time-points
(24, 48, 72 or 96 h). Dimethyl sulfoxide (DMSO) (150 µl) was used
to stop the reaction, followed by measurement of the absorbance at
490 nm.
Wound-healing and Transwell invasion
assays
For the wound-healing assay, 1×106 cells
were seeded into 6-well plates. A wound was made by scraping the
cell monolayer with a 200-µl pipette tip. Cell motility was
evaluated by observation at intervals of 0 and 24 h. For the cell
invasion assay, cells (1×105) in serum-free medium were
placed into the upper chamber of a 24-well Transwell chamber (8-µm
pore size; Corning, Cambridge, MA, USA) coated with Matrigel (BD
Biosciences, San Jose, CA, USA). The bottom chamber was filled with
medium containing 10% FBS. Cells that had invaded through the
membrane to the lower surface were fixed, stained and counted.
Luciferase activity assay
Both the wild-type (WT) and mutated (Mut) 3′-UTRs of
LARP1 mRNA were subcloned into the XhoI and NotI
sites of the psiCHECK-2 vector (Promega, Madison, WI, USA). The
mutant constructs were generated using a QuikChange Site-Directed
Mutagenesis kit (Stratagene, La Jolla, CA, USA). For the luciferase
reporter assay, 293 cells (1×105 cells/well) were
cultured and co-transfected with 50 nM miR-374a mimics or negative
control (Mim-NC), 200 ng of WT or Mut vector, and 2 ng of
Renilla luciferase-expressing vector pRL-TK (Promega). Then,
48 h later, the cells were harvested and assayed using the
Dual-Luciferase Reporter Assay system (Promega).
The fragment from XIST containing the predicted
miR-374a binding site was amplified by PCR and cloned into a
pmirGLO vector (Promega) to form the reporter vector XIST wild-type
(XIST-WT). The mutant was generated by mutating the miR-374a seed
region binding sequence, which was named XIST-Mut. 293T cells were
co-transfected with the pmirGLO vector with either wild-type
fragments or mutation fragments and miR-374a using Lipofectamine
2000. Luciferase activity was detected as aforementioned.
Western blotting
Total protein was extracted by lysing cells in RIPA
buffer (Beyotime Institute of Biotechnology, Haimen, China). Cell
protein lysates were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then
transferred onto a polyvinylidene difluoride (PVDF) membrane
(Millipore, Boston, MA, USA). After blocking with 5% fat-free milk,
the membranes were incubated with primary antibodies for LARP1
(ab86359; Abcam, Cambridge, MA, USA), p-STAT3 (Y705, ab76315),
cyclin D1 (ab134175), E-cadherin (ab40772) and N-cadherin (ab18203)
overnight at 4°C. PVDF membranes were washed in TBST and incubated
with horseradish peroxidase-conjugated secondary antibodies.
Animal experiments
All of the animal protocols were approved by the
Institutional Animal Care and Use Committee of Zhejiang University
School of Medicine. Male BALB/c nude mice (4–6 weeks old) were
purchased from SLAC Laboratory Animal Ltd. (Shanghai, China). The
H520 cells (2×106) were subcutaneously implanted into
the right flank (control shRNA) and left flank (shLARP1) of nude
mice. Five weeks later, tumours were harvested, weighed, excised,
fixed and embedded in paraffin and sectioned for histological
examination of p-STAT3 and Ki67 expression.
Immunohistochemistry (IHC)
Mouse tumour tissues were fixed and embedded in
paraffin, treated for 2 h at 65°C and then deparaffinized. In
addition to standard haematoxylin and eosin (H&E) staining, the
tumour sections were subjected to IHC staining to detect Ki67
(ab92742) and p-STAT3. The sections were incubated with
biotinylated secondary antibody, followed by incubation with
diaminobenzidine and counterstaining with haematoxylin.
Statistical analysis
Graphical depictions and statistical analyses were
conducted using GraphPad Prism 5.0 (GraphPad Software Inc., San
Diego, CA, USA) and SPSS 16.0 (SPSS, Inc., Chicago, IL, USA).
One-way ANOVA was performed for data involving three or more
groups, and sets of two groups were analysed using Student's t-test
(two-tailed). The Kaplan-Meier method was used for overall survival
analysis. A P-value of <0.05 was considered to be statistically
significant.
Results
Silenced LARP1 inhibits cell growth
and motility
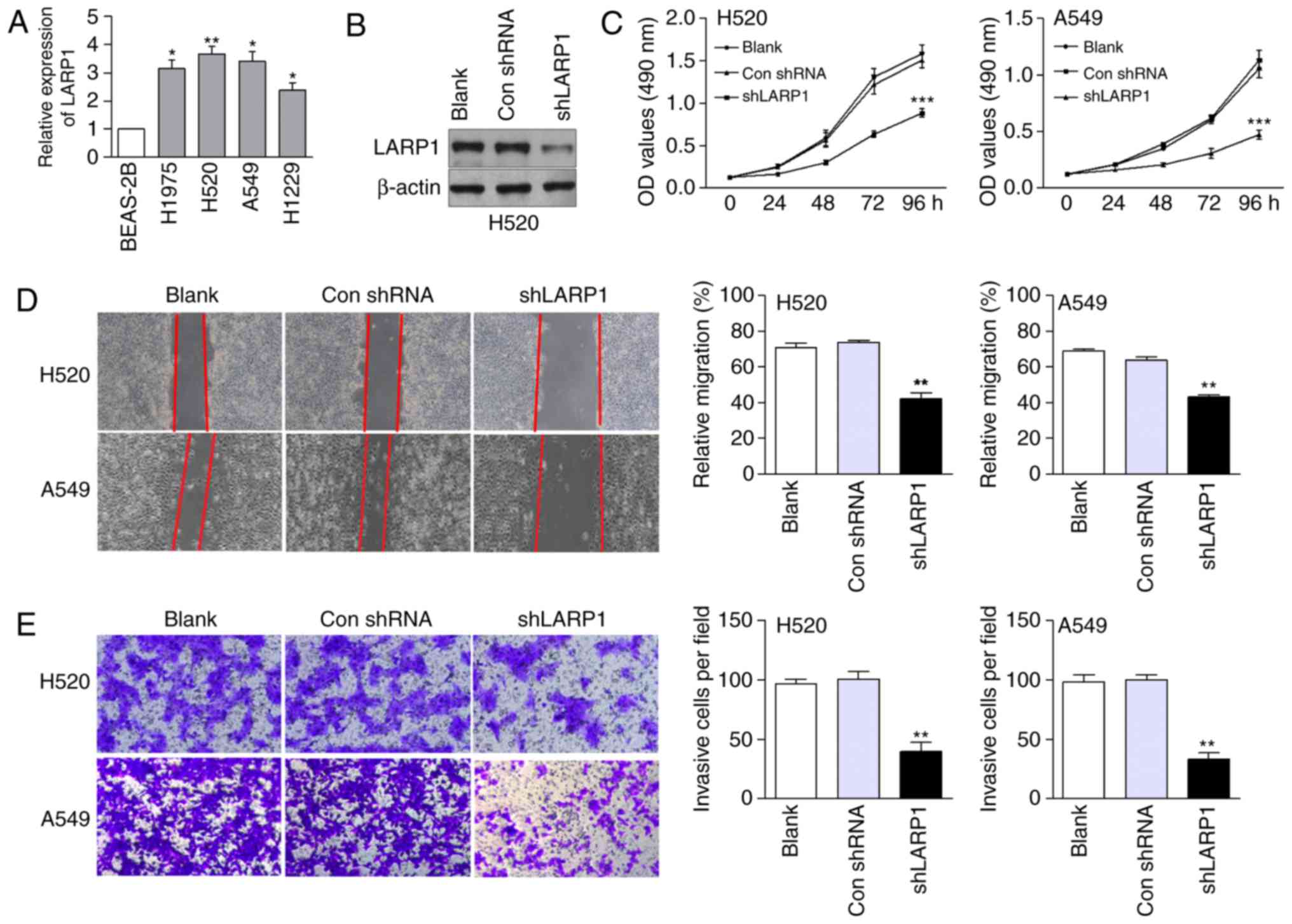
To explore the relationship between LARP1 and the
biological behaviour of NSCLC cells, we first detected endogenous
LARP1 in NSCLC and BEAS-2B cells. The mRNA levels of LARP1 were
higher in NSCLC cells than those in the normal control cells
(Fig. 1A). H520 and A549 cells,
which expressed relatively high levels of LARP1, were used in the
knockdown analysis. Compared with the control cells,
shLARP1-transfected H520 cells expressed low levels of LARP1
protein (Fig. 1B). Compared with
the negative control group, cell growth was markedly suppressed in
the LARP1-silenced group (Fig. 1C).
In addition, LARP1 downregulation decreased the migration of H520
and A549 cells compared with that in the control-transfected cells
(Fig. 1D). shLARP1-transfected
NSCLC cells also exhibited a significant reduction in invasive
ability (Fig. 1E).
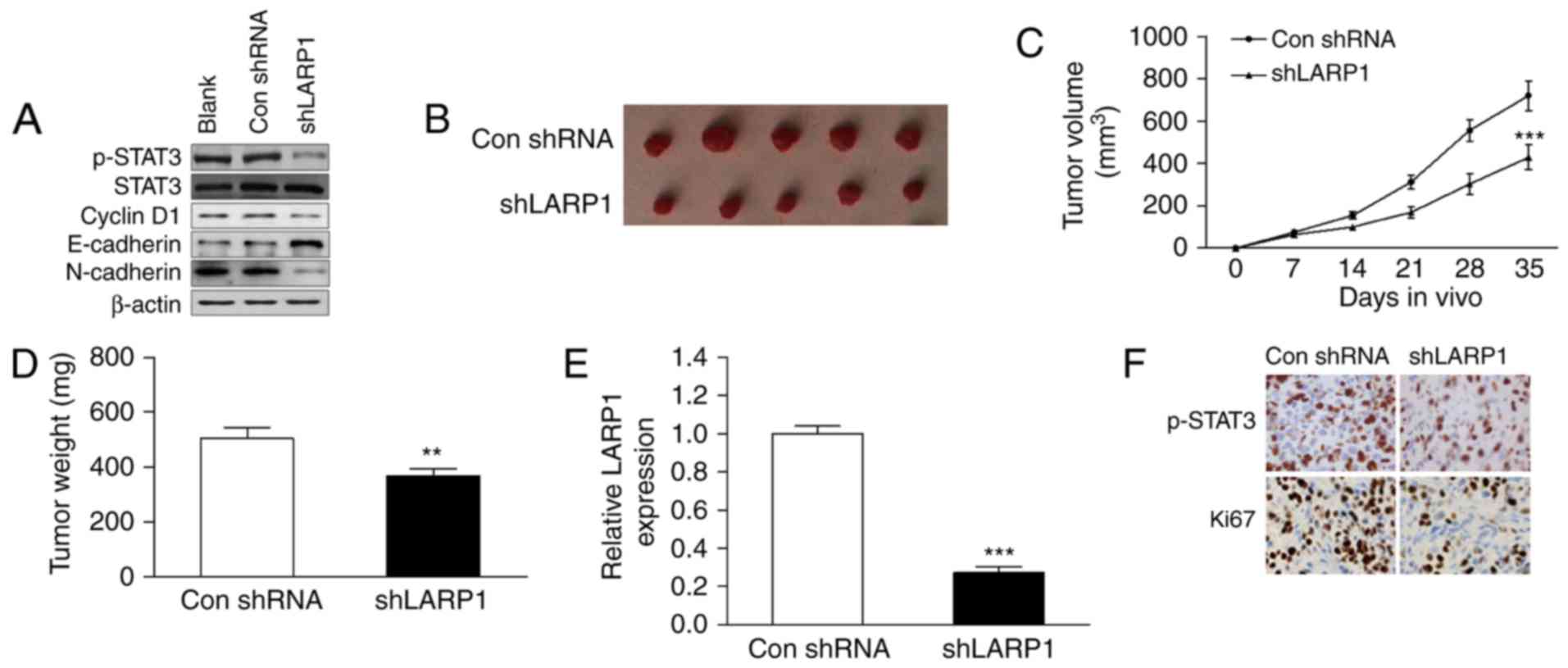
Knockdown of LARP1 inhibits tumour
growth in vivo via the STAT3 pathway
As the activated STAT3 pathway is known to be an
effector of proliferation and metastasis, we used western blotting
to explore the relationship between LARP1 and STAT3 signalling. We
found that the protein levels of p-STAT3, cyclin D1 (a cell growth
marker), and N-cadherin (an EMT marker) were significantly
decreased in shLARP1-transfected cells (Fig. 2A), while the protein level of
E-cadherin was upregulated. These data indicated that inactivated
STAT3 signalling and EMT were involved in shLARP1-induced cell
growth and invasion inhibition.
H520 cells infected with shLARP1 were injected into
the flanks of nude mice. Mouse tumours derived from the
shLARP1-transfected cells were significantly smaller than those
derived from the vector control (Fig.
2B and C). The weights of the tumours induced by LARP1
downregulation were significantly lower than those induced by the
control (Fig. 2D). To determine
whether the reduction of tumour growth was a result of the ablation
of LARP1, the xenograft tumours were subjected to qRT-PCR for LARP1
mRNA expression. As shown in Fig.
2E, the expression of LARP1 was significantly reduced in
shLARP1-treated mice. The expression levels of Ki67 (a cell
proliferation marker) and p-STAT3 were decreased in tumours treated
with shLARP1 (Fig. 2F).
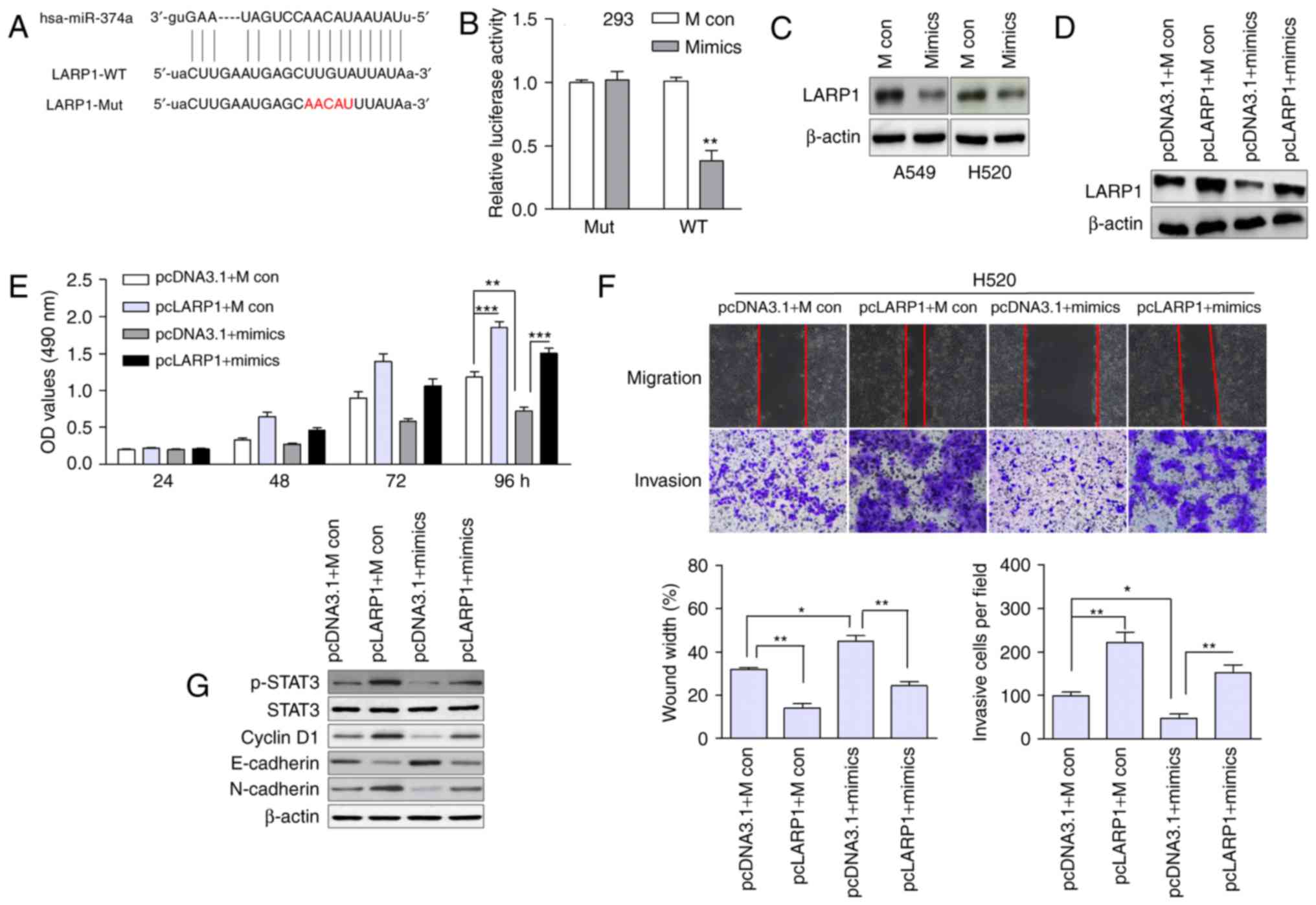
LARP1 is a direct target of
miR-374a
We used publicly available databases (TargetScan,
microRNA, miRDB) to identify candidate miRNAs that regulated LARP1.
miR-374a had a binding site in the 3′-UTR of LARP1 mRNA (Fig. 3A). The luciferase reporter assay
revealed that the overexpression of miR-374a significantly
suppressed the luciferase activity of the wild-type LARP1 3′-UTR,
but failed to affect the mutant 3′-UTR (Fig. 3B). The protein expression of LARP1
was also decreased in miR-374a-overexpressing cells (Fig. 3C).
LARP1 is involved in miR-374a-induced
inhibition of cell growth and motility
To ascertain whether miR-374a regulated cell growth
and migration through LARP1, we rescued LARP1 expression by
introducing the pcDNA3.1-LARP1 plasmid without the 3′-UTR (pcLARP1)
or the empty vector (pcDNA3.1) in the presence or absence of
ectopic miR-374a expression in H520 cells. The expression of LARP1
was confirmed by western blotting (Fig.
3D). The restoration of LARP1 markedly abrogated the
miR-374a-induced proliferation, migration and invasion inhibition
in H520 cells (Fig. 3E and F). The
protein levels of p-STAT3, cyclin D1 and N-cadherin were
upregulated in LARP1-overexpressing cells, while E-cadherin was
downregulated (Fig. 3G). miR-374a
mimics led to downregulated p-STAT3, cyclin D1 and N-cadherin and
enhanced E-cadherin, which was inconsistent with the phenomenon
induced by shLARP1. These data suggested that LARP1 was a
functional target of miR-374a.
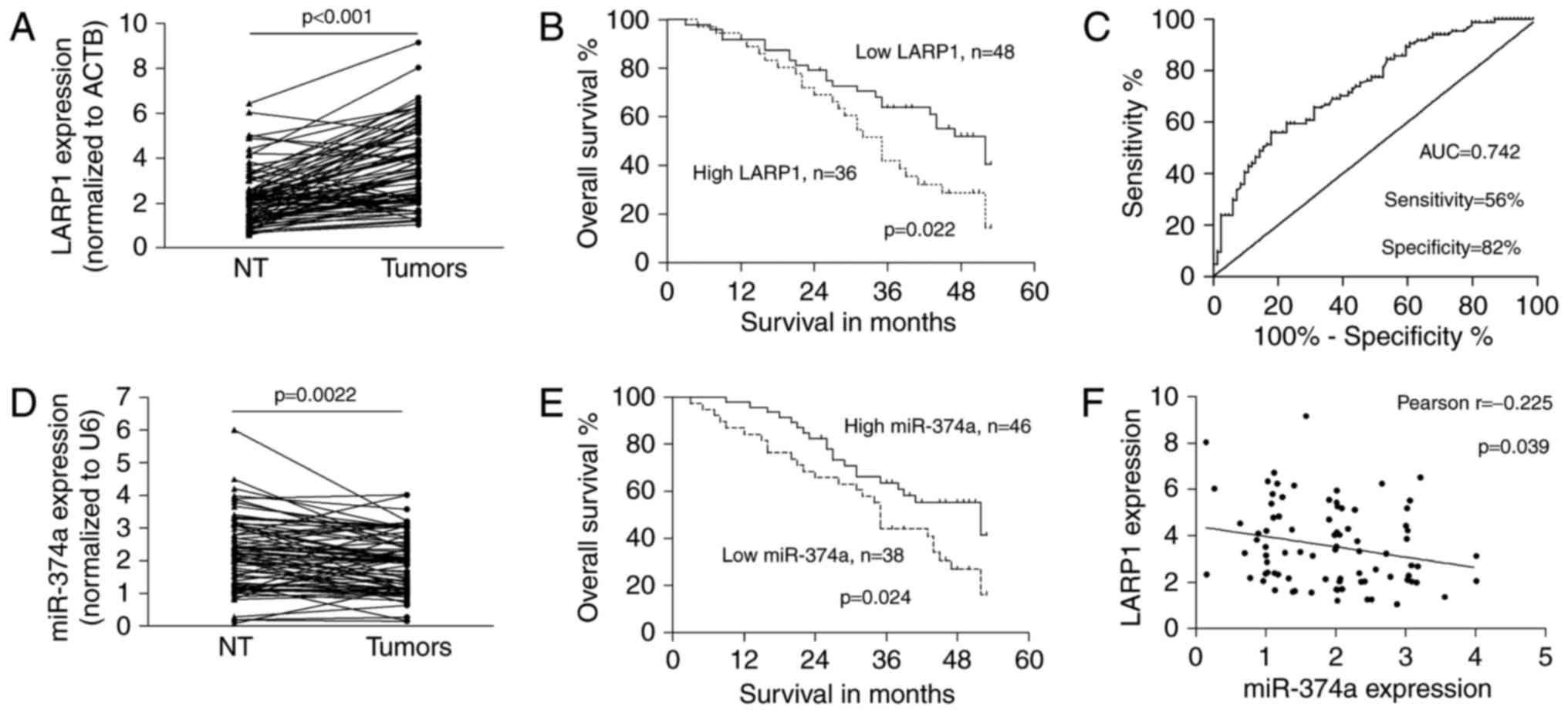
Upregulated LARP1 indicates the
advanced phenotype of NSCLC and was negatively associated with
miR-374a
The expression of LARP1 was significantly
upregulated in NSCLC (3.567±1.743) tumours compared with that in
matched non-tumour tissues (2.210±1.217; P<0.001; Fig. 4A). Upregulated LARP1 was
significantly correlated with the advanced tumour-node-metastasis
(TNM) stage (P=0.001), lymph node (P=0.017) and distant metastases
(P=0.039; Table I). According to
the mean level of LARP1 in NSCLC tumours, we separated cases into
low or high expression groups. NSCLC patients with low LARP1
expression had better overall survival than those with high LARP1
expression (P=0.022; Fig. 4B). In
addition, to determine the appropriate cut-off for a potential
biomarker application, we performed a receiver operating
characteristic curve (ROC) analysis. The area under the ROC (AUC)
was 0.742 (P<0.001). The optimal cut-off provided 56%
sensitivity and 82% specificity for the LARP1 overexpression status
as a diagnostic marker for NSCLC (Fig.
4C).
 | Table I.Correlations between the expression
of LARP1/miR-374a and clinicopathological characteristics of NSCLC
patients. |
Table I.
Correlations between the expression
of LARP1/miR-374a and clinicopathological characteristics of NSCLC
patients.
|
Characteristics | Cases (n=84) | LARP1
expression | P-value | miR-374a
expression | P-value |
|---|
| Sex |
|
| 0.396a |
| 0.516a |
|
Female | 30 | 3.445±1.661 |
| 1.839±0.964 |
|
|
Male | 54 | 3.634±1.799 |
| 1.921±0.835 |
|
| Age (years) |
|
| 0.754a |
| 0.779a |
|
<60 | 39 | 3.540±1.779 |
| 2.075±0.934 |
|
|
≥60 | 45 | 3.590±1.732 |
| 1.734±0.805 |
|
| Smoking status |
|
| 0.089b |
| 0.152b |
|
Never | 32 | 3.330±1.445 |
| 1.795±1.064 |
|
| Current
smoker | 42 | 3.626±2.028 |
| 2.000±0.939 |
|
| Former
smoker | 10 | 4.075±1.267 |
| 1.660±0.581 |
|
|
Differentiation |
|
| 0.263a |
| 0.686a |
|
Well | 29 | 3.319±1.526 |
| 1.894±1.746 |
|
|
Moderate, poor | 55 | 3.697±1.847 |
| 1.891±0.947 |
|
| TNM stage |
|
| 0.001a |
| 0.012a |
|
I,II | 37 | 2.718±1.188 |
| 2.220±0.883 |
|
| III,
IV | 47 | 4.235±1.829 |
| 1.633±0.792 |
|
| LNM status |
|
| 0.017a |
| 0.096a |
|
Negative | 23 | 2.557±1.025 |
| 2.285±0.980 |
|
|
Positive | 61 | 3.948±1.811 |
| 1.744±0.796 |
|
| Distant
metastasis |
|
| 0.039a |
| 0.005a |
| No | 78 | 3.407±1.656 |
| 1.986±0.833 |
|
|
Yes | 6 | 5.645±1.624 |
| 0.667±0.416 |
|
| Histological
type |
|
| 0.436a |
| 0.644a |
|
SCC | 53 | 3.569±1.781 |
| 1.798±0.840 |
|
| AC | 31 | 3.564±1.705 |
| 2.052±0.933 |
|
Moreover, the expression of miR-374a was
significantly decreased in primary tumours compared to that in
non-tumour tissues (1.892±0.878 vs. 2.164±1.073; P=0.0022; Fig. 4D). Downregulated miR-374a was
related to advanced tumour stage and distant metastasis (P=0.012,
and 0.005, respectively; Table I).
The mean level of miR-374a in NSCLC tissues was used as a threshold
to classify NSCLC cases into low or high expression groups; and low
expression of miR-374a indicated poor survival in patients
(P=0.024; Fig. 4E). There was an
inverse correlation between the expression of miR-374a and LARP1 in
tumour tissues (Pearson r=−0.225; P=0.039; Fig. 4F), suggesting that the
miR-374a/LARP1 regulatory pathway was related to the progression of
NSCLC.
Reciprocal repression of lncRNA XIST
and miR-374a expression
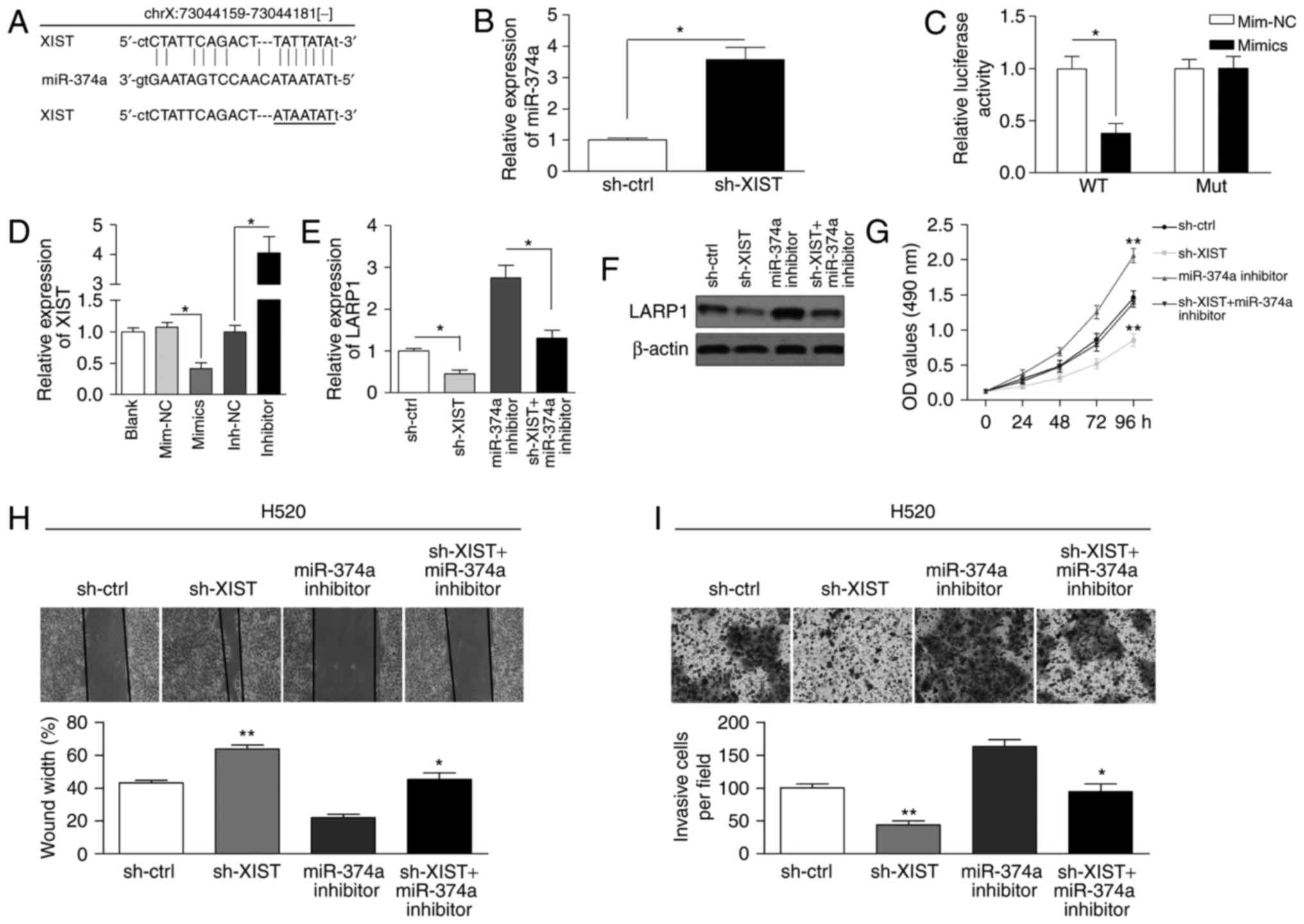
Using the starBase v2.0 database, miR-374a was found
to potentially bind to XIST (Fig.
5A). The qRT-PCR assay showed that miR-374a expression was
increased in the sh-XIST group (Fig.
5B). We cloned the predicted miR-374a binding site of XIST
(XIST-WT) and a mutated binding site (XIST-Mut) into a reporter
plasmid. The results showed that co-transfection of miR-374a mimics
and XIST-WT decreased the luciferase activity (Fig. 5C). As shown in Fig. 5D, XIST expression was reduced in
cells treated with miR-374a mimics, whereas the expression in cells
treated with the miR-374a inhibitor was increased. Finally, we
determined whether XIST can regulate the expression of LARP1.
Knockdown of XIST reduced the mRNA and protein levels that were
increased by the miR-374a inhibitor (Fig. 5E and F). Functionally, the knockdown
of XIST significantly inhibited proliferation in H520 cells and
abolished the miR-374a inhibition-induced increase in cell
proliferation (Fig. 5G). Likewise,
knockdown of XIST suppressed the migration and invasion ability
increased by miR-374a inhibition in H520 cells (Fig. 5H and I). These results indicated
that XIST regulates miR-374a to modulate cell viability and
motility in NSCLC cells. We also measured the expression of XIST in
H1975 cells derived from female and in H520, A549 and H1299 cells
derived from male. There was no significant difference between the
expression of XIST in female and male cancer cells (data not
shown).
Discussion
LARP1 has been reported as a key regulator of the
mTOR signalling pathway and is involved in cancer progression
(5). The mRNA and protein levels of
LARP1 were increased in hepatocellular carcinoma (HCC), and
upregulated LARP1 indicated large tumour sizes and poor survival
(6). In colorectal cancer (CRC),
the increased expression of LARP1 was associated with T, N and M
stage and was independently correlated with patient survival
(20). In the present study, we
found similar results. LARP1 was overexpressed in NSCLC tumours and
increased LARP1 was related to cancer progression and worse
survival. Further studies with larger groups of patients are needed
to confirm these findings. By analysing data sets from Oncomine
(www.oncomine.org), we found that the expression
of LARP1 was increased in lung and bladder cancer, CRC and HCC, as
well as in myeloma (data not shown), supporting the role of LARP1
as an important oncogene that may be commonly involved in various
types of human malignancies. In addition, since LARP1 could
distinguish normal tissues from tumours with 82% specificity in the
present study, it may be used as a novel diagnostic marker.
For the biological function of LARP1, we observed
the inhibitory effects of LARP1 silencing on cell growth,
migration, invasion and tumourigenicity. In previous studies, LARP1
promoted cervical cancer cell migration and invasion, while
knockdown of LARP1 attenuated the invasive ability of prostate
cancer cells (5,7). LARP1 partially abolished
miR-374a-induced suppression of cell growth, a finding that was
consistent with previous studies revealing that LARP1 promoted cell
proliferation (21). Thus, LARP1
functioned as an oncogene by promoting cell growth and motility,
which was consistent with the observations revealing that LARP1 was
related to metastasis in clinical samples. Collectively, our
findings expanded on the knowledge of LARP1 in NSCLC and suggested
the cancer-promoting activity of LARP1.
Tumour metastasis has been widely accepted as a
major obstacle to successful cancer treatment and ultimately leads
to poor prognosis in NSCLC patients (22). Previous studies have documented that
constitutive STAT3 activation contributed to metastasis in lung
cancer (23,24). Additionally, activated STAT3
signalling has been observed in breast cancer and HCC (25,26).
In the present study, we found that p-STAT3 proteins were
attenuated in cells and mouse tumours with LARP1 silencing.
Upregulated LARP1 in cells activated the expression of p-STAT3,
which could be reversed by miR-374a overexpression. miR-374a
inhibited cell proliferation and motility, as well as protein
levels of cyclin D1 and N-cadherin. All these results revealed that
miR-374a may influence the biological behaviour of NSCLC cells by
regulating LARP1 and downstream STAT3 signalling.
Accumulating evidence has indicated that
dysregulation of miRNAs promotes the pathogenesis of human
malignancies. Herein, we found that downregulated miR-374a was
associated with advanced tumour stage and poor survival. Similarly,
downregulated miR-374a was significantly associated with poor
survival in lung cancer patients (14). In addition, miR-374a abrogated the
promotor effect of LARP1 on cell proliferation, migration and
invasion in vitro, suggesting a tumour-suppressive role for
miR-374a in NSCLC. A previous study reported that the expression
level of miR-374a was suppressed in invasive adenocarcinoma
(27), and overexpression of
miR-374a inhibited cell growth and metastasis (15). All these studies suggest that
miR-374a may function as a tumour suppressor and that the
miR-374a/LARP1 axis may play a role in pulmonary
tumourigenesis.
Recently, numerous studies have shown that lncRNAs
can act as ceRNA for miRNAs; they function as molecular sponges to
competitively inhibit miRNAs. For example, XIST modulated gastric
cancer development by acting as a molecular sponge of miR-101 to
regulate EZH2 expression (28).
XIST could bind to miR-320b and suppress miR-320b, which was
involved in the dysregulation of RAP2B (29). From a publicly available database,
we found that XIST could potentially bind to miR-374a. A
dual-luciferase reporter assay was used to ascertain this
relationship. The results revealed that the luciferase activity of
the reporter containing XIST-WT was decreased in cells transfected
with the miR-374a mimic. XIST and miR-374a could inhibit each
other. Furthermore, the miR-374a inhibitor restored the inhibition
effect of sh-XIST on the expression of LARP1. Former studies
demonstrated that XIST contributed to NSCLC via the targeting of
different miRNAs, such as miR-449a and miR-186 (30,31),
implying that XIST may regulate the progression of NSCLC by
targeting multiple miRNAs. Herein, we found that the knockdown of
XIST exerted a tumour-suppressive role by inhibiting cell growth,
migration and invasion, which was inconsistent with previous
studies (28,32). However, XIST cannot only function as
an oncogene in tumourigenesis, but also function as a master
regulator of X chromosome inactivation (XCI) initiation. There is
no corresponding evidence to show that LARP1 is linked to XCI.
However, mmu-miR-374 was found to be mapped adjacent to XIST that
were predominantly expressed in female blastocysts (33). LncRNA FTX was reported as a positive
regulator of XIST and to be involved in XCI (34). Notably, FTX suppressed cell growth,
metastasis, Wnt/β-catenin signalling by competitively sponging
miR-374a. Thus, we speculated that miR-374a may be related with XCI
by interacting with XIST and/or FTX.
In conclusion, knockdown of LARP1 inhibited
proliferation and motility in NSCLC cells, phenomena that
contributed to NSCLC progression. Upregulated LARP1 may be used as
a potential prognostic and diagnostic factor. Our findings
highlighted that LARP1 was post-transcriptionally regulated by
miR-374a, and the XIST/miR-374a/LARP1 axis provided novel insight
into the molecular mechanisms of NSCLC oncogenesis.
Acknowledgements
The present study was supported by the Foundation of
Huzhou Science and Technology Bureau Project (grant no.
2015GY33).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen WQ, Zheng RS, Zhang SW, Li N, Zhao P,
Li GL, Wu LY and He J: Report of incidence and mortality in china
cancer registries, 2008. Chin J Cancer Res. 24:171–180. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stavraka C and Blagden S: The La-related
proteins, a family with connections to cancer. Biomolecules.
5:2701–2722. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burrows C, Latip Abd N, Lam SJ, Carpenter
L, Sawicka K, Tzolovsky G, Gabra H, Bushell M, Glover DM, Willis
AE, et al: The RNA binding protein Larp1 regulates cell division,
apoptosis and cell migration. Nucleic Acids Res. 38:5542–5553.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mura M, Hopkins TG, Michael T, Abd-Latip
N, Weir J, Aboagye E, Mauri F, Jameson C, Sturge J, Gabra H, et al:
LARP1 post-transcriptionally regulates mTOR and contributes to
cancer progression. Oncogene. 34:5025–5036. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xie C, Huang L, Xie S, Xie D, Zhang G,
Wang P, Peng L and Gao Z: LARP1 predict the prognosis for
early-stage and AFP-normal hepatocellular carcinoma. J Transl Med.
11:2722013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kato M, Goto Y, Matsushita R, Kurozumi A,
Fukumoto I, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M,
Ichikawa T, et al: MicroRNA-26a/b directly regulate
La-related protein 1 and inhibit cancer cell invasion in prostate
cancer. Int J Oncol. 47:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li XJ, Luo XQ, Han BW, Duan FT, Wei PP and
Chen YQ: MicroRNA-100/99a, deregulated in acute lymphoblastic
leukaemia, suppress proliferation and promote apoptosis by
regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J
Cancer. 109:2189–2198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe
TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al:
Genomic profiling of microRNA and messenger RNA reveals deregulated
microRNA expression in prostate cancer. Cancer Res. 68:6162–6170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Munker R and Calin GA: MicroRNA profiling
in cancer. Clin Sci. 121:141–158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu X, Wang W, Su N, Zhu X, Yao J, Gao W,
Hu Z and Sun Y: miR-374a promotes cell proliferation, migration and
invasion by targeting SRCIN1 in gastric cancer. FEBS Lett.
589:407–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan
J, Wu J and Li M: MicroRNA-374a activates Wnt/β-catenin signaling
to promote breast cancer metastasis. J Clin Invest. 123:566–579.
2013.PubMed/NCBI
|
|
13
|
Võsa U, Vooder T, Kolde R, Fischer K, Välk
K, Tõnisson N, Roosipuu R, Vilo J, Metspalu A and Annilo T:
Identification of miR-374a as a prognostic marker for survival in
patients with early-stage nonsmall cell lung cancer. Genes
Chromosomes Cancer. 50:812–822. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu H, Liu Y, Shu XO and Cai Q: MiR-374a
suppresses lung adenocarcinoma cell proliferation and invasion by
targeting TGFA gene expression. Carcinogenesis. 37:567–575.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Riemenschneider MJ, Betensky RA, Pasedag
SM and Louis DN: AKT activation in human glioblastomas enhances
proliferation via TSC2 and S6 kinase signaling. Cancer Res.
66:5618–5623. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu Z, Huang C, Sun J, Qiu W, Zhang J, Li
H, Jiang T, Huang K and Cao J: RNA interference-mediated signal
transducers and activators of transcription 3 gene silencing
inhibits invasion and metastasis of human pancreatic cancer cells.
Cancer Sci. 98:1099–1106. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takemoto S, Ushijima K, Kawano K,
Yamaguchi T, Terada A, Fujiyoshi N, Nishio S, Tsuda N, Ijichi M,
Kakuma T, et al: Expression of activated signal transducer and
activator of transcription-3 predicts poor prognosis in cervical
squamous-cell carcinoma. Br J Cancer. 101:967–972. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei R, Yang Q, Han B, Li Y, Yao K, Yang X,
Chen Z, Yang S, Zhou J, Li M, et al: microRNA-375 inhibits
colorectal cancer cells proliferation by downregulating JAK2/STAT3
and MAP3K8/ERK signaling pathways. Oncotarget. 8:16633–16641.
2017.PubMed/NCBI
|
|
19
|
Cheng N and Wang GH: miR-133b, a microRNA
targeting S1PR1, suppresses nasopharyngeal carcinoma cell
proliferation. Exp Ther Med. 11:1469–1474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ye L, Lin ST, Mi YS, Liu Y, Ma Y, Sun HM,
Peng ZH and Fan JW: Overexpression of LARP1 predicts poor prognosis
of colorectal cancer and is expected to be a potential therapeutic
target. Tumour Biol. 37:14585–14594. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tcherkezian J, Cargnello M, Romeo Y,
Huttlin EL, Lavoie G, Gygi SP and Roux PP: Proteomic analysis of
cap-dependent translation identifies LARP1 as a key regulator of
5′TOP mRNA translation. Genes Dev. 28:357–371. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sandler AB: Molecular targeted agents in
non-small-cell lung cancer. Clin Lung Cancer. 5 Suppl 1:S22–S28.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song L, Rawal B, Nemeth JA and Haura EB:
JAK1 activates STAT3 activity in non-small-cell lung cancer cells
and IL-6 neutralizing antibodies can suppress JAK1-STAT3 signaling.
Mol Cancer Ther. 10:481–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chiu HC, Chou DL, Huang CT, Lin WH, Lien
TW, Yen KJ and Hsu JT: Suppression of Stat3 activity sensitizes
gefitinib-resistant non small cell lung cancer cells. Biochem
Pharmacol. 81:1263–1270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee C, Dhillon J, Wang MY, Gao Y, Hu K,
Park E, Astanehe A, Hung MC, Eirew P, Eaves CJ, et al: Targeting
YB-1 in HER-2 overexpressing breast cancer cells induces apoptosis
via the mTOR/STAT3 pathway and suppresses tumor growth in mice.
Cancer Res. 68:8661–8666. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin YH, Wu MH, Liao CJ, Huang YH, Chi HC,
Wu SM, Chen CY, Tseng YH, Tsai CY, Chung IH, et al: Repression of
microRNA-130b by thyroid hormone enhances cell motility. J Hepatol.
62:1328–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sato T, Shiba-Ishii A, Kim Y, Dai T, Husni
RE, Hong J, Kano J, Sakashita S, Iijima T and Noguchi M: miR-3941:
A novel microRNA that controls IGBP1 expression and is associated
with malignant progression of lung adenocarcinoma. Cancer Sci.
108:536–542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL,
Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al: Long non-coding
RNA XIST regulates gastric cancer progression by acting as a
molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin
Cancer Res. 35:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lv GY, Miao J and Zhang XL: Long
non-coding RNA XIST promotes osteosarcoma progression by targeting
Ras-related protein RAP2B via miR-320b. Oncol Res. Apr
12–2017.(Epub ahead of print). doi:
10.3727/096504017X14920318811721. View Article : Google Scholar :
|
|
30
|
Zhang YL, Li XB, Hou YX, Fang NZ, You JC
and Zhou QH: The lncRNA XIST exhibits oncogenic properties via
regulation of miR-449a and Bcl-2 in human non-small cell lung
cancer. This article has been corrected since Advanced Online
Publication, and an erratum is also printed in this issue. Acta
Pharmacol Sin. 38:371–381. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang H, Shen Q, Zhang X, Yang C, Cui S,
Sun Y, Wang L, Fan X and Xu S: The long non-coding RNA XIST
controls non-small cell lung cancer proliferation and invasion by
modulating miR-186-5p. Cell Physiol Biochem. 41:2221–2229. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J,
Chen L, Xi Z, Teng H, Wang Z, et al: Knockdown of long non-coding
RNA XIST exerts tumor-suppressive functions in human glioblastoma
stem cells by up-regulating miR-152. Cancer Lett. 359:75–86. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kobayashi S, Totoki Y, Soma M, Matsumoto
K, Fujihara Y, Toyoda A, Sakaki Y, Okabe M and Ishino F:
Identification of an imprinted gene cluster in the X-inactivation
center. PLoS One. 8:e712222013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chureau C, Chantalat S, Romito A, Galvani
A, Duret L, Avner P, Rougeulle C, Okabe M and Ishino F: Ftx
is a non-coding RNA which affects Xist expression and
chromatin structure within the X-inactivation center region. Hum
Mol Genet. 20:705–718. 2011. View Article : Google Scholar : PubMed/NCBI
|