Introduction
Nasopharyngeal carcinoma (NPC), which is a form of
squamous cell carcinoma (1), is a
head and neck malignant tumour. NPC is endemic in southeast Asia
and southern areas of China (2).
For the treatment of NPC, radiotherapy can be used alone or in
combination with chemotherapy and surgery (3). NPC is characterised by early cervical
lymph node and distant metastases. The probabilities of treatment
failure and relapse in NPC patients with distant metastasis have
not been significantly reduced to date. Therefore, comprehensive
studies on the causes and biological processes of NPC metastasis
are needed to improve the therapeutic efficacy for this
malignancy.
Yeast two-hybrid (Y2H) is a method to screen unknown
proteins that interact with known proteins (4, 5). In our previous
study, we discovered that flotillin2 (Flot2) can promote the
progression and metastasis of NPC and revealed the molecular
mechanisms by which it exerts this function (6–8). In
the present study, we aimed to identify the proteins that interact
with Flot2 in NPC using a Y2H system. Among the five obtained
proteins, PLCD3 attracted our attention since it is located on the
cell membrane similar to Flot2.
Flot2 is a lipid raft marker protein, and directly
interacts with signaling molecules such as receptors, kinases,
adhesion molecules and G proteins. It also serves as a tumour
regulator by regulating cell proliferation, differentiation,
apoptosis, adhesion and invasion. It has been reported that Flot2
participates in the development of several types of malignant
tumours such as breast cancer, melanoma, gastric and cervical
cancer, and NPC (9).
PLCD3 is a member of the phospholipase C (PLC)
family. PLC is a pivotal enzyme in the phosphoinositide pathway and
is involved in eukaryotic signal transduction through the
generation of two second messengers: diacylglycerol (DAG) and
inositol 1,4,5-trisphosphate (IP3). The former mediates the
activation of protein kinase C (PKC) (10), and the latter regulates the release
of Ca2+ from intracellular stores (11,12).
Since the 1950s, 13 isozymes of six PLC family members in mammals
have been reported, including PLCβ1-4, PLCγ1-2, PLCδ1-4, PLC-ε,
PLC-ζ and PLC-η (13–16). The core structures of these proteins
include a PH domain in N-terminus, a catalytic centre formed by X
and Y regions, an EF-hands motif and a C2 domain in the C-terminal
region (17). PLC isozymes have
both common and characteristic features that reflect their
physiological and pathological functions, so each isozyme may be
associated with specific human disease (18). PLC-β1 is involved in schizophrenia
(19,20), myelodysplastic syndromes (MDS)
(21–23), leukemogenesis (24) and diabetes (25). PLC-γ1 is implicated in epilepsy
(26), cancer cell invasion and
metastasis in brain tumours (27)
and breast tumour formation (28).
PLC-δ1 is associated with obesity (29), neurodegenerative disorders (30), and Alzheimer disease (31) and may be a tumour suppressor
(32,33). PLC-ε may function as a tumour
suppressor (34,35). PLC-ζ is related to male infertility
(36), whereas PLC-β3 is associated
with lymphomas and other tumours (37).
In the present study, we identified a
Flot2-interacting protein, PLCD3, which may play an important role
in NPC progression.
Materials and methods
Cell culture
Briefly, 5-8F NPC cells were maintained by our
laboratory and grown in RPMI-1640 media (Invitrogen, Carlsbad, CA,
USA) supplemented with 10% fetal bovine serum (FBS) in a humidified
atmosphere with 5% CO2 at 37°C.
Yeast two-hybrid
The culture conditions and treatment methods of
yeast strains Y2Gold and Y187 were similar to those described
previously (38). PMD18-T vector
(kept by our laboratory) was ligated to the open reading frame
(ORF) of Flot2, which was amplified by PCR using a sense primer
(5′-CGAATTCATGAGTAGCTCCCACTCTC-3′) and an antisense primer
(5′-CGGATCCTCAGGGCACCACAACGC-3′) from 5-8F cDNA. Then, the yielded
product and pGBKT7-BD were double digested by EcoRI and
BamHI (both from Takara, Shiga, Japan), respectively. The
digested DNA fragment was recombined in vitro. The obtained
yeast bait vector pGBKT7-Flot2 was transfected into yeast bait
strain Y2Gold.
The cDNA of 5–8F was reverse transcribed using
SMART™ technology and co-transfected into yeast library strain Y187
(both from Clontech, Shiga, Japan) with pGADT7-Rec vector, which
was then plated on SD/-leu agar medium. After 4–5 days, the
colonies were pooled in rich broth medium, divided into 1
ml-aliquots and cryopreserved. The cDNA library screening was
performed according to our previous protocol (38).
Co-immunoprecipitation (Co-IP)
The ORF of Flot2 was cloned into a BamHI- and
NotI-digested pEF1/myc-His B vector (Invitrogen), which is a
6.2-kb expression vector used to overexpress recombinant protein.
pFlag-CMV-3 (Sigma, Rockford, IL, USA) is a 6.2-kb expression
vector transiently or stably expressed in mammals, in which the
N-terminal Flag can form fusion protein with a correctly inserted
ORF. The ORF of PLCD3 was amplified from 5–8F cDNA by PCR using the
sense primer (5′-CAAGCTTATGCTGTGCGGCCGCTGGA-3′) and the antisense
primer (5′-CGGATCCTCAGGAGCGCTGGATGCGGAT-3′), and then subcloned
into pFlag-CMV-3. Then, pEF1/myc-His-Flot2 and pFlag-CMV-PLCD3 were
transfected into 293T cells with Lipofectamine 2000™ (Invitrogen).
All experiments were performed according to a previously published
protocol (38). Anti-Flag (Sigma)
was used for immunoprecipitation. Anti-His (ProteinTech, Wuhan,
China) was used for immunoblotting.
siRNA treatment
Cells were transfected with RNAiMax (Invitrogen) and
PLCD3 siRNA or control siRNA was synthesised by RiboBio Co.
(Guangzhou, China). The siRNA complex was formed in Opti I medium
(Invitrogen) following the manufacturer's recommendations. The
optimal concentrations tested to effectively reduce expression of
PLCD3 24 h after seeding 2×105 cells in 60-mm dishes
were 7.5 pmol siRNA and 5 µl of RNAiMax/1 ml of growth medium.
Human PLCD3 siRNA and control siRNA were purchased from RiboBio Co.
The siRNA sequences are as follows: si-1, GGATGAACT CAGCCAACT;
si-2, GCCCACT ACTTCATCTCTT; and si-3, GGAGCCCGTCATCTATCAT. In our
experiments, si-1 and si-2 were most effective on suppressing PLCD3
expression.
Real-time qPCR
Real-time qPCR was performed as previously described
(39,40). Briefly, 48 h after treatment, each
60-mm dish of monolayer cells was washed twice with 2 ml D-Hank's
solution. Then, total RNA was isolated. qPCR reactions were
performed as previously described. Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was used as an internal control. PLCD3 was
amplified using the forward primer
(5′-CAAGCTTATGCTGTGCGGCCGCTGGA-3′) and the reverse primer
(5′-CGGATCCTCAGGAGCGCTGGATGCGGAT-3′). This experiment was
independently repeated three times.
Western blot analysis
Western blotting was performed according to a
previous protocol (2). Rabbit
polyclonal anti-PLCD3, anti-MMP2, anti-MMP9, anti-Snai1,
anti-E-cadherin, anti-vimentin, anti-N-cadherin and anti-Flot2 were
purchased from ProteinTech. Mouse anti-β-actin was purchased from
Sigma. Rabbit anti-mouse IgG was purchased from Abcam (1:40,000;
Cambridge, MA, USA). Quantification of signal intensity [integral
optical density (IOD)] was performed with Gel-Pro Analyzer software
(version 4.0). Expression changes in protein levels were assessed
by IOD, and the intensity was normalised to the β-actin signal.
This experiment was independently repeated three times.
Colony formation assay
Twenty-four hours after treatment, the transfected
cells were digested with trypsin and seeded in 60-mm dishes at
1,000 cells/well. Colony formation assays were performed according
to a published manual (41). The
data are expressed as the means ± SD of colony numbers. This
experiment was independently repeated three times.
In vitro cell proliferation assay
Twenty-four hours after treatment, the transfected
cells were digested with trypsin and seeded in 96-well dishes at
3,000 cells/well. MTT assays were performed to assess the effect of
PLCD3 on 5–8F proliferation in accordance with a published protocol
(42). The experiment was
independently repeated three times.
Fluorescence-activated cell sorting
(FACS)
Twenty-four to forty-eight hours after treatment,
the transfected cells were digested with trypsin, and then, FACS
analysis was performed as previously described (43). The experiment was independently
repeated three times.
Matrigel invasion assay
Twenty-four hours after treatment, Matrigel invasion
assays were performed according to a published protocol (8). The experiment was independently
repeated three times.
Wound healing assay
Forty-eight hours after treatment, wound healing
assays were performed according to a previous protocol (43). The experiment was independently
repeated three times.
Statistical analysis
Statistical analyses were executed using SPSS
version 18.0 statistical software (SPSS, Inc., Chicago, IL, USA).
Differences were analysed using Student's t-test. A P-value ≤0.05
was indicative of statistical significance.
Results
No autonomous activity and toxicity of
the pGBKT7-Flot2 yeast bait vector
Y2Hgold yeast cells transfected with pGBKT7-Flot2
were plated on SD/-Trp/X-α-Gal, SD/-His/-Trp/X-α-Gal or
SD/-Ade/-Trp/X-α-Gal for 3–5 days. pGBKT7-Flot2-Y2Hgold yeast can
proliferate on SD/-Trp/X-α-Gal (Fig.
1A, below), but not on SD/-His/-Trp/X-α-Gal (Fig. 1A, upper left) or
SD/-Ade/-Trp/X-α-Gal (Fig. 1A,
upper right), suggesting the lack of autonomous activity of the
yeast bait vector in the tested yeast. Y2Hgold yeast transfected
with pGBKT7-Flot2 or pGBKT7-BD were individually plated on SD/-Trp
plates. No obvious differences in the growth rate and colony size
were noted between these two groups (Fig. 1B). The results above demonstrate
that the Y2H system could be used in the following experiments.
Four candidate Flot2-interacting
proteins are identified
The quality of the 5–8F cDNA library was verified by
calculating the titre of the library (Fig. 1C). In total, 65 positive candidate
clones were observed on QDO (SD/-Ade/-His/-Leu/-Trp) plates
(Fig. 1D, upper panel), and 32
positive clones with blue colour appeared on
SD/-Ade/-His/-Leu/-Trp/ABA/X-α-Gal plates (Fig. 1D, lower panel). The positive
candidate clones were amplified. The plasmids were extracted, and
the target cDNAs were sequenced. Finally, five candidate
Flot2-interacting proteins were obtained (Table I), including Copine-II PCDH7, PCBP1,
PTPRJ and PLCD3. After further co-transfection experiments, only
PCDH7, PCBP1, PTPRJ and PLCD3 proteins were confirmed to interact
with Flot2. Only yeasts transfected with pGBKT7-Flot2 and
pGADT7-candidate vectors encoding PCDH7, PCBP1, PTPRJ and PLCD3 was
able to grow on QDO plates, and further proliferate and appear blue
when grown on SD/-Ade/-His/-Leu/-Trp/ABA/X-α-Gal plates, whereas
yeasts transfected with pGBKT7-BD and pGADT7-Copine-II vectors
could not (Fig. 1E and F),
indicating that these four candidate proteins interact with
Flot2.
 | Table. I.BLAST results of positive clones in
the Yeast two-hybrid analysis. |
Table. I.
BLAST results of positive clones in
the Yeast two-hybrid analysis.
| Protein no. | Protein name | Gene | NCBI protein
accession no. | Max identify
(%) |
|---|
| 1 | CPNE2 | Copine-II | XP_003823796 | 99 |
| 2 |
Protocadherin-7 | PCDH7 | XP_006184446 | 99 |
| 3 | Poly(C)-binding
protein 1 | PCBP1 | XP_003922705 | 100 |
| 4 | PTPRJ protein | PTPRJ | AAH63417 | 100 |
| 5 | PLCD3 protein,
partial | PLCD3 | AAH10668 | 99 |
Interaction between Flot2 and PLCD3 is
further confirmed by Co-IP
To further verify the interaction between Flot2 and
PLCD3, a mammal expression plasmid encoding His-Flot2 (ORF) and a
plasmid encoding Flag-PLCD3 were co-transfected into HEK-293T
cells. The lysates were immunoprecipitated with anti-Flag antibody,
and then immunoblotted with an anti-His antibody. The His tag could
be detected (Fig. 2), which
confirmed the interaction between PLCD3 and Flot2.
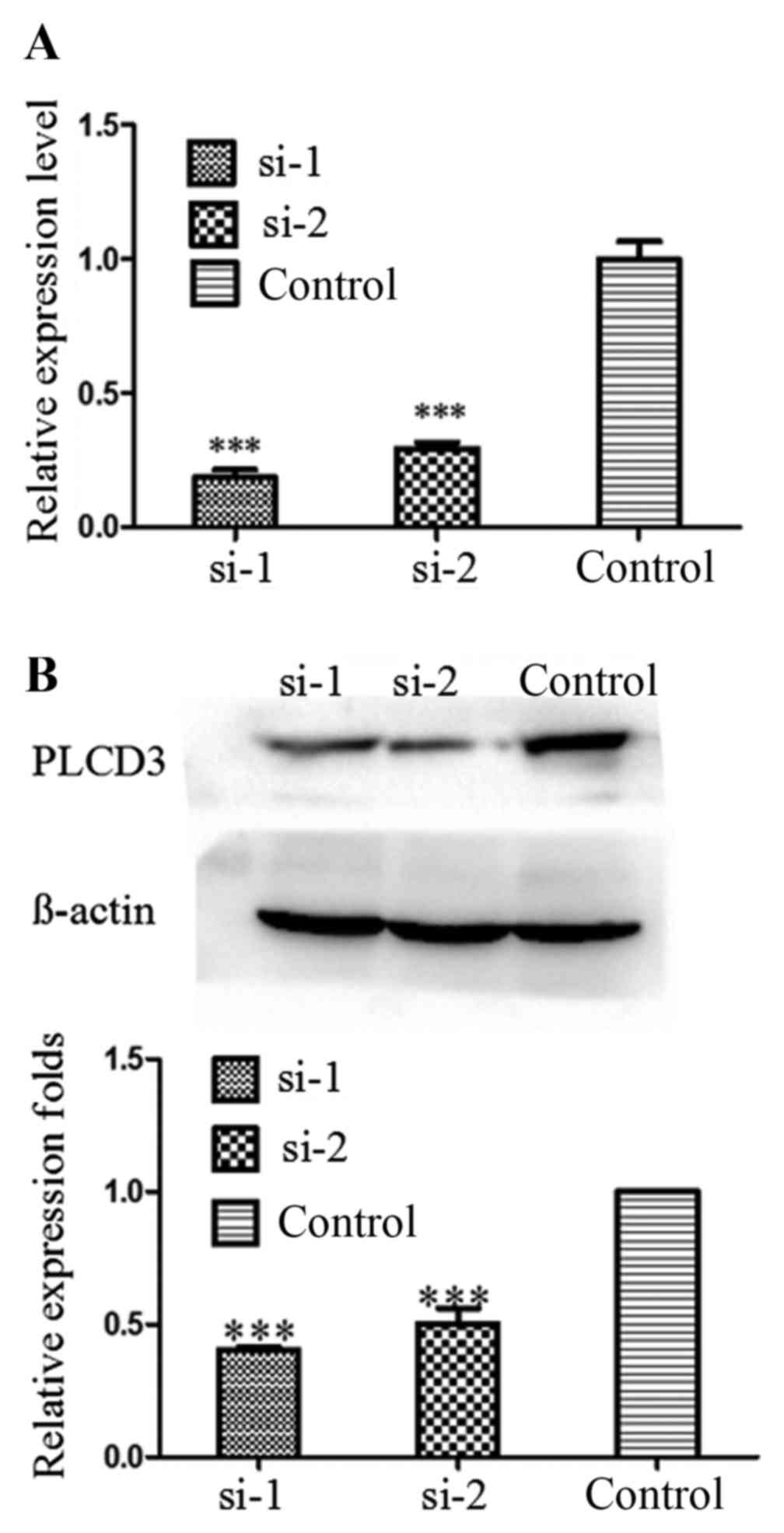
PLCD3 expression is silenced in 5–8F
cells by siRNA transfection
To verify the knockdown efficiency after
transfection with PLCD3 siRNAs (si-1 or si-2), total RNA and
protein of 5–8F cells were extracted at 24 and 48 h after
treatment, separately. Then, qPCR and western blotting were
performed. As shown in Fig. 3A and
B, the knockdown efficiency of PLCD3 mRNA was ~80%, and that of
PLCD3 protein was ~50–60%.
5-8F cell proliferation is inhibited
by PLCD3 silencing
MTT assay results showed that the proliferation rate
of the 5–8F cells transfected with si-1 and si-2 was significantly
decreased compared with the control group (Fig. 4). The results demonstrated that
downregulation of PLCD3 inhibited the proliferation of 5–8F
cells.
PLCD3 silencing inhibits the colony
formation ability of 5–8F cells
The colony formation ability of 5–8F cells after
transfection with PLCD3 siRNA was detected by colony formation
assay. Six days after transfection with PLCD3 siRNA, as shown in
Fig. 5, the number of colonies that
formed in the si-1 and si-2 groups were significantly reduced
compared with this number noted in the control group.
PLCD3 silencing suppresses 5–8F cell
migration
The migration of 5–8F cells after transfection with
PLCD3 siRNA was detected by wound healing assays. Twenty-four to
forty-eight hours after treatment, the migration was measured. As
shown in Fig. 6A, the relative
blank areas in the si-1 and si-2 groups were larger compared with
the control group. Statistical analysis showed that transfection
with si-1 and si-2 significantly inhibited 5–8F cell migration
(Fig. 6B)
PLCD3 silencing impairs the invasive
ability of 5–8F cells
The invasive ability of 5–8F cells after
transfection with siRNA was detected by Matrigel invasion assay.
Forty-eight hours after treatment, the number of cells passing
through the Matrigel was counted. As shown in Fig. 7, the number of cells passing through
the Matrigel in the si-1 and si-2 groups was obviously decreased
compared with the control group. Statistical analysis revealed that
transfection with si-1 and si-2 significantly inhibited the
invasive ability of the 5–8F cells (Fig. 7B).
PLCD3 silencing influences the cell
cycle of 5–8F cells
The cell cycle of 5–8F cells after transfection with
siRNA was detected by FACS (Fig.
8A). Compared with the control group, the number of cells in
the G2/M phase was increased significantly, and those in S and G1
phase were decreased in the si-1 group. Compared with the control
group, the number of cells in the G2/M phase was significantly
increased. The number of S phase cells was decreased, and the
number of G1 phase cells was not significantly altered in the si-2
group (Fig. 8B).
PLCD3 silencing influences the
expression of Flot2 and several proteins related to EMT and
invasion
We found that the invasion and migration ability of
the 5–8F cells was inhibited after silencing of PLCD3, thus we
detected the expression of several proteins related to EMT and
invasion including MMP2, MMP9, Snai1, E-cadherin, N-cadherin and
vimentin. At the same time, we detected the expression of the
interaction protein Flot2. After silencing the expression of PLCD3,
the expression levels of MMP2, MMP9, Snai1, vimentin, N-cadherin
were decreased, and the expression level of E-cadherin was
increased. However, the expression of Flot2 was not significantly
altered (Fig. 9).
Discussion
In the present study, we found that PLCD3 interacts
with Flot2. Flot2 is highly expressed and associated with tumour
progression and metastasis in a variety of tumours (8,44–53),
suggesting that it may be an independent diagnostic marker and
potential therapeutic target. Our previous research revealed that
downregulation of Flot2 in 5–8F cells led to reductions in colony
formation, proliferation, migration and invasion; and inhibited
cell cycle progression. In addition, it is associated with NPC
metastasis (8).
Phosphoinositide-specific phospholipase C (PLC)
catalyses phosphatidylinositol 4,5-bisphosphate (PIP2) into two
second messengers, which play important roles in cell movement,
growth and diseases. To date, the roles of PLC in the progression
of cancers have rarely been investigated. PLC-γ1 may play a role in
cancer cell invasion, metastasis and breast tumour formation
(27,28). PLC-δ1 and PLC-ε are regarded as
tumour suppressors (32–35), whereas PLC-β3 is associated with
lymphomas and other tumours, such as myeloid malignancies (37). PLCD3 plays important roles in
numerous biological processes, such as survival of cardiomyocytes
and trophoblasts, maintaining normal heart function and promoting
neurite expansion (54–56). Studies concerning the roles of PLCD3
in tumours are rare. To the best of our knowledge, there is only
one study demonstrating that PLCD3 can promote the proliferation
and migration of neoplastic mammary epithelial cells (57). The role of PLCD3 in NPC has not been
studied to date. Thus, we ascertained whether PLCD3 plays a certain
role in NPC. In the present study, PLCD3 silencing in 5–8F cells by
siRNA inhibited colony formation, proliferation, migration and
invasion and blocked cell cycle progression. These results are
consistent with previous findings.
We also analysed the effects of PLCD3 silencing on
the expression of several proteins related to EMT and invasion
including MMP2, MMP9, Snai1, E-cadherin, N-cadherin and vimentin.
After silencing the expression of PLCD3, the expression levels of
MMP2, MMP9, Snai1, vimentin and N-cadherin were apparently
decreased, and the expression level of E-cadherin was apparently
increased, which is in accordance with our previous study on Flot2
(8).
In conclusion, we first discovered the interaction
between Flot2 and PLCD3 using the Y2H system and verified this
interaction by co-immunoprecipitation. The roles of PLCD3 in
promoting proliferation, migration, and invasion of NPC cells were
also demonstrated by silencing its expression with siRNAs. Flot2
plays an important role in the progression of NPC, which may be
partially related to its interaction with PLCD3.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (nos. 81773179 and 81272972),
the Natural Science Foundation of Hunan Province of China (nos.
2016JJ2172), Mittal Student Innovation Project, the National Basic
Research Program of China (2010CB833605), the Hunan Provincial
Science and Technology Department (nos. 2016JC2049, 2014FJ6006 and
2013FJ4010), and the Open-End Fund for the Valuable and Precision
Instruments of Central South University.
References
|
1
|
Sham JS, Wei WI, Zong YS, Choy D, Guo YQ,
Luo Y, Lin ZX and Ng MH: Detection of subclinical nasopharyngeal
carcinoma by fibreoptic endoscopy and multiple biopsy. Lancet.
335:371–374. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feng X, Ren C, Zhou W, Liu W, Zeng L, Li
G, Wang L, Li M, Zhu B, Yao K, et al: Promoter hypermethylation
along with LOH, but not mutation, contributes to inactivation of
DLC-1 in nasopharyngeal carcinoma. Mol Carcinog. 53:858–870. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang L, Chen QY, Liu H, Tang LQ and Mai
HQ: Emerging treatment options for nasopharyngeal carcinoma. Drug
Des Devel Ther. 7:37–52. 2013.PubMed/NCBI
|
|
4
|
Brückner A, Polge C, Lentze N, Auerbach D
and Schlattner U: Yeast two-hybrid, a powerful tool for systems
biology. Int J Mol Sci. 10:2763–2788. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
James P, Halladay J and Craig EA: Genomic
libraries and a host strain designed for highly efficient
two-hybrid selection in yeast. Genetics. 144:1425–1436.
1996.PubMed/NCBI
|
|
6
|
Yang XY, Ren CP, Wang L, Li H, Jiang CJ,
Zhang HB, Zhao M and Yao KT: Identification of differentially
expressed genes in metastatic and non-metastatic nasopharyngeal
carcinoma cells by suppression subtractive hybridization. Cell
Oncol. 27:215–223. 2005.PubMed/NCBI
|
|
7
|
Wen Q, Li J, Wang W, Xie G, Xu L, Luo J,
Chu S, She L, Li D, Huang D, et al: Increased expression of
flotillin-2 protein as a novel biomarker for lymph node metastasis
in nasopharyngeal carcinoma. PLoS One. 9:e1016762014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Huang W, Ren C, Wen Q, Liu W, Yang
X, Wang L, Zhu B, Zeng L, Feng X, et al: Flotillin-2 promotes
metastasis of nasopharyngeal carcinoma by activating NF-κB and
PI3K/Akt3 signaling pathways. Sci Rep. 5:116142015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao K, Xie D, Cao P, Zou Q, Lu C, Xiao S,
Zhou J and Peng X: SiRNA-mediated flotillin-2 (Flot2)
downregulation inhibits cell proliferation, migration, and invasion
in gastric carcinoma cells. Oncol Res. 21:271–279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nishizuka Y: Protein kinase C and lipid
signaling for sustained cellular responses. FASEB J. 9:484–496.
1995.PubMed/NCBI
|
|
11
|
De Smedt H and Parys JB: Molecular and
functional diversity of inositol triphosphate-induced
Ca2+ release. Verh K Acad Geneeskd Belg. 57:423–458.
1995.(In Dutch). PubMed/NCBI
|
|
12
|
Berridge MJ: Inositol trisphosphate and
calcium signalling. Nature. 361:315–325. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cockcroft S and Thomas GM:
Inositol-lipid-specific phospholipase C isoenzymes and their
differential regulation by receptors. Biochem J. 288:1–14. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rhee SG and Bae YS: Regulation of
phosphoinositide-specific phospholipase C isozymes. J Biol Chem.
272:15045–15048. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kelley GG, Reks SE, Ondrako JM and Smrcka
AV: Phospholipase C(epsilon): A novel Ras effector. EMBO J.
20:743–754. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Harden TK and Sondek J: Regulation of
phospholipase C isozymes by ras superfamily GTPases. Annu Rev
Pharmacol Toxicol. 46:355–379. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Katan M and Williams RL:
Phosphoinositide-specific phospholipase C: Structural basis for
catalysis and regulatory interactions. Semin Cell Dev Biol.
8:287–296. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhee SG: Reflections on the days of
phospholipase C. Adv Biol Regul. 53:223–231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
García del Caño G, Montaña M, Aretxabala
X, González-Burguera I, López de Jesús M, Barrondo S and Sallés J:
Nuclear phospholipase C-β1 and diacylglycerol LIPASE-α in brain
cortical neurons. Adv Biol Regul. 54:12–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koh HY: Phospholipase C-β1 and
schizophrenia-related behaviors. Adv Biol Regul. 53:242–248. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Follo MY, Faenza I, Fiume R, Ramazzotti G,
McCubrey JA, Martelli AM, Manzoli FA and Cocco L: Revisiting
nuclear phospholipase C signalling in MDS. Adv Biol Regul. 52:2–6.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Follo MY, Marmiroli S, Faenza I, Fiume R,
Ramazzotti G, Martelli AM, Gobbi P, McCubrey JA, Finelli C, Manzoli
FA, et al: Nuclear phospholipase C β1 signaling, epigenetics and
treatments in MDS. Adv Biol Regul. 53:2–7. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manzoli L, Mongiorgi S, Clissa C, Finelli
C, Billi AM, Poli A, Quaranta M, Cocco L and Follo MY: Strategic
role of nuclear inositide signalling in myelodysplastic syndromes
therapy. Mini Rev Med Chem. 14:873–883. 2014. View Article : Google Scholar
|
|
24
|
Zaidi SK, Trombly DJ, Dowdy CR, Lian JB,
Stein JL, van Wijnen AJ and Stein GS: Epigenetic mechanisms in
leukemia. Adv Biol Regul. 52:369–376. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barker CJ, Li L, Köhler M and Berggren PO:
β-Cell Ca2+ dynamics and function are compromised in
aging. Adv Biol Regul. 57:112–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jang HJ, Yang YR, Kim JK, Choi JH, Seo YK,
Lee YH, Lee JE, Ryu SH and Suh PG: Phospholipase C-γ1 involved in
brain disorders. Adv Biol Regul. 53:51–62. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lattanzio R, Piantelli M and Falasca M:
Role of phospholipase C in cell invasion and metastasis. Adv Biol
Regul. 53:309–318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arteaga CL, Johnson MD, Todderud G, Coffey
RJ, Carpenter G and Page DL: Elevated content of the tyrosine
kinase substrate phospholipase C-gamma 1 in primary human breast
carcinomas. Proc Natl Acad Sci USA. 88:pp. 10435–10439. 1991;
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakamura Y, Kanemarum K and Fukami K:
Physiological functions of phospholipase Cδ1 and phospholipase Cδ3.
Adv Biol Regul. 53:356–362. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shimohama S, Perry G, Richey P, Takenawa
T, Whitehouse PJ, Miyoshi K, Suenaga T, Matsumoto S, Nishimura M
and Kimura J: Abnormal accumulation of phospholipase C-delta in
filamentous inclusions of human neurodegenerative diseases.
Neurosci Lett. 162:183–186. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shimohama S, Homma Y, Suenaga T, Fujimoto
S, Taniguchi T, Araki W, Yamaoka Y, Takenawa T and Kimura J:
Aberrant accumulation of phospholipase C-delta in Alzheimer brains.
Am J Pathol. 139:737–742. 1991.PubMed/NCBI
|
|
32
|
Fu L, Qin YR, Xie D, Hu L, Kwong DL,
Srivastava G, Tsao SW and Guan XY: Characterization of a novel
tumor-suppressor gene PLC delta 1 at 3p22 in esophageal squamous
cell carcinoma. Cancer Res. 67:10720–10726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu XT, Zhang FB, Fan YC, Shu XS, Wong AH,
Zhou W, Shi QL, Tang HM, Fu L, Guan XY, et al: Phospholipase C
delta 1 is a novel 3p22.3 tumor suppressor involved in cytoskeleton
organization, with its epigenetic silencing correlated with
high-stage gastric cancer. Oncogene. 28:2466–2475. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chan JJ and Katan M: PLCε and the RASSF
family in tumour suppression and other functions. Adv Biol Regul.
53:258–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Zbou C, Qiu G, Fan J, Tang H and
Peng Z: Screening of new tumor suppressor genes in sporadic
colorectal cancer patients. Hepatogastroenterology. 55:2039–2044.
2008.PubMed/NCBI
|
|
36
|
Amdani SN, Jones C and Coward K:
Phospholipase C zeta (PLCζ): Oocyte activation and clinical links
to male factor infertility. Adv Biol Regul. 53:292–308. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiao W, Hong H, Kawakami Y, Kato Y, Wu D,
Yasudo H, Kimura A, Kubagawa H, Bertoli LF, Davis RS, et al: Tumor
suppression by phospholipase C-beta3 via SHP-1-mediated
dephosphorylation of Stat5. Cancer Cell. 16:161–171. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi J, Ren C, Liu H, Wang L, Zhu B, Huang
W, Liu W, Liu J, Liu Y, Xia X, et al: An ESRG-interacting protein,
COXII, is involved in pro-apoptosis of human embryonic stem cells.
Biochem Biophys Res Commun. 460:130–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wanggou S, Jiang X, Li Q, Zhang L, Liu D,
Li G, Feng X, Liu W, Zhu B, Huang W, et al: HESRG: A novel
biomarker for intracranial germinoma and embryonal carcinoma. J
Neurooncol. 106:251–259. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang G, Liu D, Xiao G, Liu Q and Yuan J:
Transcriptional repression of FOXO1 by KLF4 contributes to glioma
progression. Oncotarget. 7:81757–81767. 2016.PubMed/NCBI
|
|
41
|
Zhou W, Feng X, Ren C, Jiang X, Liu W,
Huang W, Liu Z, Li Z, Zeng L, Wang L, et al: Over-expression of
BCAT1, a c-Myc target gene, induces cell proliferation, migration
and invasion in nasopharyngeal carcinoma. Mol Cancer. 12:532013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang H, Feng X, Liu W, Jiang X, Shan W,
Huang C, Yi H, Zhu B, Zhou W, Wang L, et al: Underlying mechanisms
for LTF inactivation and its functional analysis in nasopharyngeal
carcinoma cell lines. J Cell Biochem. 112:1832–1843. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng X, Li C, Liu W, Chen H, Zhou W, Wang
L, Zhu B, Yao K, Jiang X and Ren C: DLC-1, a candidate tumor
suppressor gene, inhibits the proliferation, migration and
tumorigenicity of human nasopharyngeal carcinoma cells. Int J
Oncol. 42:1973–1984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hazarika P, McCarty MF, Prieto VG, George
S, Babu D, Koul D, Bar-Eli M and Duvic M: Up-regulation of
Flotillin-2 is associated with melanoma progression and modulates
expression of the thrombin receptor protease activated receptor 1.
Cancer Res. 64:7361–7369. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gómez V, Sesé M, Santamaría A, Martínez
JD, Castellanos E, Soler M, Thomson TM and Paciucci R: Regulation
of aurora B kinase by the lipid raft protein flotillin-1. J Biol
Chem. 285:20683–20690. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Satyamoorthy K, Li G, Gerrero MR, Brose
MS, Volpe P, Weber BL, Van Belle P, Elder DE and Herlyn M:
Constitutive mitogen-activated protein kinase activation in
melanoma is mediated by both BRAF mutations and autocrine growth
factor stimulation. Cancer Res. 63:756–759. 2003.PubMed/NCBI
|
|
47
|
Liu Y, Lin L, Huang Z, Ji B, Mei S, Lin Y
and Shen Z: High expression of flotillin-2 is associated with poor
clinical survival in cervical carcinoma. Int J Clin Exp Pathol.
8:622–628. 2015.PubMed/NCBI
|
|
48
|
Berger T, Ueda T, Arpaia E, Chio II,
Shirdel EA, Jurisica I, Hamada K, You-Ten A, Haight J, Wakeham A,
et al: Flotillin-2 deficiency leads to reduced lung metastases in a
mouse breast cancer model. Oncogene. 32:4989–4994. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang YL, Yao WJ, Guo L, Xi HF, Li SY and
Wang ZM: Expression of flotillin-2 in human non-small cell lung
cancer and its correlation with tumor progression and patient
survival. Int J Clin Exp Pathol. 8:601–607. 2015.PubMed/NCBI
|
|
50
|
Lin C, Wu Z, Lin X, Yu C, Shi T, Zeng Y,
Wang X, Li J and Song L: Knockdown of FLOT1 impairs cell
proliferation and tumorigenicity in breast cancer through
upregulation of FOXO3a. Clin Cancer Res. 17:3089–3099. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yan Y, Yang FQ, Zhang HM, Che J and Zheng
JH: Up-regulation of flotillin-2 is associated with renal cell
carcinoma progression. Tumour Biol. 35:10479–10486. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao L, Lin L, Pan C, Shi M, Liao Y, Bin J
and Liao W: Flotillin-2 promotes nasopharyngeal carcinoma
metastasis and is necessary for the epithelial-mesenchymal
transition induced by transforming growth factor-β. Oncotarget.
6:9781–9793. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Takano N, Iizuka N, Hazama S, Yoshino S,
Tangoku A and Oka M: Expression of estrogen receptor-alpha and
-beta mRNAs in human gastric cancer. Cancer Lett. 176:129–135.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nakamura Y, Hamada Y, Fujiwara T, Enomoto
H, Hiroe T, Tanaka S, Nose M, Nakahara M, Yoshida N, Takenawa T, et
al: Phospholipase C-delta1 and -delta3 are essential in the
trophoblast for placental development. Mol Cell Biol.
25:10979–10988. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nakamura Y, Kanemaru K, Kojima R,
Hashimoto Y, Marunouchi T, Oka N, Ogura T, Tanonaka K and Fukami K:
Simultaneous loss of phospholipase Cδ1 and phospholipase Cδ3 causes
cardiomyocyte apoptosis and cardiomyopathy. Cell Death Dis.
5:e12152014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kouchi Z, Igarashi T, Shibayama N, Inanobe
S, Sakurai K, Yamaguchi H, Fukuda T, Yanagi S, Nakamura Y and
Fukami K: Phospholipase Cdelta3 regulates RhoA/Rho kinase signaling
and neurite outgrowth. J Biol Chem. 286:8459–8471. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rebecchi MJ, Raghubir A, Scarlata S,
Hartenstine MJ, Brown T and Stallings JD: Expression and function
of phospholipase C in breast carcinoma. Adv Enzyme Regul. 49:59–73.
2009. View Article : Google Scholar : PubMed/NCBI
|