Introduction
The ubiquitin proteasome system is the main pathway
for protein degradation in eukaryotic organisms. Proteins are first
ubiquitylated through 3 main steps: activation, conjugation and
ligation, performed by ubiquitin-activating enzymes (E1s),
ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s),
respectively; ubiquitylated proteins are then degraded by
proteasomes. E3 ubiquitin ligases catalyze the final, but key step
of the ubiquitination cascade through the specific recognition of
the substrate target protein and E2 (1,2). It
has been found that the RING-finger family is a major member of the
E3 family. Most members of the RING-finger E3 ligase family are
complexes of multiple molecules, in which the cullin-RING-based E3
ligases (CRLs) form the main body of such ubiquitin ligases
(3). Cullin is a ‘scaffold’ of a
CRL, which is linked to E2 through its C terminal binding to the
Roc1 protein and linked to the substrate protein through its N
terminal binding to different F-box proteins. The cullin-E3 ligase
family can recognize a variety of substrates including molecules
involved in signal transduction (SMAD3/4 and Notch1/4),
transcriptional regulation (E2F1 and HIF1), DNA replication (CDT1
and ORC1) and growth and development (E2A); it plays an important
role in maintaining normal and steady cell growth (4).
Numerous studies have found that the abnormal
expression of cullin protein family members are closely related to
the occurrence, development, metastasis and recurrence of various
malignant tumors (5). For example,
Min et al study demonstrated that the expression of cullin 1
in breast cancer cells is positively correlated with the expression
of p53 and regulates cell apoptosis (6). Cullin 3 accelerates the progression of
breast cancer by regulating the effect of speckle-type POZ protein
on the expression of breast cancer metastasis suppressor 1
(7). The overexpression of cullin
4A promotes growth and metastasis of basal-like breast tumors
(8). Cullin 7 is one of the
structural components of E3 ubiquitin ligases and functions as an
oncogene to play a critical role in the proliferation and
differentiation of pancreatic cancer cells (9). Cullin 7 inhibits Myc-induced apoptosis
and promotes Myc-mediated malignant transformation of cells
(10). Cullin 7 inhibits
p53-dependent DNA repair function (11) and activates EMT in choriocarcinoma
(12).
Our previous study using whole genome exon
sequencing found that cullin 7 was one of the 12 metastatic
candidate genes (13). Guo et
al reported high cullin 7 protein expression in breast cancer
specimen, but its clinical significance was not addressed (11). They also reported that forced
expression of cullin 7 enhances cell migration and invasion in
human breast cancer cells (11),
but the mechanisms were not addressed. In the present study, we
detected cullin 7 protein expression in normal, benign and
malignant breast tissues, and then analyzed the correlation of
cullin 7 expression in breast cancer tissues in regards to various
clinicopathological characteristics and estrogen receptor (ER),
progesterone receptor (PR) and human epidermal growth factor
receptor-2 (HER-2) expression. The present study also investigated
the effects and mechanism of cullin 7 in breast cancer cell
proliferation and invasion.
Materials and methods
Samples
The use of tissue specimens was approved by the
Ethics Committee of the Affiliated Cancer Hospital, Guangzhou
Medical University. The specimens of 13 normal breast tissues, 20
benign breast tumors and 93 breast cancer tissues were used for the
present study. Paraffin-embedded tissue samples were obtained from
the Department of Pathology, Affiliated Tumor Hospital, Guangzhou
Medical University. Of the 93 breast cancer patients, 52 had lymph
node metastasis and 41 had no lymph node metastasis.
Immunohistochemistry
Cullin 7 protein expression in tissues was measured
by immunohistochemical staining. Briefly, tissue sections were
dewaxed, rehydrated, followed by incubation with 3% hydrogen
peroxide for 10 min and antigen retrieval in 100 mM Tris (pH 10.0)
at 98°C for 30 min. The slices were then blocked with 2.5% horse
serum and incubated with biotin-labeled cullin 7 primary antibody
(1:200 dilution; Abcam, Guangzhou, China) overnight at 4°C followed
by incubation with the avidin-biotin complex according to the user
manual (Vector Laboratories, Guangzhou, China). After
counterstaining with hematoxylin, the staining was scored: 0 score
for negative staining, 1 for weak staining, 2 for moderate
staining, and 3 for strong staining.
Cell culture
MDA-MB-231 and BT549 cells were cultured in
RPMI-1640 medium containing 10% fetal calf serum (FCS) (Invitrogen,
Carlsbad, CA, USA). HS578T, MCF7, T47D, SKBR3 and BT474 cells were
cultured in Dulbeccos modified Eagles medium (DMEM) containing 10%
FCS. Cells were cultured at 37°C in 5% CO2.
Establishment of stable cells
expressing cullin 7 siRNA
MDA-MB-231 and BT549 stable cells expressing cullin
7 siRNA were established by lentivirus infections by following the
manufacturers instructions; the produced stable cells were called
231-siCul7 and 549-siCul7, respectively. MDA-MB-231 and BT549
control stable cells (231-siCtrl and 549-siCtrl, respectively were
produced to express control small interference RNA (siRNA) not
targeting any gene using lentivirus infection. The cullin 7 and
control lentiviral particles were purchased from Shanghai GeneChem
Co., Ltd. (Shanghai, China).
Western blot analysis
Cells (1×106) were homogenized in 100 µl
of RIPA buffer on ice for 30 min and mixed for 15 sec every 5 min.
After centrifuging at 12,000 rpm for 10 min at 4°C, the supernatant
was harvested and protein concentration was measured using a Pierce
BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
Total protein (10 µg) was separated by electrophoresis on 10%
sodium dodecyl sulfate-polyacrylamide gel. After being transferred
to polyvinylidene difluoride membranes, the membranes were blocked
with 5% non-fat milk in Tris-buffered saline (TBS) buffer at room
temperature (RT) for 1 h, followed by incubation with anti-cullin
7, anti-cyclin A and anti-p21 antibody (Abcam) overnight at 4°C.
After washing with 1X TBS + 0.1% Tween-20 (TBST) buffer for 3×10
min, the membranes were incubated with secondary antibody for 1 h
at RT. The immune reaction was visualized using enhanced
chemiluminescence (Pierce, Waltham, MA, USA). The bands on X-ray
film were scanned using Quantity One software.
Cell viability assay
Cell viability was measured using the Cell Counting
Κit-8 (CCK-8) kit by following the manufacturers instructions
(Sigma-Aldrich, Beijing, China). Briefly, 1,000 of 231-siCul7,
549-siCul7, 231-siCtrl and 549-siCtrl cells in 0.1 ml medium
containing 10% FCS were seeded in each well of 96-well plates.
Twenty-four hours later, 10 µl of CCK-8 solution was added to each
well and cells were continuously incubated for 2 h. The absorbance
was measured at a wavelength of 450 nm.
Flow cytometry
231-siCul7, 549-siCul7, 231-siCtrl and 549-siCtrl
cells were seeded in 6-well plates and treated for 24 h after
having attached. The cells were then harvested by a centrifuge at
1,000 rpm for 5 min. After washing the pellets with 1 ml of
phosphate-buffered saline (PBS), cells were re-suspended with 3 ml
of pre-cooled anhydrous ethanol and placed at −20°C overnight.
After washing with 1 ml PBS at 4°C, cells were re-suspended in 1 ml
of pre-cooled PBS and 400 µl of propidium iodide (PI) staining
solution (Sigma-Aldrich). After incubation for 30 min at 4°C, cells
were subjected to flow cytometry assay (BD Biosciences, Franklin
Lakes, NJ, USA).
In vitro tumor invasion assay
231-siCul7, 549-siCul7, 231-siCtrl and 549-siCtrl
cells were cultured in 10-cm dish to 80% confluency and digested
with 0.25% trypsin and re-suspended in complete medium at
5×104 cells/ml. Of cells (0.5 ml) was transferred into
Transwell covered with Matrigel and continuously cultured for 24 h.
The cells that migrated to the membranes at the lower chamber were
fixed with ice pre-cooled methanol for 30 min, and stained with 1%
crystal violet for 10 min. Cells on the membranes were observed
under a microscope.
Cell microtubule regeneration
analysis
The adherent cultured 231-siCul7, 549-siCul7,
231-siCtrl and 549-siCtrl cells were incubated with a medium
containing 10 mM nocodazole for 2 h to completely depolymerize the
microtubules in the cells. The medium containing nocodazole was
then replaced with fresh medium and incubated in a CO2
incubator at 37°C. The microtubules in the cells were
re-polymerized in a process called microtubule regeneration
(14). Ten minutes after
microtubule regeneration, cells were treated with PEMT buffer (100
mM PIPES, 1 mM EGTA, 0.5 mM MgCl2, 0.5% Triton X-100, pH
6.9) for 30 min and then fixed with 4% paraformaldehyde at RT for
30 min or fixed with pre-cooled methanol for 5 min. After blocking
with 2% bovine serum albumin for 1 h at RT, cells were incubated
with anti-α-tubulin antibody (Cell Signaling Technology, Guangzhou,
China) for 2 h followed by secondary antibody for 1 h at RT after
washing with PBS. Cells were then incubated with
4′,6-diamidino-2-phenylindole (1:500 dilution) for 5 min at RT, and
then observed under a fluorescence microscope.
Tumor growth
Animal experiments were approved by the Animal
Ethics Committee of Guangzhou Medical University. Male nude mice at
6 weeks old were inoculated with 0.5×106 of 231-siCul7
and 231-siCtrl cells on the left back proximal axillary. Tumor
growth was observed every 3 days. Nude mice were sacrificed, tumors
were excised on the 48th day after inoculation, and the tumor
volume and weight were assessed.
Statistical analysis
The correlations of positive cullin 7 protein
expression in breast cancer tissues with the clinicopathological
characteristics of patients were analyzed using χ2 test.
The association of immunohistochemical staining with the patients
prognosis was analyzed using Kaplan-Meier survival analysis. The
correlation between cullin 7 protein expression and ER, PR and
HER-2 expression in breast cancer tissues was analyzed using
Spearman correlation analysis. Measurement of data between 2 groups
of samples was carried out using the Students t-test. P<0.05 was
considered statistically significant.
Results
Expression of cullin 7 is positively
correlated with the malignancy of breast cancer
The expression of cullin 7 protein in normal breast
tissues, breast benign legions, breast cancer tissues, and axillary
lymph nodes of breast cancer patients was detected by
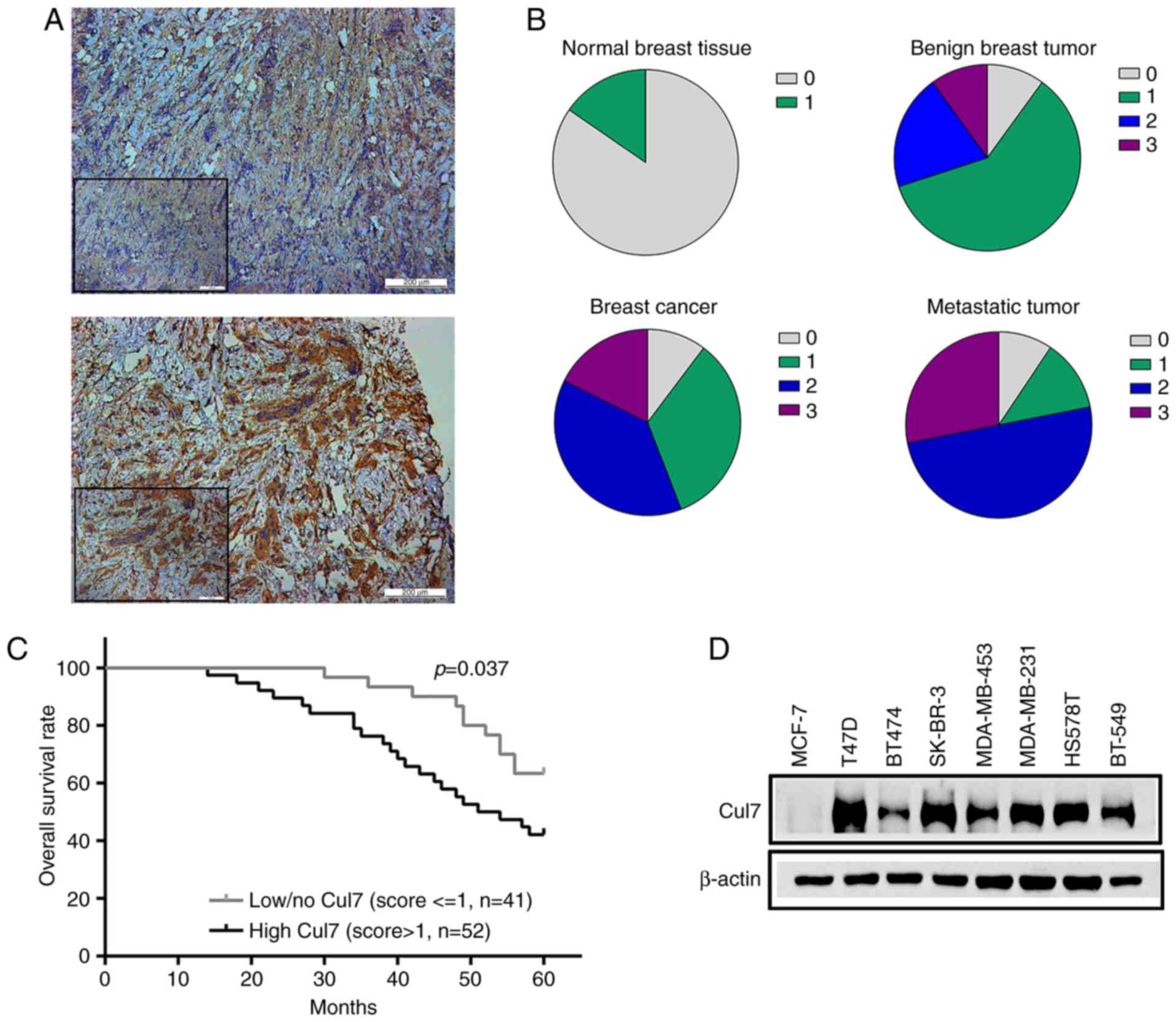
immunohistochemistry (Fig. 1A).
Cullin 7 was negatively or weakly expressed in normal breast
tissues, but its expression in benign breast tumor tissues was
increased (30.9% positive) compared to the normal breast tissues.
The percentage of positive cullin 7 protein expression was 55.9% in
breast cancer tissues and 78.1% in the axillary lymph nodes of
breast cancer patients (Fig.
1B).
Positive cullin 7 expression in breast cancer
patients was significantly associated with the tumor, lymph node,
metastasis (TNM) staging, such as that the percentage of positive
cullin 7 expression was significantly lower in tumor tissues in
patients with early breast cancer than that of patients with
advanced disease. The expression of cullin 7 was significantly
higher in poorly differentiated tumor tissues than that in well
differentiated tumor tissues. The expression of cullin 7 was
positively associated with histological grade of breast cancer
(P=0.013) and axillary lymph node metastasis in breast cancer
patients (P=0.022). No significant correlations were observed
between cullin 7 expression and age, tumor size and pathological
type (P>0.05), as well as ER, PR and HER-2 expression in breast
cancer tissues (P>0.05) (Table
I).
 | Table I.Cullin 7 staining and
clinicopathological characteristics of the 93 breast cancer
patients. |
Table I.
Cullin 7 staining and
clinicopathological characteristics of the 93 breast cancer
patients.
|
| Cullin 7
staining |
|
|
|---|
|
|
|
|
|
|---|
| Variables | Negative or low
(%) | High positive
(%) | Total | P-valuea |
|---|
| Age (years) |
|
|
| 0.536 |
| ≤50 | 20 (40.8) | 29 (59.2) | 49 |
|
|
>50 | 21 (47.7) | 23 (52.3) | 44 |
|
| Tumor size (cm) |
|
|
| 0.493 |
| T1
(<2) | 10 (37.1) | 17 (62.9) | 27 |
|
| T2
(2–5) | 20 (48.7) | 21 (51.3) | 41 |
|
| T3
(>5) | 9
(36.0) | 16 (64.0) | 25 |
|
| Lymph node
metastasis |
|
|
| 0.022a |
|
Negative | 24 (58.5) | 17 (41.5) | 41 |
|
|
Positive | 18 (34.6) | 34 (65.4) | 52 |
|
| Histologic
grade |
|
|
| 0.013a |
| I | 19 (79.2) | 8
(20.8) | 24 |
|
| II | 25 (65.8) | 14 (34.2) | 38 |
|
|
III | 10 (32.2) | 19 (67.8) | 31 |
|
| Histologic
type |
|
|
| 0.990 |
|
Ductal | 35 (45.4) | 42 (54.6) | 77 |
|
|
Lobular | 4
(44.4) | 5
(55.6) | 9 |
|
|
Other | 3
(42.8) | 4
(57.2) | 7 |
|
| ER status |
|
|
| 0.336 |
|
Negative | 22 (56.4) | 17 (43.6) | 39 |
|
|
Positive | 25 (46.3) | 29 (53.7) | 54 |
|
| PR status |
|
|
| 0.808 |
|
Negative | 20 (47.6) | 22 (52.4) | 42 |
|
|
Positive | 23 (45.1) | 28 (54.9) | 51 |
|
| HER-2 status |
|
|
| 0.384 |
|
Negative | 30 (56.6) | 23 (43.4) | 53 |
|
|
Positive | 19 (47.5) | 21 (52.5) | 40 |
|
Kaplan-Meier survival analysis showed that the
expression of cullin 7 was negatively correlated with the overall
survival rate of breast cancer patients. The 5-year survival rate
of patients with high cullin 7 expression was significantly lower
than that of patients with low cullin 7 expression (P<0.05)
(Fig. 1C). In addition, cullin 7
was highly expressed in breast cancer cell lines with high
metastatic capacity, such as MDA-MB-231, BT549 and HS578T and lowly
expressed in poorly metastatic breast cancer cell lines, such as
MCF-7 and BT474 (Fig. 1D). These
results suggest that cullin 7 plays an important role in the
metastasis and progression of breast cancer.
Cullin7 is involved in the
proliferation of breast cancer cells
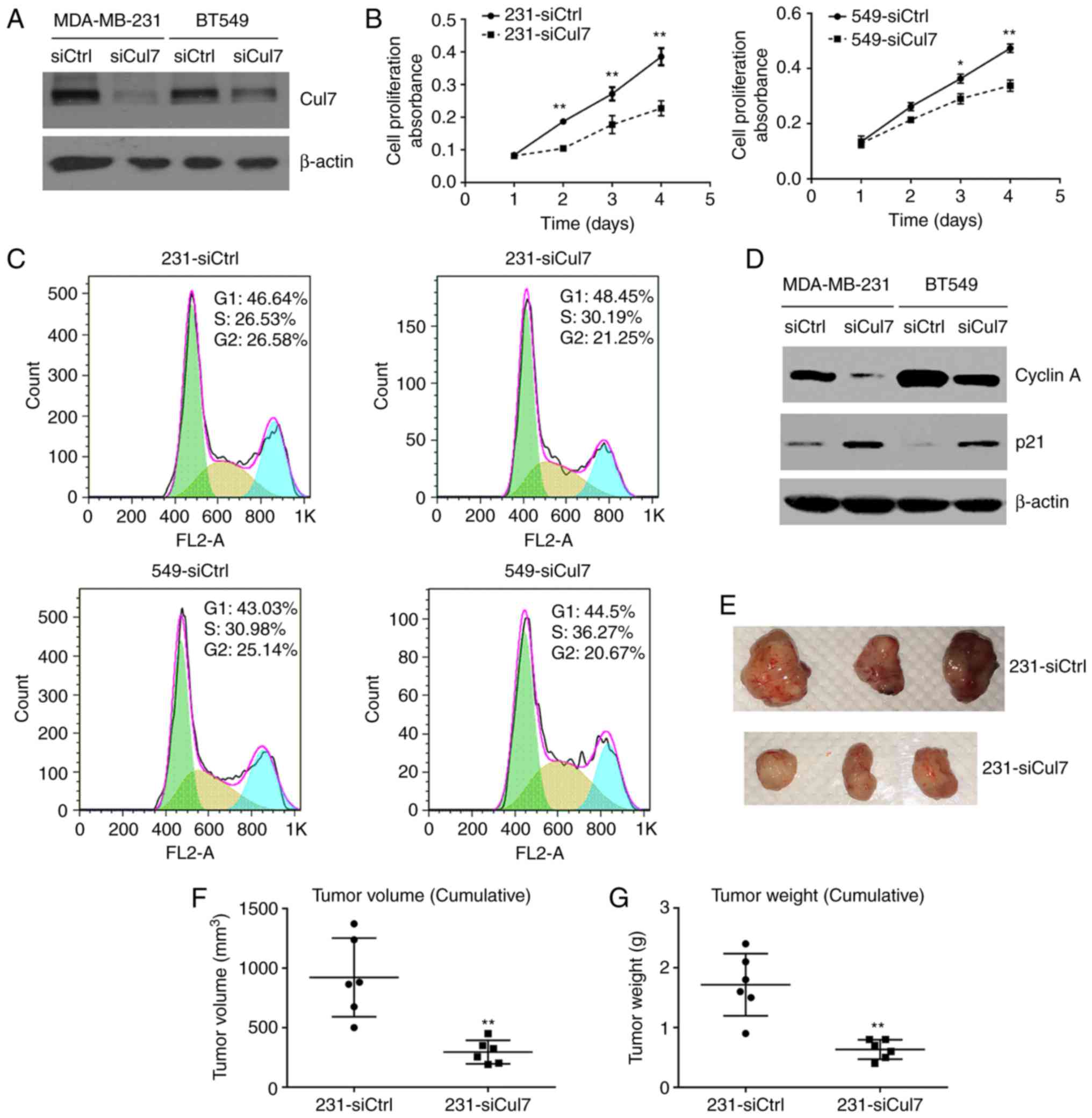
Western blotting showed that cullin 7 protein
expression was significantly decreased in 231-siCul7 and 549-siCul
stable cells compared to that in 231-siCtrl and 549-siCtrl stable
control cells, respectively (Fig.
2A). Cell viability assay using the CCK-8 kit showed that the
cell proliferation ability was significantly reduced in the
231-siCul7 and 549-siCul stable cells compared to that noted in the
control cells (Fig. 2B). Cell
cytometric assay showed that the percentage of cells in the S1
phase was significantly increased, but the percentage of cells in
the G2 phase was significantly decreased in the 231-siCul7 and
549-siCul stable cells compared to these populations in the control
cells (P<0.05) (Fig. 2C).
Western blotting showed that the expression of cyclin A protein was
significantly decreased, while p21 protein expression was
significantly increased in the 231-siCul7 and 549-siCul7 stable
cells compared to levels noted in the control cells (P<0.05)
(Fig. 2D). These results suggest
that the proliferation of breast cancer cells is associated with
decreased cell cycle arrest.
The study of xenograft 231-siCul7 and 231-siCtrl
cells in nude mice showed that silencing of cullin 7 expression
(231-siCul7) significantly decreased the tumor volume and weight
compared to these parameters in the control group (231-siCtrl)
(Fig. 2E-G).
Silencing of cullin 7 expression
changes cell morphology
231-siCul7, 549-siCul7, 231-siCtrl and 549-siCtrl
cells were plated in 10-cm plates and allowed to form small
colonies for 6 days and were then observed to ascertain whether
colonies maintained compact, loose or scattered contact with
neighboring cells. After inhibiting cullin 7 expression, the growth
of 231-siCul7 and 549-siCul7 cells changed from scatter growth to
compact growth. The proportion of cells with compact growth was
significantly increased in the 231-siCul7 (60±2.5%) and 549-siCul7
(61.67±3.4%) cells compared to the 231-siCtrl and 549-siCtrl cells
(10±2.7 and 10.33±2.8%, respectively, P<0.05). On the contrary,
the proportion of cells with scatter growth was significantly
decreased in the 231-siCul7 (20±3.1%) and 549-siCul7 (12±2.5%)
cells compared to the 231-siCtrl and 549-siCtrl cells (69±3.8 and
57.6±2.9%, respectively, P<0.05) (Fig. 3A).
Silencing of cullin 7 expression
inhibits cell invasion
In vitro invasion assay showed that the
number of 231-siCul7 and 549-siCul7 cells that passed the Matrigel
was significantly less than the number of 231-siCtrl and 549-siCtrl
cells (P<0.01) (Fig. 3B). These
results suggest that cullin 7 is involved in the invasion of breast
cancer cells.
Silencing of cullin 7 expression
affects microtubule regeneration
The immunofluorescence of α-tubulin showed that the
morphology of 231-siCul7 and 549-siCul7 cells was transformed from
normal fuselage into polygons, the cytoskeleton clarity decreased,
the microtubule tissue around the nucleus was partially
disappeared, and the microtubule was obviously disturbed (Fig. 4A). This suggests that silencing of
cullin 7 expression affected the formation of pseudopodia and
subsequently decreased the migration capacity. After removing
nocodazole, the α-tubulin fluorescence in the 231-siCul7 and
549-siCul7 cells was significantly weaker than that in the
231-siCtrl and 549-siCtrl cells (Fig.
4B), suggesting that silencing of cullin 7 expression increased
microtubule stability and inhibited microtubule regeneration.
Discussion
A recent study demonstrated that cullin 7 is highly
expressed in hepatocellular carcinoma (HCC) tumor tissues,
particularly in metastatic HCC, which is associated with shorter
survival in HCC patients (15).
Cullin 7 expression was increased in primary lung cancer tissues of
humans (16), and the overexpressed
cullin 7 mRNA was found to be significantly associated with poor
prognosis in patients with non-small cell lung carcinoma (17). The expression of cullin 7 mRNA was
significantly higher in epithelial ovarian cancer compared to that
noted in normal ovarian surface tissues, which is related to high
International Federation of Gynecology and Obstetrics (FIGO) stage
and lymph node metastasis (18). In
breast cancer tissues, cullin 7 protein was found highly expressed,
but its clinical significance was not documented (11). Forced expression of cullin 7 was
demonstrated to enhance cell migration, invasion and/or metastasis
in human choriocarcinoma cells (12), breast cancer (11) and liver cancer cells (15) with the mechanisms not being fully
elucidated. Our results suggest that positive cullin 7 expression
is positively correlated with the malignant phenotypes of breast
cancer and lower 5-year survival rate. In vitro cell
experiments further confirmed that silencing of cullin 7 expression
inhibited the proliferation and invasion of breast cancer cells, by
affecting the cell cycle and microtubule stability.
Cyclin A is a member of the cyclin family, which
forms a complex with CDK1 to regulate the initiation and completion
of DNA replication in S phase (19,20).
Cyclin A also forms a complex with CDK2, which is exclusively
involved in S phase progression. In late S phase, cyclin A is
involved in the activation and stabilization of cyclin B/CDK1
complex (21,22). After cyclin B is activated, cyclin A
is subsequently degraded through the ubiquitin pathway (21,23).
The present study first revealed that silencing of cullin 7
expression significantly decreased cyclin A protein expression in
breast cancer cells. p21, also called CDK-interacting protein 1, is
a cyclin-dependent kinase inhibitor that inhibits the activity of
cyclin-CDK1, -CDK2 and -CDK4/6 complexes, and is responsible for
arrest of cell cycle progression at G1 and S phase (24). The present study showed that
silencing of cullin 7 expression significantly increased p21
protein expression in breast cancer cells. Thus, cullin 7 may be
involved in the proliferation of breast cancer cells by increasing
cyclin A, but decreasing p21 protein expression and subsequently
enhancing cell progression from S1 to G2 phase.
It is widely known that tumor cells need a structure
called ‘invasive pseudopodia’ to penetrate the basement membrane of
tissues to establish local or distant metastases. The extension of
invasive pseudopods needs to be achieved by the extension of the
cell microtubules. Microtubules are intracellular networks
assembled by tubulin and tubulin-bound heterodimeric protein
subunits. The depolymerization and polymerization of microtubules
maintain a dynamic equilibrium, and the dynamic characteristic of
microtubules is critical for the migration of cells (25–27). A
previous study found that imbalance of polymerization and
depolymerization dynamics inhibits the ability of tumor cells to
form invasive pseudopodia, leading to a decrease in invasion and
metastasis (28). It has been
reported that abnormal function of tubulin cofactors can induce
cell cycle arrest and apoptosis (29,30).
For example, the study on 3M syndrome found that cullin 7 mutation
increased the stability of cell microtubules, prolonged or even
stagnationed cell mitosis, and induced cell growth stagnation or
aging (31). The present study
revealed that silencing of cullin 7 expression in breast cancer
cells decreased the cell proliferation and induced cell cycle
arrest, such as increasing the proportion of cells in the S phase
while reducing the proportion of cells in the G2 phase. The present
study also showed that silencing of cullin 7 expression in
MDA-MB-231 and BT549 cells changed the mode of cell growth and
morphology with obvious disorder of microtubule dynamics,
suggesting that cullin 7 promotes tumor cell invasion by affecting
the cytoskeleton.
In conclusion, the present study suggests that
positive cullin 7 expression is associated with the malignant
phenotype of breast cancer and is a predictor of poor prognosis in
breast cancer patients. Cullin 7 is involved in cell proliferation
and invasion by regulating the cell cycle and microtubule
stability. Therefore, cullin 7 can be used as a new biological
marker for the early diagnosis and treatment of breast cancer.
Acknowledgements
The present study was supported by a grant from the
Science Foundation of Guangdong Province (2013B021800301). We thank
Professor Zhiming He for providing the reagents and breast cancer
cell lines. We are also grateful to Dr Zhijie Zhang for providing
the cell flow cytometry and cell cycle analysis.
References
|
1
|
Ding F, Xiao H, Wang M, Xie X and Hu F:
The role of the ubiquitin-proteasome pathway in cancer development
and treatment. Front Biosci. 19:886–895. 2014. View Article : Google Scholar
|
|
2
|
Jain CK, Arora S, Khanna A, Gupta M,
Wadhwa G and Sharma SK: The ubiquitin-proteasome pathway an
emerging anticancer strategy for therapeutics: A patent analysis.
Recent Patents Anticancer Drug Discov. 10:201–213. 2015. View Article : Google Scholar
|
|
3
|
Kitagawa K and Kitagawa M: The SCF-type E3
ubiquitin ligases as cancer targets. Curr Cancer Drug Targets.
16:119–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sarikas A, Hartmann T and Pan ZQ: The
cullin protein family. Genome Biol. 12:2202011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Y and Sun Y: Cullin-RING ligases as
attractive anti-cancer targets. Curr Pharm Des. 19:3215–3225. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Min KW, Kim DH, Do SI, Sohn JH, Chae SW,
Pyo JS, Park CH, Oh YH, Jang KS, Kim HL, et al: Diagnostic and
prognostic relevance of Cullin1 expression in invasive ductal
carcinoma of the breast. J Clin Pathol. 65:896–901. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim B, Nam HJ, Pyo KE, Jang MJ, Kim IS,
Kim D, Boo K, Lee SH, Yoon JB, Baek SH, et al: Breast cancer
metastasis suppressor 1 (BRMS1) is destabilized by the Cul3-SPOP E3
ubiquitin ligase complex. Biochem Biophys Res Commun. 415:720–726.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saucedo-Cuevas LP, Ruppen I, Ximénez-Embún
P, Domingo S, Gayarre J, Muñoz J, Silva JM, García MJ and Benítez
J: CUL4A contributes to the biology of basal-like breast tumors
through modulation of cell growth and antitumor immune response.
Oncotarget. 5:2330–2343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Chen Y, Lin P, Li L, Zhou G, Liu
G, Logsdon C, Jin J, Abbruzzese JL, Tan TH, et al: The CUL7/F-box
and WD repeat domain containing 8 (CUL7/Fbxw8) ubiquitin ligase
promotes degradation of hematopoietic progenitor kinase 1. J Biol
Chem. 289:4009–4017. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu X, Sarikas A, Dias-Santagata DC, Dolios
G, Lafontant PJ, Tsai SC, Zhu W, Nakajima H, Nakajima HO, Field LJ,
et al: The CUL7 E3 ubiquitin ligase targets insulin receptor
substrate 1 for ubiquitin-dependent degradation. Mol Cell.
30:403–414. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo H, Wu F, Wang Y, Yan C and Su W:
Overexpressed ubiquitin ligase Cullin7 in breast cancer promotes
cell proliferation and invasion via down-regulating p53. Biochem
Biophys Res Commun. 450:1370–1376. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu J, Lv X, Lin H, Wu L, Wang R, Zhou Z,
Zhang B, Wang YL, Tsang BK, Zhu C, et al: Ubiquitin ligase cullin 7
induces epithelial-mesenchymal transition in human choriocarcinoma
cells. J Biol Chem. 285:10870–10879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Yang B, Xing K, Yuan N, Wang B, Chen
Z, He W and Zhou J: A preliminary study of the relationship between
breast cancer metastasis and loss of heterozygosity by using exome
sequencing. Sci Rep. 4:54602014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu J, He Z, Wang DL and Sun FL: Depletion
of JMJD5 sensitizes tumor cells to microtubule-destabilizing agents
by altering microtubule stability. Cell Cycle. 15:2980–2991. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang D, Yang G, Li X, Xu C and Ge H:
Inhibition of liver carcinoma cell invasion and metastasis by
knockdown of Cullin7 in vitro and in vivo. Oncol Res. 23:171–181.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Men X, Wang L, Yu W and Ju Y: Cullin7 is
required for lung cancer cell proliferation and is overexpressed in
lung cancer. Oncol Res. 22:123–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim SS, Shago M, Kaustov L, Boutros PC,
Clendening JW, Sheng Y, Trentin GA, Barsyte-Lovejoy D, Mao DY, Kay
R, et al: CUL7 is a novel antiapoptotic oncogene. Cancer Res.
67:9616–9622. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xi J, Zeng ST, Guo L and Feng J: High
expression of Cullin7 correlates with unfavorable prognosis in
epithelial ovarian cancer patients. Cancer Invest. 34:130–136.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bendris N, Lemmers B, Blanchard JM and
Arsic N: Cyclin A2 mutagenesis analysis: A new insight into CDK
activation and cellular localization requirements. PLoS One.
6:e228792011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pagano M, Pepperkok R, Verde F, Ansorge W
and Draetta G: Cyclin A is required at two points in the human cell
cycle. EMBO J. 11:961–971. 1992.PubMed/NCBI
|
|
21
|
Yam CH, Fung TK and Poon RY: Cyclin A in
cell cycle control and cancer. Cell Mol Life Sci. 59:1317–1326.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De Boer L, Oakes V, Beamish H, Giles N,
Stevens F, Somodevilla-Torres M, Desouza C and Gabrielli B: Cyclin
A/cdk2 coordinates centrosomal and nuclear mitotic events.
Oncogene. 27:4261–4268. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Henglein B, Chenivesse X, Wang J, Eick D
and Bréchot C: Structure and cell cycle-regulated transcription of
the human cyclin A gene. Proc Natl Acad Sci USA. 91:pp. 5490–5494.
1994; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gartel AL and Radhakrishnan SK: Lost in
transcription: p21 repression, mechanisms, and consequences. Cancer
Res. 65:3980–3985. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amos LA: What tubulin drugs tell us about
microtubule structure and dynamics. Semin Cell Dev Biol.
22:916–926. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nogales E: An electron microscopy journey
in the study of microtubule structure and dynamics. Protein Sci.
24:1912–1919. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szarama KB, Gavara N, Petralia RS, Kelley
MW and Chadwick RS: Cytoskeletal changes in actin and microtubules
underlie the developing surface mechanical properties of sensory
and supporting cells in the mouse cochlea. Development.
139:2187–2197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Carranza G, Castaño R, Fanarraga ML,
Villegas JC, Gonçalves J, Soares H, Avila J, Marenchino M,
Campos-Olivas R, Montoya G, et al: Autoinhibition of TBCB regulates
EB1-mediated microtubule dynamics. Cell Mol Life Sci. 70:357–371.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bendre S, Rondelet A, Hall C, Schmidt N,
Lin YC, Brouhard GJ and Bird AW: GTSE1 tunes microtubule stability
for chromosome alignment and segregation by inhibiting the
microtubule depolymerase MCAK. J Cell Biol. 215:631–647. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gergely ZR, Crapo A, Hough LE, McIntosh JR
and Betterton MD: Kinesin-8 effects on mitotic microtubule dynamics
contribute to spindle function in fission yeast. Mol Biol Cell.
27:3490–3514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan J, Yan F, Li Z, Sinnott B, Cappell KM,
Yu Y, Mo J, Duncan JA, Chen X, Cormier-Daire V, et al: The 3M
complex maintains microtubule and genome integrity. Mol Cell.
54:791–804. 2014. View Article : Google Scholar : PubMed/NCBI
|