Introduction
Thyroid cancer (TC) is the most common malignancy of
the endocrine system and is classified as well differentiated
(WDTC), poorly differentiated (PDTC) and anaplastic thyroid
carcinoma (ATC) (1,2). ATC, which is composed of
undifferentiated cells arising from thyroid follicular epithelium,
represents the least common but the most lethal thyroid cancer
sub-type (3), since it is
unresponsive to standard treatments.
An innovative approach employs compounds that target
chromatin readers and, so far, BET proteins are the best
characterized ones. BET is the acronym for bromodomain and
extra-terminal domain family of proteins (4) that act as acetyl-lysine readers, which
are known to be associated with the transfer of epigenetic
information by the so-called ‘write-read-erase’ concept (5). BET inhibitors (BETi) have recently
attained the consensus of researchers as well established
anti-neoplastic agents in different cancer models (3,6–8), and
JQ1, I-BET762 and I-BET151 are the better characterized drugs
(9). A consensus in epigenetic drug
mechanisms of action is that their anti-neoplastic activity is due
both by direct and indirect effects. We have already established
that BET inhibition impairs different key biological pathways in
human ATC cell lines, deeming these epigenetic drugs as promising
therapeutic tools for thyroid cancer (3).
MicroRNAs (miRNAs) are 20–22 nucleotide endogenous
non-coding RNA molecules that regulate transcription and/or
translation by controlling the expression of a large number of
genes, binding to specific sites (called seeds) in the target mRNA,
i.e., 3′UTR (untranslated region). This interaction is generally
involved in mRNA cleavage/degradation or translational suppression
(1). Therefore, these small
molecules play important roles in development, metabolism,
proliferation, differentiation and apoptosis. Consequently, due to
their leading role, miRNAs are promising therapeutic targets in
many diseases, including thyroid cancer (1,10).
Several miRNAs were found to be dysregulated in thyroid cancer
(11,12). For these many reasons, the aim of
our study was to demonstrate a link between JQ1 treatment and miRNA
regulation in thyroid cancer.
Materials and methods
Human cell lines
SW1736 (obtained from Cell Lines Service GmbH,
Eppelheim, Germany) and 8505c (purchased from Sigma-Aldrich, St.
Louis, MO, USA) are human cell lines derived from ATC (13,14),
while Nthy-ori 3-1 (purchased from Sigma-Aldrich) is a human
thyroid follicular epithelial cell line immortalized by the SV40
large T gene. Cell lines were tested and confirmed to be
mycoplasma-free and authenticated by STR analysis as appropriate
cell lines of thyroid cancer origin. Cell lines were grown in
RPMI-1640 medium (EuroClone S.p.A, Pero MI, Italy) supplemented
with 10% fetal bovine serum (Gibco Invitrogen, Milan, Italy) and 50
mg/ml gentamicin (Gibco Invitrogen), in a humidified incubator (5%
CO2 in air at 37°C) (Eppendorf AG, Hamburg, Germany).
Cultured cells were treated with vehicle (DMSO, Sigma-Aldrich) or
JQ1 (5 µM in DMSO) (Cayman Chemical, Ann Arbor, MI, USA) for either
48 or 72 h.
miRNA expression and
normalization
Expression analysis of 812 miRNAs was performed by
the genetic laboratory of Pharmadiagen Srl, using the NanoString
nCounter v2 miRNA Assay kit (NanoString Technologies, Seattle, WA,
USA). The nCounter miRNA expression assay was used to detect and
count miRNAs through hybridization with fluorescently-labeled
barcoded probes. A total of 280 ng of RNA was processed according
to the manufacturer's instructions (NanoString Technologies),
including preparation and the hybridization protocol performed at
65°C for 16 h. Subsequent purification and detection of miRNA
target were carried out using the nCounter Analysis system.
The total miRNA counts obtained were elaborated and
normalized following the Nanostring's Data Analysis Guide. The
background correction was calculated as the geometric mean of
negative control plus 2X standard deviation and subtracted to each
count. miRNAs expressing less than the background were setting as
not expressed to overcome basal noise. The technical normalization
was performed using the positive control spike counts. We
calculated the normalization factor as the ratio between the
average of geometric means and the geometric mean of each sample.
The counts of every miRNAs were multiply by the normalization
factor. To be more conservative and avoid false positive miRNA, we
set an additional cut off (15 counts) after the above
normalization. Fold change was, then, calculated as JQ1-treated vs.
vehicle or ATC cell line vs. NThy ori 3.1. Only miRNA showing a
deregulation greater than two log2-fold change were taken into
consideration.
MicroRNA expression validation
Briefly, 200 ng of total RNA, treated with either
JQ1 5 µM or vehicle, was extracted and reverse-transcribed using
miRCURY LNA™ Universal cDNA Synthesis kit II (Exiqon, Vedbaek,
Denmark) as described by the manufacturer. Before reverse
transcription, 108 copies of UniSP6 synthetic spike-in were added
to each condition, as a quality control for subsequent analyses.
Real-time PCRs were performed using ExiLENT SYBR® Green
master mix (Exiqon) with the ABI Prism 7300 Sequence Detection
System (Applied Biosystems, Foster City, CA, USA). SNORD44 was used
as an inter-plate calibrator. The ∆∆CT method, by means of the SDS
software (Applied Biosystems), was used to calculate microRNA
levels. All oligonucleotide primers were purchased from Exiqon.
Each sample was run in triplicate and experiments were repeated at
least three times using at least two independent samples.
Protein extraction and western
blotting
Total protein extraction was performed as previously
described (15). Briefly, SW1736
and 8505c cells, incubated in vehicle-treated medium (NT, untreated
cultures) or with JQ1 5 µM, were harvested by scraping and lysed
with total lysis buffer (Tris-HCl 50 mM pH 8.0, NaCl 120 mM, EDTA 5
mM, Triton 1%, NP40 1%, protease inhibitors). For western blot
analysis, the proteins were electrophoresed on 10% SDS-PAGE and
then transferred to nitrocellulose membranes, which were saturated
with 5% non-fat dry milk in PBS/0.1% Tween-20. The membranes were
then incubated overnight with rabbit polyclonal anti-p21 antibody
1:500 (Santa Cruz Biotechnology, Inc., Heidelberg, Germany), rabbit
polyclonal anti-p27 antibody 1:500 (Santa Cruz Biotechnology,
Inc.), rabbit monoclonal anti-phoshpo-STAT3 (Tyr705) antibody
1:10,000 [Merck Millipore, Vimodrone (MI), Italy], mouse monoclonal
anti-STAT3 antibody 1:500 (Santa Cruz Biotechnology, Inc.) or
rabbit anti-actin antibody 1:1,000 (Abcam, Cambridge, UK). The
following day, the membranes were incubated for 2 h with
anti-rabbit immunoglobulin coupled to peroxidase 1:4,000
(Sigma-Aldrich). The blots were developed using UVItec Alliance LD
(UVItec Ltd., Cambridge, UK) with SuperSignal Technology (Thermo
Fisher Scientific Inc., Waltham, MA, USA).
Statistical analysis
MicroRNA and protein levels were expressed as the
means ± SD, and significances were determined using a one-way ANOVA
followed by Dunnett's test performed with GraphPad Software for
Science (San Diego, CA, USA). P-value <0.05 was considered to
indicate a statistically significant difference.
Results
JQ1 treatment modifies miRNA
expression in ATC
Our previously published data demonstrated the
anti-neoplastic activity of 5 µM JQ1 in anaplastic thyroid cancer
cells as indicated by a decrease in cell viability coupled by an
increase in cell death phenomena and cell cycle arrest (3). Moreover, our RNA-seq analysis revealed
the complexity of the JQ1 mechanism of action, which takes
advantage of hundreds of direct and indirect targets.
To elucidate the possible role of miRNAs in ATC
therapeutic response after JQ1 treatment, we used the nCounter
miRNA Expression Assay kit, a novel digital color-coded barcode
technology based on direct multiplexed assessment of gene
expression. Two ATC cell lines, SW1736 and 8505c, JQ1- or
vehicle-treated for either 48 or 72 h, were run on the NanoString
platform. We chose these time-points as it was demonstrated that
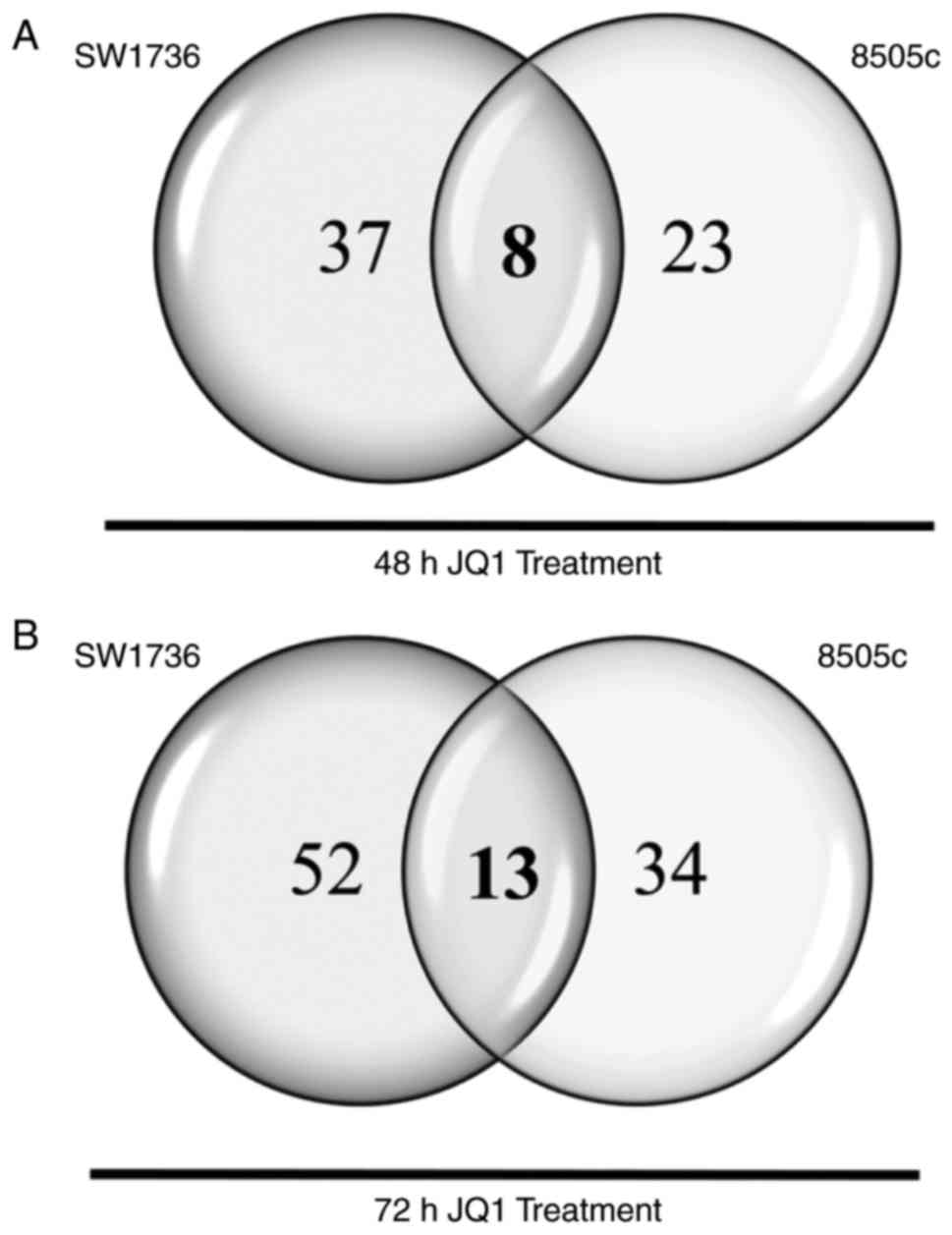
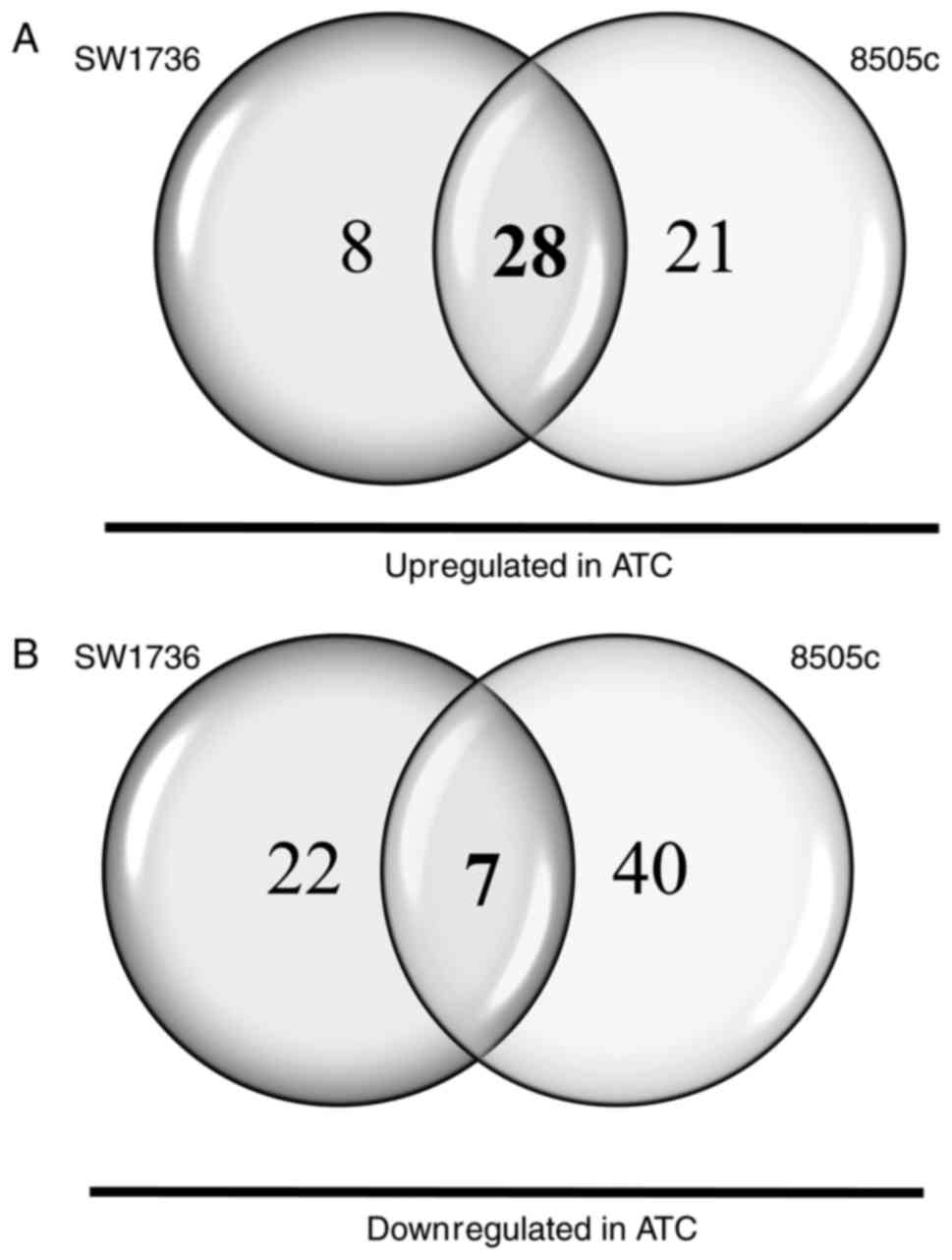
miRNA half-lives usually exceeded 24 h (16). Of the 812 miRNAs analyzed, 45 and 31
miRNAs turned out to be dysregulated after 48 h of JQ1 treatment in
SW1736 and 8505c cells, respectively. As shown in Fig. 1A, 8 miRNAs displayed a common
upregulation in both cell lines. After 72 h of JQ1 treatment,
conversely, 65 and 47 miRNAs were demonstrated to be dysregulated
in SW1736 and 8505c cells, respectively. Thirteen miRNAs exhibited
a common upregulation in the two cell lines (Fig. 1B). In order to delineate central
miRNAs in JQ1-mediated effects, we compared common miRNAs at 48 and
72 h, highlighting a pool of 7 miRNAs commonly upregulated in both
SW1736 and 8505c after both 48 and 72 h of JQ1 treatment (Table I).
 | Table I.Shared miRNAs altered after both 48
and 72 h of JQ1 5-µM treatment in ATC cells. |
Table I.
Shared miRNAs altered after both 48
and 72 h of JQ1 5-µM treatment in ATC cells.
|
|
| Fold change |
|---|
|
|
|
|
|---|
| miRNAs | Cells | 48 h | 72 h |
|---|
| hsa-miR-182-5p | SW1736 | 2.97 | 4.07 |
|
| 8505c | 7.99 | 9.34 |
| hsa-miR-4516 | SW1736 | 3.03 | 3.58 |
|
| 8505c | 3.44 | 3.41 |
| hsa-miR-1234 | SW1736 | 4.26 | 3.82 |
|
| 8505c | 2.75 | 2.26 |
| hsa-miR-30c-5p | SW1736 | 4.50 | 5.26 |
|
| 8505c | 7.70 | 8.15 |
| hsa-miR-4488 | SW1736 | 9.39 | 9.58 |
|
| 8505c | 4.30 | 4.32 |
| hsa-miR-4532 | SW1736 | 9.01 | 9.10 |
|
| 8505c | 8.96 | 9.73 |
|
hsa-miR-548t-5p | SW1736 | 7.54 | 7.78 |
|
| 8505c | 2.83 | 3.21 |
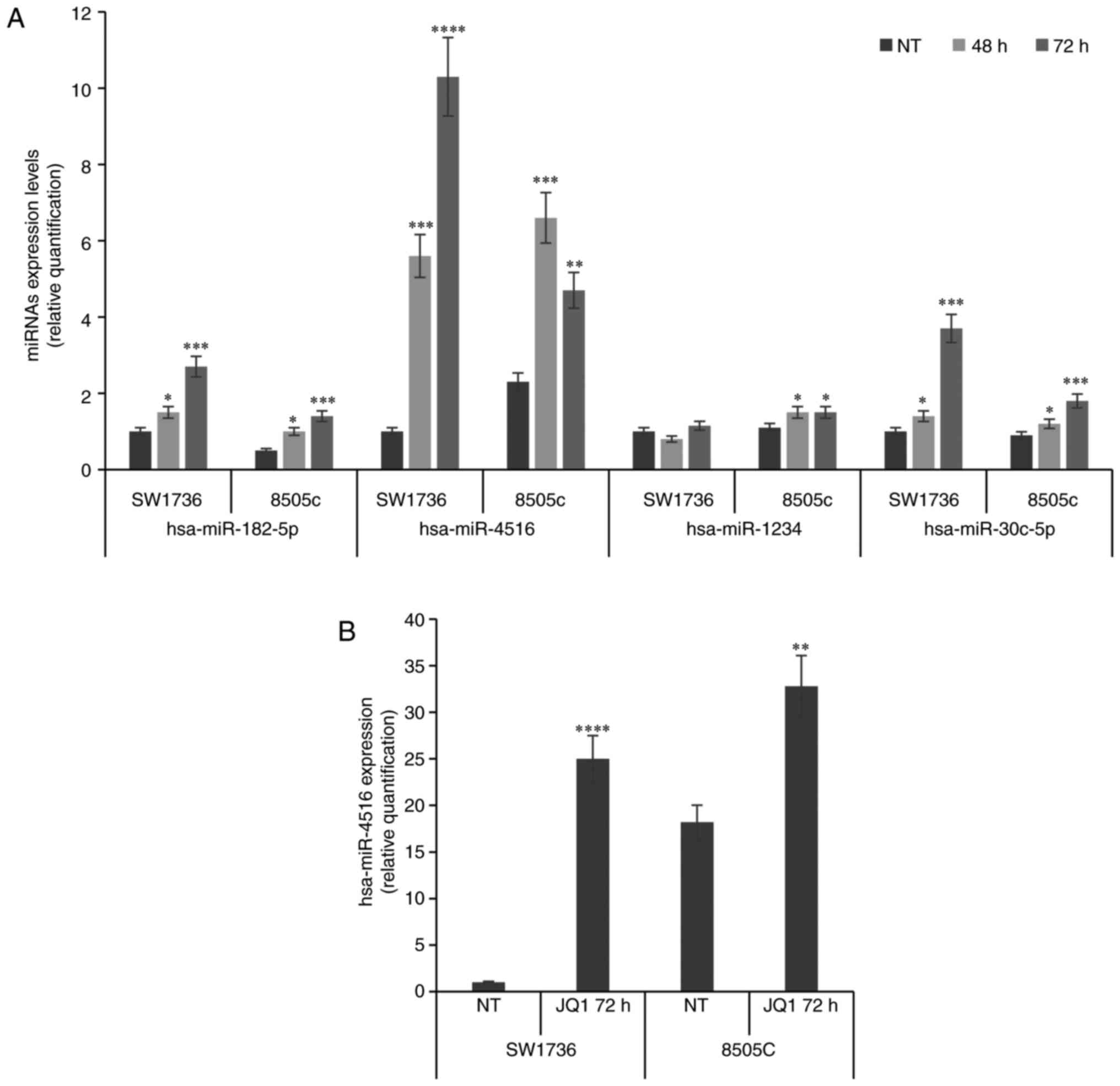
To validate the miRNA expression data, we performed
qPCR analysis of these seven differentially expressed miRNAs. Out
of the seven microRNAs, significant changes were observed only for
three miRNAs (miR-182-5p, miR-30c-5p and miR-4516), as shown in
Fig. 2A. Notably, validation of
miR-4488/4532/548t-5p was not possible due to technical issues.
Among the validated microRNAs, after further validation in
biological replicates in which a consistent increase in miR-4516
levels after JQ1 treatment was demonstrated (Fig. 2B), we focused our attention on
miR-4516 and its downstream pathway, since this microRNA was
previously linked to apoptotic phenomena (17). JQ1 treatment increased both
apoptotic and necrotic phenomena (represented by caspase-3/7
activity levels and PARP to cleaved-PARP ratio) in both ATC cell
lines, as demonstrated in our previously published study (3).
JQ1 effects on miR-4516 levels and
STAT3-dependent pathway
We exploited the target prediction database miRDB
(http://mirdb.org/miRDB/) to delineate miR-4516
potential mRNA targets. Of the 1545 hits, the signal transducer and
activator of transcription 3 (STAT3) was already characterized as a
miR-4516 downstream effector. STAT3 is a transcription factor that,
when phosphorylated, relocates into the nucleus where it acts as a
transcription activator. Besides its normal functions, this protein
plays an important role in cellular transformation and
tumorigenesis and it was revealed to be overexpressed and
hyper-activated in thyroid cancer (18–20).
Once activated, STAT3 induces the expression of SKP2, a member of
the F-box protein family, an oncogene that mediates the
ubiquitination of p21 (CDKN1A) and p27 (CDKN1B) decreasing the
tumorigenic potential of cells (21,22).
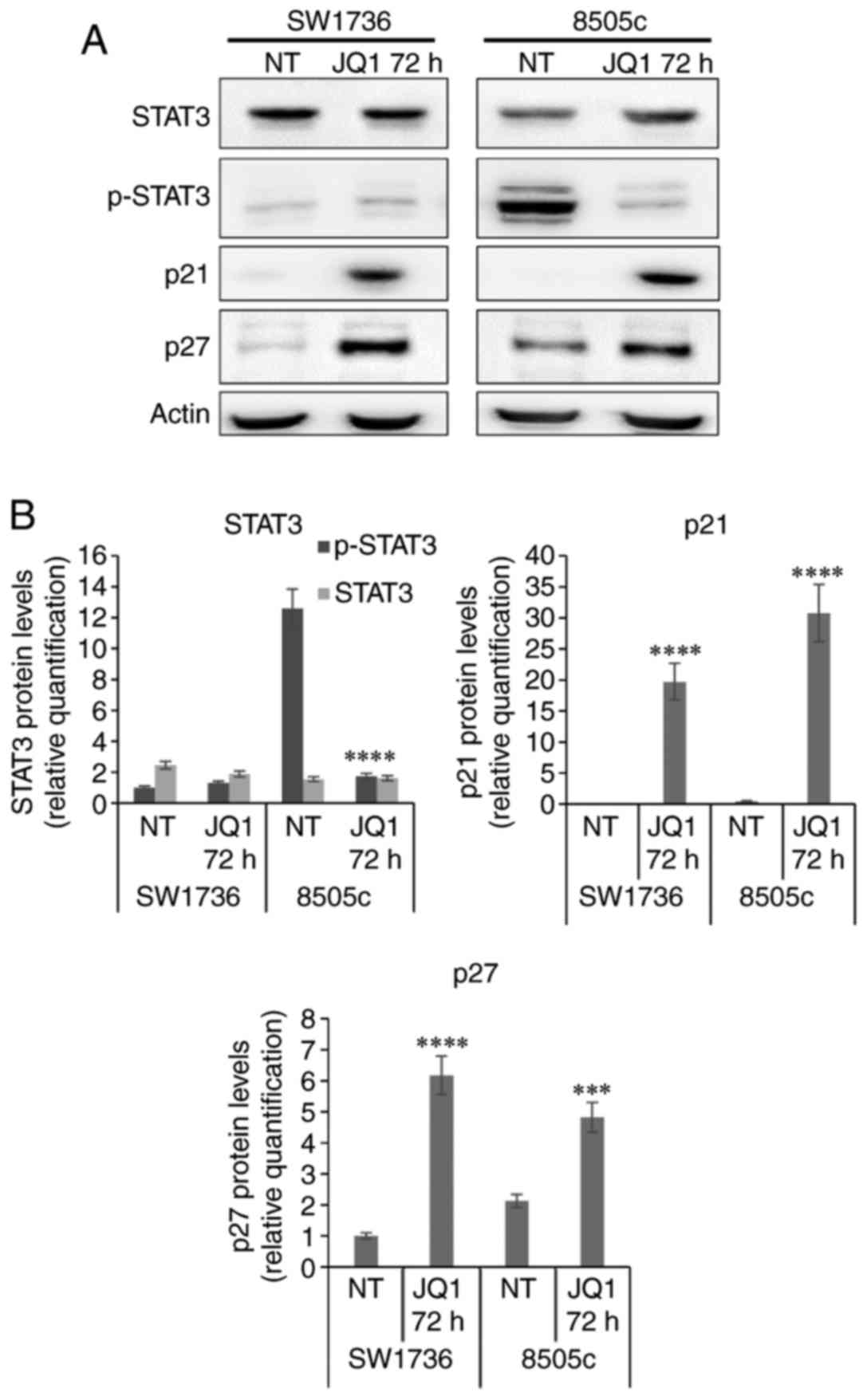
Cell line 8505c treated with 5 µM JQ1 displayed a
strong decrease in phospho-STAT3 (p-STAT3) protein levels when
compared to the vehicle-treated cells. SW1736, conversely, did not
display any detectable variation in p-STAT3 protein levels after
treatment; this was possibly due to a diverse site of
phosphorylation (i.e., Ser727), different from the one detected
with our antibody (Tyr705) (Fig. 3A and
B). Moreover, Sos et al reported a delayed STAT3
activation by Tyr705 phosphorylation in SW1736 cells, hypothesizing
an IL6-related autocrine loop that may be responsible for the
time-dependent activation of this signaling pathway (23). We, then, evaluated whether a
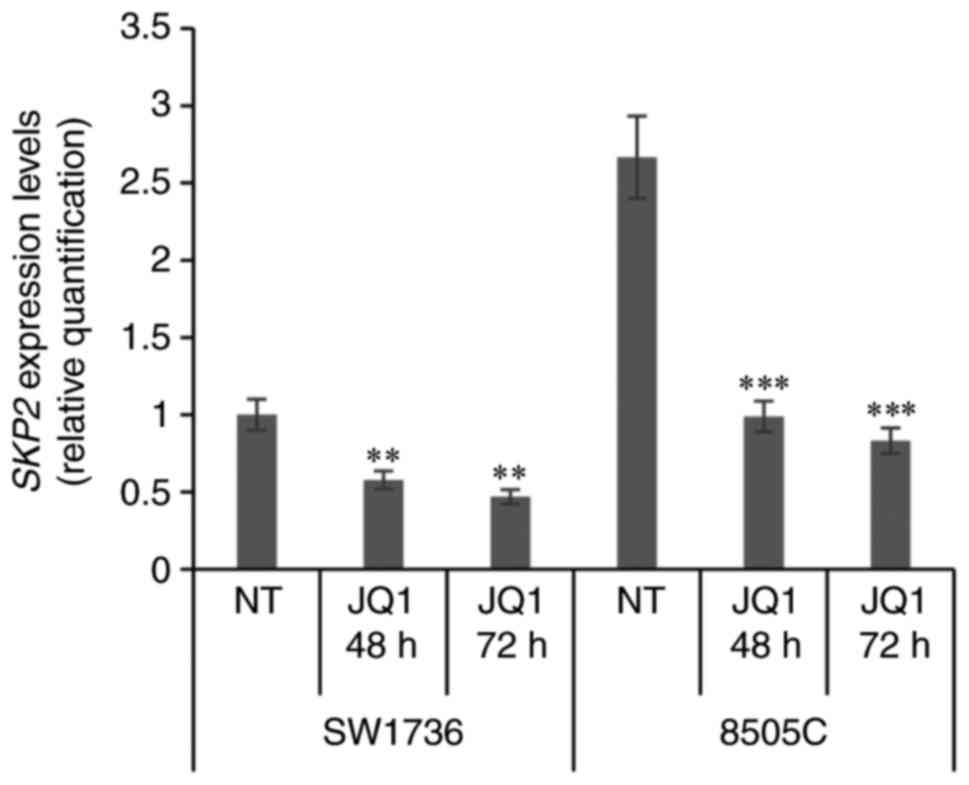
reduction in p-STAT3 corresponded in a downregulation of
SKP2 expression. After both 48 h and 72 h of JQ1 treatment,
ATC cells displayed a significant decrease in SKP2 mRNA
levels (Fig. 4), corroborating a
possible STAT3-dependent cascade alteration. We next evaluated if
the ultimate targets of this signaling cascade were upregulated
after JQ1 treatment. Both SW1736 and 8505c exhibited a striking
increase in both p21 and p27 protein levels after 5 µM JQ1
treatment (Fig. 3A and B).
Differentially expressed miRNAs
between ATC and normal thyroid tissue
Finally, to identify chief miRNAs differentially
expressed between ATC cells and non-tumorigenic ones, SW1736, 8505c
and Nthy-ori 3-1 RNAs were run on the NanoString platform.
Considering the 812 miRNAs analyzed on the NanoString platform, 65
and 96 miRNAs turned out to be dysregulated in SW1736 and 8505c
cells, respectively, when compared to Nthy-ori 3-1. Among them,
miR-146a-5p and miR-221-3p were overexpressed in the two ATC cell
lines confirming data already enlisted in literature reviews
(21,24). Thirty-five miRNAs exhibited a common
dysregulation in the two ATC cell lines when compared to Nthy-ori
3-1 (Fig. 5 and Table II). Several of these have been
already described as upregulated in thyroid cancer (24,25)
and miR-221/222 have undoubtedly been correlated to cancer
aggressiveness (11). Crosschecking
Tables I and II, we could highlight that three miRNAs
(miR-4516, miR-1234 and miR-4488) exhibited aberrant behavior: they
were downregulated in ATC cells when compared to Nthy-ori 3-1 and
turned out to be upregulated in ATC after both 48 and 72 h of JQ1
treatment.
 | Table II.Altered miRNAs in ATC cells compared
to non-tumorigenic ones. |
Table II.
Altered miRNAs in ATC cells compared
to non-tumorigenic ones.
| miRNAs commonly
dysregulated in ATC cell lines |
|---|
|
|---|
| Upregulated | Downregulated |
|---|
| hsa-miR-1 | hsa-miR-1234 |
| hsa-miR-100-5p | hsa-miR-1268a |
|
hsa-miR-1226-3p | hsa-miR-127-3p |
| hsa-miR-1257 |
hsa-miR-193b-3p |
|
hsa-miR-125b-5p | hsa-miR-34a-5p |
| hsa-miR-139-3p | hsa-miR-4488 |
|
hsa-miR-146a-5p | hsa-miR-4516 |
| hsa-miR-183-5p |
|
| hsa-miR-221-3p |
|
| hsa-miR-29b-3p |
|
| hsa-miR-302f |
|
| hsa-miR-31-5p |
|
| hsa-miR-335-5p |
|
| hsa-miR-337-3p |
|
| hsa-miR-337-5p |
|
| hsa-miR-33a-5p |
|
| hsa-miR-412 |
|
| hsa-miR-4286 |
|
|
hsa-miR-450a-5p |
|
| hsa-miR-502-5p |
|
| hsa-miR-504 |
|
| hsa-miR-512-3p |
|
| hsa-miR-512-5p |
|
|
hsa-miR-548a-5p |
|
|
hsa-miR-548am-3p |
|
|
hsa-miR-548d-3p |
|
| hsa-miR-7-5p |
|
| hsa-miR-936 |
|
Discussion
Recently, the investigation of BET inhibitor-derived
miRNA regulation has gained the attention of researchers. Hitherto,
only focused studies, aimed to investigate the modulation of one
miRNA or one miRNA subfamily after BET inhibition, are available
(26). Based on this, our data, for
the first time, explored the link between JQ1 and global miRNA
expression. In thyroid cancer, miRNA dysregulation has been
reported to contribute to tumor progression (27). ATC cells treated with 5 µM JQ1
exhibited substantial miRNA dysregulation at both 48 and 72 h of
exposure. We identified several miRNAs substantially upregulated
after sustained JQ1 exposure in ATC cells. Some of these miRNAs
have already been associated to the inhibition of proliferation,
apoptosis induction and have been correlated with survival in
different solid tumor models (28,29).
The signal transducer and activator of transcription
STAT3 is a transcription factor that gets phosphorylated upon
activation, then relocates into the nucleus and activates target
genes (30). This protein plays an
important role in cellular transformation and tumorigenesis and is
constitutively activated in approximately 70% of solid and
hematological cancers (31,32). Increased expression and activation
of STAT3 has been described in thyroid cancer tissues and
inhibition of the STAT3 signaling pathway has been considered as a
novel therapeutic approach to treat human cancers that harbor
aberrantly active STAT3 (18).
Noteworthy, several miRNAs included in our analysis turned out to
be associated to STAT3 signaling pathways, miR-4516 being the most
characterized (17,33). STAT3 is known to positively regulate
SPK2 gene expression, which in turn mediates the
ubiquitination of p21 and p27 (34). SKP2 is an F-box protein belonging to
the SCF E3 ubiquitin ligase complex; it is a known oncogene
regulating cellular proliferation, cancer progression and
metastasis by inducing the degradation of the cyclin-dependent
kinase (Cdk) inhibitors p21Cip1, p27Kip1 and
p57 (21). However, upstream
regulators of SKP2 in the progression of human cancer are not yet
thoroughly known. Many genes and regulatory pathways have been
discovered (STAT3, BCR-ABL, PI3K/AKT, ERK, PPARγ, PTEN and mTOR)
making SKP2 regulation a complex issue in cancer research (22). Nevertheless, several studies have
demonstrated a direct and solid link between STAT3 phosphorylation
and the SKP2/p27 axis. JQ1 treatment undeniably produced a decrease
in SKP2 expression correlated to an increase in p21 and p27
protein levels, in both ATC cell lines. STAT3 phosphorylation was
subjected to a strong decrease in cell line 8505c, while it was
undetectable in SW1736 cells, probably due to a diverse site of
phosphorylation from the one detected in our analysis.
In conclusion, our results highlighted that a
JQ1-regulated miRNA impaired STAT3 transduction pathways leading to
antineoplastic activities in ATC cell lines. Both ATC cell lines
displayed an upregulation of some miRNAs enlisted in multiple ATC
literature reviews (i.e., miR-146a and miR-221). An interesting
event could be disclosed comparing JQ1-regulated and ATC-linked
miRNAs: three miRNAs (miR-4516, miR-1234, miR-4488) were determined
to be downregulated in both ATC cell lines when compared to
Nthy-ori 3-1 and, then, turned out to be upregulated by JQ1
treatment, after both 48 and 72 h of exposure. In brief, these data
revealed the antineoplastic activity of JQ1 in thyroid cancer
cells.
Acknowledgements
We thank Dr Emanuele Cettul and Dr Luciana Gualdi
from Pharmadiagen S.r.l. (Pordenone, Italy) for the miRNome
profiling. This study was supported by a grant to GD from Ministero
degli Affari Esteri of Italy (Progetti grande rilevanza 2016,
project no. PGR02954) and from MIUR (PRIN 2015, project no.
2015HPMLFY-011).
Glossary
Abbreviations
Abbreviations:
|
ATC
|
anaplastic thyroid cancer
|
|
BET
|
bromodomain and extra-terminal
protein
|
|
DMSO
|
dimethyl sulphoxide
|
|
miRNA
|
microRNA
|
|
PDTC
|
poorly differentiated thyroid
cancer
|
|
RNA-seq
|
RNA sequencing
|
|
STAT
|
signal transducer and activator of
transcription
|
|
STR
|
single tandem repeats
|
|
UTR
|
untranslated region
|
|
WDTC
|
well-differentiated thyroid cancer
|
References
|
1
|
Gu L and Sun W: MiR-539 inhibits thyroid
cancer cell migration and invasion by directly targeting CARMA1.
Biochem Biophys Res Commun. 464:1128–1133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gao X, Wu X, Zhang X, Hua W and Zhang Y,
Maimaiti Y, Gao Z and Zhang Y: Inhibition of BRD4 suppresses tumor
growth and enhances iodine uptake in thyroid cancer. Biochem
Biophys Res Commun. 469:679–685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mio C, Lavarone E, Conzatti K, Baldan F,
Toffoletto B, Puppin C, Filetti S, Durante C, Russo D, Orlacchio A,
et al: MCM5 as a target of BET inhibitors in thyroid cancer cells.
Endocr Relat Cancer. 23:335–347. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nicodeme E, Jeffrey KL, Schaefer U, Beinke
S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H,
et al: Suppression of inflammation by a synthetic histone mimic.
Nature. 468:1119–1123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fu LL, Tian M, Li X, Li JJ, Huang J,
Ouyang L, Zhang Y and Liu B: Inhibition of BET bromodomains as a
therapeutic strategy for cancer drug discovery. Oncotarget.
6:5501–5516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce
LA, Thompson RC, Muller S, Knapp S and Wang J: Inhibition of BET
bromodomain targets genetically diverse glioblastoma. Clin Cancer
Res. 19:1748–1759. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sahni JM, Gayle SS, Bonk KLW, Vite LC,
Yori JL, Webb B, Ramos EK, Seachrist DD, Landis MD, Chang JC, et
al: Bromodomain and extraterminal protein inhibition blocks growth
of triple-negative breast cancers through the suppression of aurora
kinases. J Biol Chem. 291:23756–23768. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dawson MA, Prinjha RK, Dittmann A,
Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C,
Savitski MM, et al: Inhibition of BET recruitment to chromatin as
an effective treatment for MLL-fusion leukaemia. Nature.
478:529–533. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Filippakopoulos P, Qi J, Picaud S, Shen Y,
Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et
al: Selective inhibition of BET bromodomains. Nature.
468:1067–1073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L, Lv B, Chen B, Guan M, Sun Y, Li H,
Zhang B, Ding C, He S and Zeng Q: Inhibition of miR-146b expression
increases radioiodine-sensitivity in poorly differential thyroid
carcinoma via positively regulating NIS expression. Biochem Biophys
Res Commun. 462:314–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fuziwara CS and Kimura ET: MicroRNA
Deregulation in anaplastic thyroid cancer biology. Int J
Endocrinol. 2014:7434502014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Borrelli N, Denaro M, Ugolini C, Poma AM,
Miccoli M, Vitti P, Miccoli P and Basolo F: miRNA expression
profiling of ‘noninvasive follicular thyroid neoplasms with
papillary-like nuclear features’ compared with adenomas and
infiltrative follicular variants of papillary thyroid carcinomas.
Mod Pathol. 30:39–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pilli T, Prasad KV, Jayarama S, Pacini F
and Prabhakar BS: Potential utility and limitations of thyroid
cancer cell lines as models for studying thyroid cancer. Thyroid.
19:1333–1342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schweppe RE, Klopper JP, Korch C,
Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow LA, Copland
JA, Smallridge RC, et al: Deoxyribonucleic acid profiling analysis
of 40 human thyroid cancer cell lines reveals cross-contamination
resulting in cell line redundancy and misidentification. J Clin
Endocrinol Metab. 93:4331–4341. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Passon N, Gerometta A, Puppin C, Lavarone
E, Puglisi F, Tell G, Di Loreto C and Damante G: Expression of
Dicer and Drosha in triple-negative breast cancer. J Clin Pathol.
65:320–326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gatfield D, Le Martelot G, Vejnar CE,
Gerlach D, Schaad O, Fleury-Olela F, Ruskeepää AL, Oresic M, Esau
CC, Zdobnov EM, et al: Integration of microRNA miR-122 in hepatic
circadian gene expression. Genes Dev. 23:1313–1326. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chowdhari S and Saini N: hsa-miR-4516
mediated downregulation of STAT3/CDK6/UBE2N plays a role in PUVA
induced apoptosis in keratinocytes. J Cell Physiol. 229:1630–1638.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dong W, Cui J, Tian X, He L, Wang Z, Zhang
P and Zhang H: Aberrant sonic hedgehog signaling pathway and STAT3
activation in papillary thyroid cancer. Int J Clin Exp Med.
7:1786–1793. 2014.PubMed/NCBI
|
|
19
|
Yan LI, Li LI, Li Q, Di W, Shen W, Zhang L
and Guo H: Expression of signal transducer and activator of
transcription 3 and its phosphorylated form is significantly
upregulated in patients with papillary thyroid cancer. Exp Ther
Med. 9:2195–2201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Gill A, Atmore B, Johns A,
Delbridge L, Lai R and McMullen T: Upregulation of the signal
transducers and activators of transcription 3 (STAT3) pathway in
lymphatic metastases of papillary thyroid cancer. Int J Clin Exp
Pathol. 4:356–362. 2011.PubMed/NCBI
|
|
21
|
Lee J-J, Lee J-S, Cui MN, Yun HH, Kim HY,
Lee SH and Lee J-H: BIS targeting induces cellular senescence
through the regulation of 14-3-3 zeta/STAT3/SKP2/p27 in
glioblastoma cells. Cell Death Dis. 5:e15372014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bochis OV, Irimie A, Pichler M and
Berindan-Neagoe I: The role of Skp2 and its substrate CDKN1B (p27)
in colorectal cancer. J Gastrointestin Liver Dis. 24:225–234.
2015.PubMed/NCBI
|
|
23
|
Sos ML, Levin RS, Gordan JD, Oses-Prieto
JA, Webber JT, Salt M, Hann B, Burlingame AL, McCormick F,
Bandyopadhyay S, et al: Oncogene mimicry as a mechanism of primary
resistance to BRAF inhibitors. Cell Reports. 8:1037–1048. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wójcicka A, Kolanowska M and Jażdżewski K:
Mechanisms in endocrinology: MicroRNA in diagnostics and therapy of
thyroid cancer. Eur J Endocrinol. 174:R89–R98. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Leonardi GC, Candido S, Carbone M,
Colaianni V, Garozzo SF, Cinà D and Libra M: microRNAs and thyroid
cancer: Biological and clinical significance (Review). Int J Mol
Med. 30:991–999. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Z, Sharp PP, Yao Y, Segal D, Ang CH,
Khaw SL, Aubrey BJ, Gong J, Kelly GL, Herold MJ, et al: BET
inhibition represses miR17-92 to drive BIM-initiated apoptosis of
normal and transformed hematopoietic cells. Leukemia. 30:1531–1541.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Forte S, La Rosa C, Pecce V, Rosignolo F
and Memeo L: The role of microRNAs in thyroid carcinomas.
Anticancer Res. 35:2037–2047. 2015.PubMed/NCBI
|
|
28
|
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang
Z, Wang X, Lin Y, Mao Y, et al: Downregulation of microRNA-182-5p
contributes to renal cell carcinoma proliferation via activating
the AKT/FOXO3a signaling pathway. Mol Cancer. 13:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu N, Cui R-X, Sun Y, Guo R, Mao YP, Tang
LL, Jiang W, Liu X, Cheng YK, He QM, et al: A four-miRNA signature
identified from genome-wide serum miRNA profiling predicts survival
in patients with nasopharyngeal carcinoma. Int J Cancer.
134:1359–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sosonkina N, Starenki D and Park J-I: The
Role of STAT3 in Thyroid Cancer. Cancers (Basel). 6:526–544. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Frank DA: STAT3 as a central mediator of
neoplastic cellular transformation. Cancer Lett. 251:199–210. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sansone P and Bromberg J: Targeting the
interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol.
30:1005–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chowdhari S and Saini N: Gene expression
profiling reveals the role of RIG1 like receptor signaling in p53
dependent apoptosis induced by PUVA in keratinocytes. Cell Signal.
28:25–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin H-P, Lin C-Y, Huo C, Hsiao PH, Su LC,
Jiang SS, Chan TM, Chang CH, Chen LT, Kung HJ, et al: Caffeic acid
phenethyl ester induced cell cycle arrest and growth inhibition in
androgen-independent prostate cancer cells via regulation of Skp2,
p53, p21Cip1 and p27Kip1. Oncotarget. 6:6684–6707. 2015. View Article : Google Scholar : PubMed/NCBI
|