Introduction
Cervical cancer is one of the most severe malignant
tumors and it also has high rates of morbidity especially in many
developing countries (1). China's
National Cancer Center earlier published data showed that the
incidence and mortality rate of cervical cancer is
10.4/105 and 2.59/105 (2). Contemporary management of cervical
cancer involves chemotherapy, surgery and radiotherapy (3). As recommended by the National
Comprehensive Cancer Network (NCCN), Platinum-based chemotherapy is
often used in cervical cancer management (4). However, common platinum-based drugs,
for example, cisplatin, often induce resistance. Thus there is an
urgent need of novel drugs for cervical cancer management (5).
The heterogeneous nuclear ribonucleoprotein (hnRNP)
family consists of approximately 20 hnRNA-binding proteins and most
of them are related to key biogenesis of messenger RNA (mRNA)
(6). The hnRNP A2/B1 complex are
important members of hnRNP family and are made up of the proper
proportion of protein A and protein B, it has been proved that
hnRNPs regulate transportation and splicing of mRNA in cells
(7,8). Several studies have demonstrated that
hnRNP A2/B1 was highly expressed in gastric adenocarcinoma,
pancreatic cancer and glioblastoma. Overexpression of hnRNP A2/B1
in non-small cell lung cancer increased cell proliferation, while
downregulation of hnRNP A2/B1 enhanced apoptosis of breast cancer
cells. Importantly, hnRNP A2/B1 has been used as biomarker and
prognostic indicator in non-small cell lung cancer (9–13).
However, the role of hnRNP A2/B1 in cervical cancer has not been
fully studied.
Phophatidylinositol 3-kinase/protein kinase-B
(PI3K/AKT) signaling pathway and its downstream targets play
important roles in tumorigenesis. For example, PI3K/AKT pathway
trigger tumor cell death through binding to Bad/Bcl-2 complex and
inactive caspase-8/9 (14,15). Moreover, the PI3K/AKT signaling
pathway is involved in cell proliferation by activating cyclin
dependent kinase (CDK), upregulating cyclins and downregulating
p21/Waf1/Cip1 and p27/Kip2 (16). A
study based on the Cancer Genome Atlas revealed that the expression
of the subunits of PI3K/AKT varies in cervical cancer, ovarian
cancer and uterine epithelial tumor (17). However the relationship between
hnRNP A2/B1 and PI3K/AKT in cervical cancer has not been fully
clarified.
From previous research, we found that hnRNP A2/B1
was inhibited in CaSki cells after treatment with lobaplatin
(18). However the relationship
between hnRNP A2/B1 and PI3K/AKT in cervical cancer is not clear
and the underlying mechanism remains unknown. Thus, in this study,
we explored the role of hnRNP A2/B1 in proliferation, apoptosis as
well as relationship between hnRNP A2/B1 and PI3K/AKT pathway in
cervical cancer both in vitro and in vivo.
Materials and methods
Cell culture
Human cervical cancer cells HeLa and CaSki were
purchased from the cell bank of the Type Culture Collection of
Chinese Academy of Sciences (Shanghai, China). Cells were cultured
in RPMI-1640 medium (Hyclone, Logan, UT, USA) supplemented with 10%
fetal bovine serum (Gibco, Grand Island, NY, USA), 1%
penicillin-streptomycin (Solarbio, Beijing, China) and 1%
L-glutamine (Amresco, Solon, OH, USA) in a 5% CO2
incubator at 37°C. The medium was replaced every 2–3 days, and
subsequent studies were performed when cells in exponential growth
phase.
hnRNP A2/B1-shRNA design and cell
transfection
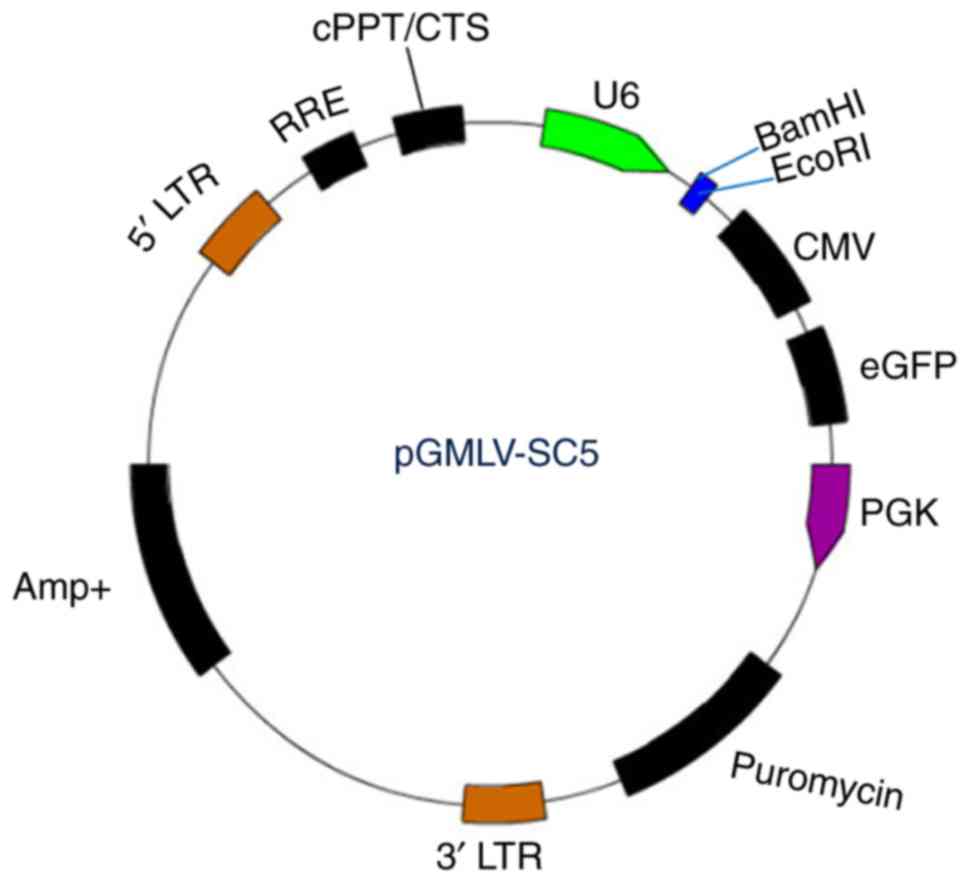
pGMLV-SC5 RNAi lentiviral vector (Fig. 1) was purchased from Genomeditech
(Shanghai, China). We followed the criteria described by Invitrogen
(Carlsbad, CA, USA) to design multiple shRNAs targeting hnRNP A2/B1
or negative control mRNA sequence. Four of the positive targeting
sequences and one negative shRNA against hnRNP A2/B1 sequence
(Table I) were specially chosen for
subsequent studies. Synthesized oligonucleotides (Table II) were annealed and ligated to the
BamHI/EcoRI sites of pGMLV-eGFP to produce
pGMLV-eGFP-shhnRNP A2/B1 or pGMLV-eGFP-shCon, eGFP expression was
used to exhibit the infection of lentiviruses. The HeLa and CaSki
cells were cultured in RPMI-1640 supplemented with 10% FBS, 1%
penicillin-streptomycin liquid and 1% L-glutamine at 6-well plate.
When the cells were in exponential growth phase, the medium of the
negative control of HeLa or CaSki cells were replaced by medium
with NC-shRNA diluent and the positive group were replaced by
medium with hnRNP A2/B1-shRNA diluent. After 24 h, the culture
media was replaced with RPMI-1640 medium, and cells were incubated
in an incubator for 72 h. HeLa and CaSki were further screened in
media consisting of puromycin (2 mg/l). Because pGMLV-SC5RNAi
lentiviral vector contains eGFP and anti-puromycin gene, the cells
transfacted with lentiviral vector can reveal green fluorescence.
Then the eGFP-positive cells can be picked up for further
analysis.
 | Table I.Four specific target sequences of
hnRNPA2/B1 gene. |
Table I.
Four specific target sequences of
hnRNPA2/B1 gene.
| No. | TargetSeq |
|---|
| 1 |
CAGAAATACCATACCATCAAT |
| 2 |
TGACAACTATGGAGGAGGAAA |
| 3 |
GGGCTCATGTAACTGTGAAGA |
| 4 |
GCTTTGTCTAGACAAGAAATG |
| NC-shRNA |
TTCTCCGAACGTGTCACGT |
 | Table II.Oligos of 4 pairs of shRNA and 1 pair
of NC-shRNA. |
Table II.
Oligos of 4 pairs of shRNA and 1 pair
of NC-shRNA.
| Oligo | Oligonucleotide DNA
sequence 5′ to 3′ |
|---|
|
3935hnRNPA2/B1-shRNA1-T
(EcoRI) |
gatccGCAGAAATACCATACCATCAATCTCGAGATTGATGGTATGGTATTTCTGTTTTTTg |
|
3935hnRNPA2/B1-shRNA1-B
(BamHI) |
aattcAAAAAACAGAAATACCATACCATCAATCTCGAGATTGATGGTATGGTATTTCTGCg |
|
3936hnRNPA2/B1-shRNA2-T
(EcoRI) |
gatccGTGACAACTATGGAGGAGGAAACTCGAGTTTCCTCCTCCATAGTTGTCATTTTTTg |
|
3936hnRNPA2/B1-shRNA2-B
(BamHI) |
aattcAAAAAATGACAACTATGGAGGAGGAAACTCGAGTTTCCTCCTCCATAGTTGTCACg |
|
3937hnRNPA2/B1-shRNA3-T
(EcoRI) |
gatccGGGCTCATGTAACTGTGAAGACTCGAGTCTTCACAGTTACATGAGCCCTTTTTTg |
|
3937hnRNPA2/B1-shRNA3-B
(BamHI) |
aattcAAAAAAGGGCTCATGTAACTGTGAAGACTCGAGTCTTCACAGTTACATGAGCCCg |
|
3938hnRNPA2/B1-shRNA4-T
(EcoRI) |
gatccGCTTTGTCTAGACAAGAAATGCTCGAGCATTTCTTGTCTAGACAAAGCTTTTTTg |
|
3938hnRNPA2/B1-shRNA4-B
(BamHI) |
aattcAAAAAAGCTTTGTCTAGACAAGAAATGCTCGAGCATTTCTTGTCTAGACAAAGCg |
| NC-shRNA1-1 |
gatccGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAACTTTTTTACGCGTg |
| NC-shRNA1-2 |
aattcACGCGTAAAAAAGTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAACg |
Quantitative real-time PCR
analysis
HeLa and CaSki cells were transfected with either
control or hnRNP A2/B1 shRNAs as described previously, total RNA
was extracted by trizol (Invitrogen), cDNAs were synthesized from
total RNAs by SuperScript VILO cDNA Synthesis kit (Invitrogen)
according to the manufacturer's protocol. Quantitative real-time
PCR was performed by SYBR-Green PCR Master Mix (Applied Biosystems,
USA). The specific primers for cDNA were as follows: hnRNP A2/B1,
sence: 5′-GATGGCAGAGAACGGTGTGAAG-3′, and antisense,
5′-AGGCATAGGTATTGGCAACTGC-3′. β-actin, sence:
5′-GTCTCCTCTGACTTCAACAGCG-3′, antisence:
5′-ACCACCCTGTTGCTGTAGCCAA-3′. β-actin was considered as an internal
control. PCR reaction conditions were perfomed as follows: 95°C for
10 min, and 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The
respective gene expression were calculated by the 2−ΔΔCt
method.
Western blot analysis
HeLa and CaSki cells and nude mouse tumor tissues
were lysed by RIPA lysis buffer containing 1% PMSF and 1% protein
phosphatase inhibitor for 30 min and then centrifuged at 14,000 × g
at 4°C for 20 min. Collecting the supernatant to store at −80°C.
The protein concentration was performed by Bradford assay kit.
Cells and tissue lysates were electrophoresed by SDS-PAGE and
proteins were transferred to PVDF membrane (Millipore) and then
blocked with Tris-buffered saline Tween-20 with 5% non-fat milk for
2 h. Next, they were incubated with primary antibodies diluted with
TBST overnight at 4°C, the primary antibodies were as follows:
Anti-hnRNP A2/B1 (Bioworld Technology, St. Louis Park, MN, USA;
1:1,000), anti-AKT (Proteintech, Wuhan, China; 1:500), anti-p-AKT
(Cell Signaling Technology, Danvers, MA, USA; 1:1,000), anti-p21
(Abcam, Cambridge, UK; 1:2,000), anti-p27 (Abcam; 1:1,000),
anti-PI3K (Wanleibio, Shenyang, China; 1:500), anti-caspase-3
(Wanleibio; 1:500), anti-cleaved caspase-3 (Wanleibio; 1:500),
β-actin (Bioss, Beijing, China; 1:6,000) was used as a loading
control and followed by incubation with secondary antibody
(ZSGB-Bio, Beijing, China; 1:90,000) at room temperature for 1 h.
Chemiluminent detection was determined by ECL kit. ImageJ software
was used to conclude data.
Cell proliferation assay
HeLa and CaSki with hnRNP A2/B1-shRNA, NC-shRNA and
control group cells were digested by 0.25% trypsin (Gibco), diluted
to 5×104/ml and the single cell was suspended in 200 µl
culture medium and then seeded in 96-well plates to culture in a 5%
CO2 incubator at 37°C overnight. Subsequently, the
medium was respectively replaced by 200 µl of new medium that
consisted of insulin-like growth factor 1 (IGF-1, Prospec-Tany
Technogene Ltd., Rehovot, Israel) and LY294002 (Beyotime
Biotechnology Corporation, Shanghai, China) or not for 12, 24, 48
h. The concentration of IGF-1 on HeLa and CaSki cells was 100
ng/ml, the concentration of LY294002 on HeLa and CaSki cells was 20
and 15 µM, respectively. Then the culture medium was removed before
the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
(MTT) solution (5 mg/ml) was added to the medium and maintained at
37°C for 4 h, the supernatant was removed and 150 µl DMSO was added
to each well under the absorbance measurement at 490 nm.
Colony formation assay
Single-cell suspensions were digested with 0.25%
trypsin, harvested and seeded into 6-well plates for 500 cells per
well and then cultured in RPMI-1640 in a 5% CO2
incubator at 37°C for two weeks. The cell supernatants were removed
and washed twice with PBS after visible colonies appeared. Cells
were cultured with 4% paraformaldehyde for 15 min then stained with
moderate gentian violet solution for 30 min before being washed
with PBS. The efficiency of the assay was estimated as follows:
Clone forming efficiency = number of colonies/number of inoculated
cells ×100%.
Cell invasion assay
The invasion of HeLa and CaSki cells was estimated
by transwell chambers. Serum free medium and matrigel (1:4)
solution (60 µl of 4°C) was added to upper chambers for 4–5 h at
37°C. The 200 µl single-cell suspensions (2×105/ml) were
seeded into upper chamber and the lower chambers were fixed with
10% medium to incubate for 24 h. The chamber which was fixed in
100% methanol was taken out and subsequently stained with 2%
crystal violet for 30 min. The cell invasion assay was performed by
the stained cells in the chamber.
Wound healing assay
Single-cell suspensions were diluted to
5×105 per well and seeded into 6-well plates
subsequently cultured with serum-free medium overnight. Then the
adherent cells were washed with PBS 3 times and exposed in
different drugs, respectively, for 0 and 24 h after the cells were
scratched by 10 µl pipette tip across the center of the well. The
gap distance was photographed by microscopy. ImageJ was used to
assess quantitative data.
Cell cycle assay
The exponential phase of HeLa and CaSki cells were
added with 0.25% trypsin for digestion, collected, centrifuged at
100 × g at 4°C for 5 min. Cells were fixed with 70% ethanol after
supernatant was removed and then suspended in 50 µg/ml RNaseA
(KeyGen Biotech Corp., Ltd., Jiangsu, China) at 37°C for 30 min and
stained in 50 µg/ml PI (KeyGen Biotech Corp., Ltd.) solution for 30
min in the dark. The single-cell suspensions were performed by flow
cytometry.
Cell apoptosis assay
Cells in the exponential phase were digested and
diluted to a 5×104/ml suspension and seeded into 6-well
plates overnight. Subsequently, 5×105 single-cell
suspension was collected with 500 µl Binding Buffer, then fixed
with 5 µl Annexin APC (KeyGen Biotech Corp., Ltd.) and 5 µl PI
(KeyGen Biotech Corp., Ltd.) staining solution in the dark for 15
min. Apoptosis of different cells were evaluated by flow
cytometry.
IC50 of HeLa and CaSki by MTT
assay
Cells (5×104/ml) were harvested and
seeded in 96-well plates, exposed in different concentrations of
lobaplatin or irinotecan (concentration was determined by
pre-experimentation and referenced to the data of published
(18–20), final concentration: 2, 4, 6, 8, 12
and 16 µg/ml or 20, 40, 60, 80, 160 and 240 µg/ml, Hainan Changan
International Pharmaceutical Co., Ltd., Hainan, China) in a 5%
CO2 incubator at 37°C for 24 h, the test wells were
six-replica. Then each well was fixed with 10 µl per well
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
(final concentration: 5 µg/ml) for 4 h. Subsequently, the media was
removed and 150 µl per well of DMSO was added before it was
measured at 490 nm by an enzyme standard instrument. This was
repeated three times and the average value was taken.
Tumor xenograft experiment
Female BALB/c nude mice at 4 weeks were purchased
from Chongqing National Bio Industrial Base Experimental Animal
Center (Chongqing, China). HeLa cells (2×106/ml) were
collected and suspended in 200 µl PBS. The cell suspensions were
injected subcutaneously into the right side near the back of the
neck, all of the nude mice were kept in a homeothermic and specific
pathogen-free room, temperature and humidity were maintained at
26–28°C and 40–60%. Nude mice were randomly divided into 3 groups
(7 mice per group), Vernier caliper was performed to measure tumor
volume every 3 days. All the nude mice were sacrificed by breaking
the neck after anesthetized by 10% chloral hydrate and tumor
tissues were collected for the next analysis after 30 days. The
tumor volume calculation method referred to the following formula:
(LxW2)/2 (21), where L
is the longest tumor diameter and W is the shortest tumor diameter.
Animal experiments were strictly as the guidelines of Guizhou
Medical University Animal Care Welfare Committee (number:
1702256).
Immunohistochemistry
The expression of PCNA and Ki-67 in nude mice
injected with HeLa cells were revealed by immunohistochemistry
assay. Fresh tissues were soaked in 4% neutral formaldehyde for 24
h and then dehydrated and paraffin-embedded, the adhesion slides
with 4-µm sections were kept at 60°C for 4 h. Deparaffinizing with
xylene and hydrating with an ethanol gradient. Followed by citrate
buffer under high pressure to repair the antigen for 3 min and then
the slides were incubated with 0.3% H2O2 for
30 min. After that, the sections were rinsed by PBS-T (PBS with 1%
Tween-20) and were blocked with 10% goat serum for 30 min at 37°C.
Subsequently, the sections were treated with rabbit polyclonal
anti-PCNA (Bioss; 1:100) and rabbit polyclonal anti-Ki-67 (Bioss;
1:100) at 4°C overnight. According to the recommendation of the
antibody specification, positive tissue sections of PCNA (rat
liver) and Ki-67 (mouse placenta tissue) were used as control, with
PBS instead of primary antibody treated as a negative control.
After successfully completing the previous steps, the sections were
incubated with a secondary antibody for 20 min at 37°C and then
3,3′-diaminobenzidine (DAB) color reagent was added followed by
hematoxylin staining. The slides were dehydrated and then mounted
in neutral resins. Image acquisition was performed by microscope
and Image-Pro Plus 6.0 software was used to analyze the Integrated
Option Density (IOD) values of the brown area and then the IOD
values of each group were statistically analyzed.
H&E staining
The pretreatment of hematoxylin and eosin (H&E)
staining was basically the same as the immunohistochemical steps.
Sections (4 µm) were treated with hematoxylin reagent for 5 min
after deparaffinization and rehydration and then treated with 1%
acid-ethanol for 1 sec. Subsequently, the sections were stained by
eosin reagent for 3 min. The slides were dehydrated and mounted
then photographed by microscopy.
Statistical analysis
The data was collected and expressed as means ± SD.
SPSS.23.0 software was used for statistical analysis. All of the
experiment were repeated three times and the average value was
taken. The comparison of the means of two groups was analyzed by
Student's t-test. For all of the differences, P<0.05 was
considered to indicate a statistically significant difference.
Results
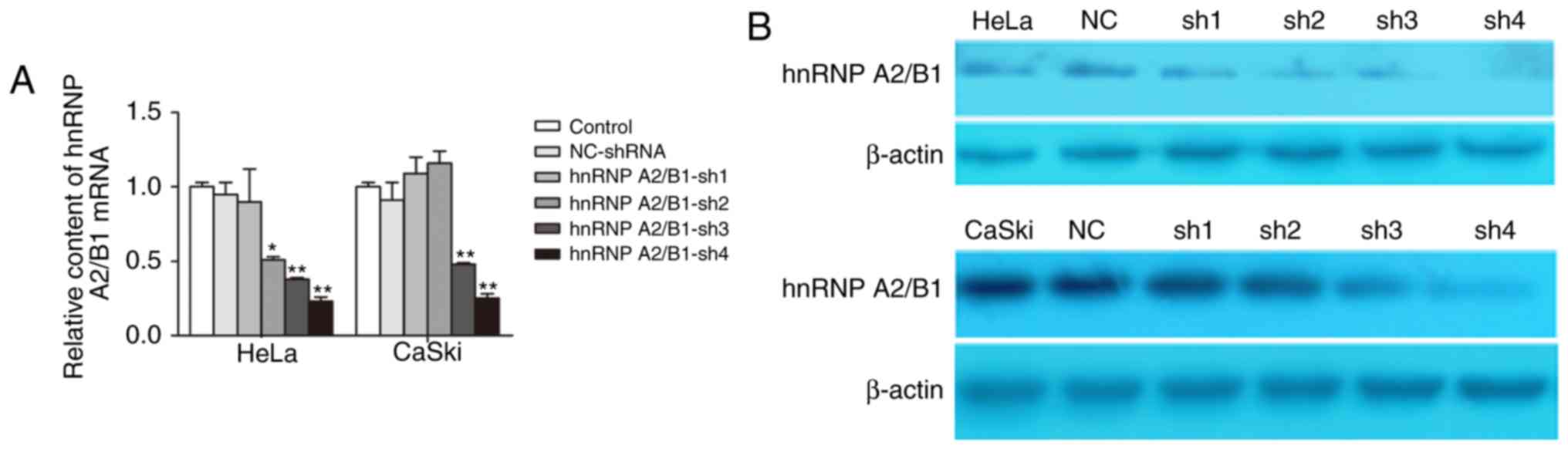
hnRNP A2/B1 is significantly
downregulated by lentivirus-mediated shRNA in HeLa and CaSki
cells
qRT-PCR and western blot were used to evaluate the
efficiency of hnRNP A2/B1 knockdown by lentivirus-mediated shRNA.
The results showed that the expression of hnRNP A2/B1 was highly
suppressed at both mRNA and protein levels. We designed 4 hnRNP
A2/B1-shRNA, the results of qRT-PCR indicted that the best
inhibition efficiency was HeLa-shRNA4 (76.58±3.55%) and
CaSki-shRNA4 (74.79±3.38%) (Fig.
2A). Western blot showed that protein expression of hnRNP A2/B1
in HeLa-shRNA4 (73.25±2.78%) and CaSki-shRNA4 (85.74±6.52%) was
markedly decreased compared to levels in other shRNA and NC-shRNA
group (Fig. 2B). These results
confirmed that the hnRNP A2/B1 was significantly knoced down in
HeLa and CaSki cells which were used for further experiments.
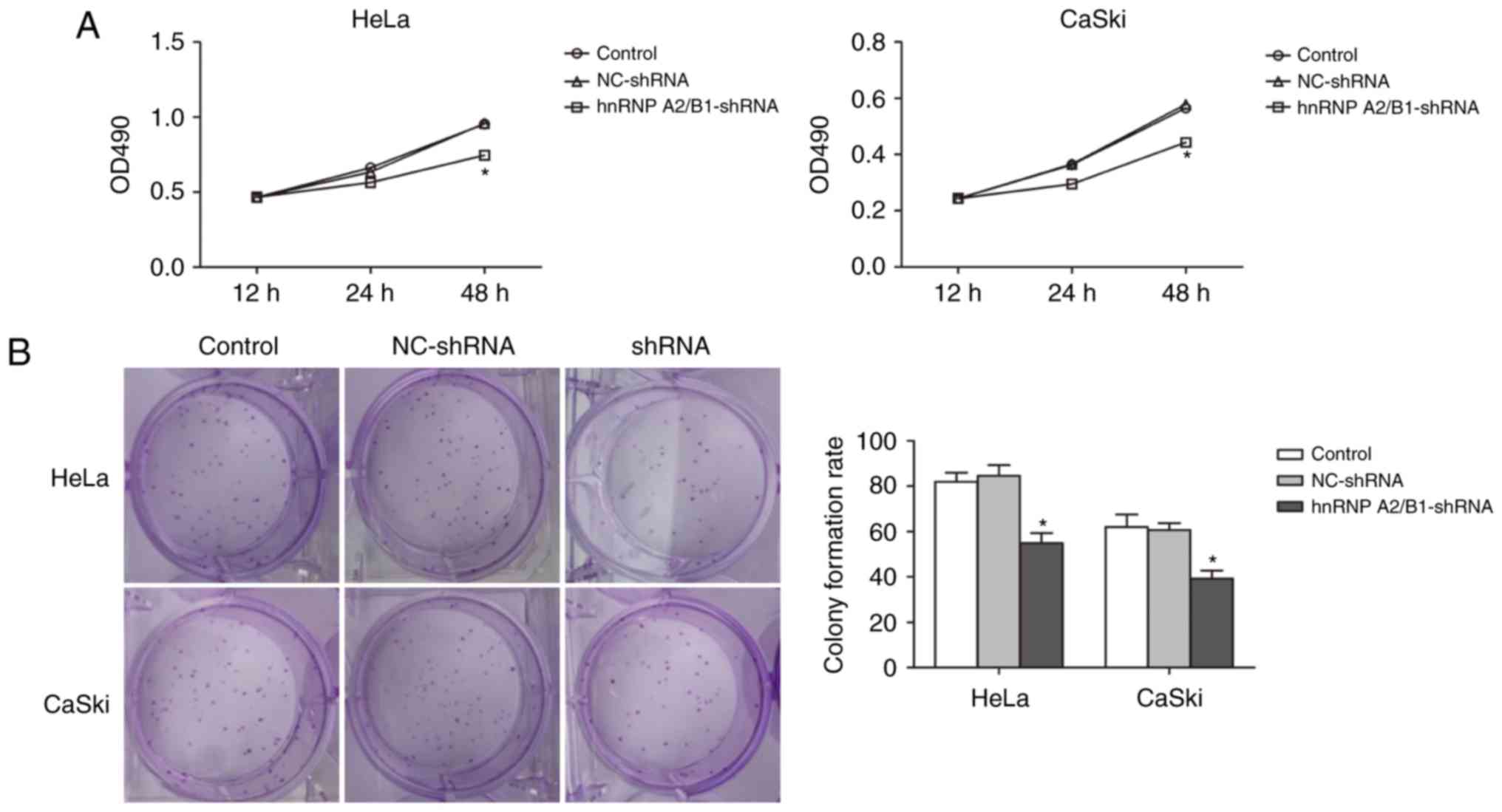
Inhibition of proliferation and colony
formation in cervical cancer cells via hnRNP A2/B1 knockdown
MTT assay was used to demonstrate the relationship
between suppression of hnRNP A2/B1 and cell proliferation (Fig. 3A). Absorbance value was measured in
HeLa and CaSki cells at 12, 24 and 48 h. The absorbance value of
hnRNP A2/B1-shRNA in HeLa and CaSki cells significantly decreased
compared with NC-shRNA and blank control group at 48 h suggesting
successful inhibition of cell proliferation in hnRNP A2/B1
knockdown cell lines.
In addition, colony formation efficiency was
decreased in both hnRNP A2/B1 shRNA-treated HeLa and CaSki cell
lines, as shown by the colony formation assay (Fig. 3B). An at least 30% drop in the
colony formation rate was observed in HeLa hnRNP A2/B1-shRNA cells
as compared to HeLa blank control or HeLa NC-shRNA cells
(55.00±4.35 vs. 82.00±4.00 or 84.67±4.61). The colony formation
rate was also decreased by more than 35% in CaSki hnRNP A2/B1-shRNA
cells as compared to CaSki blank control or CaSki NC-shRNA cells
(39.33±3.50 vs. 62.00±5.57 or 60.67±3.05). No significant
difference between NC-shRNA and blank control group was
observed.
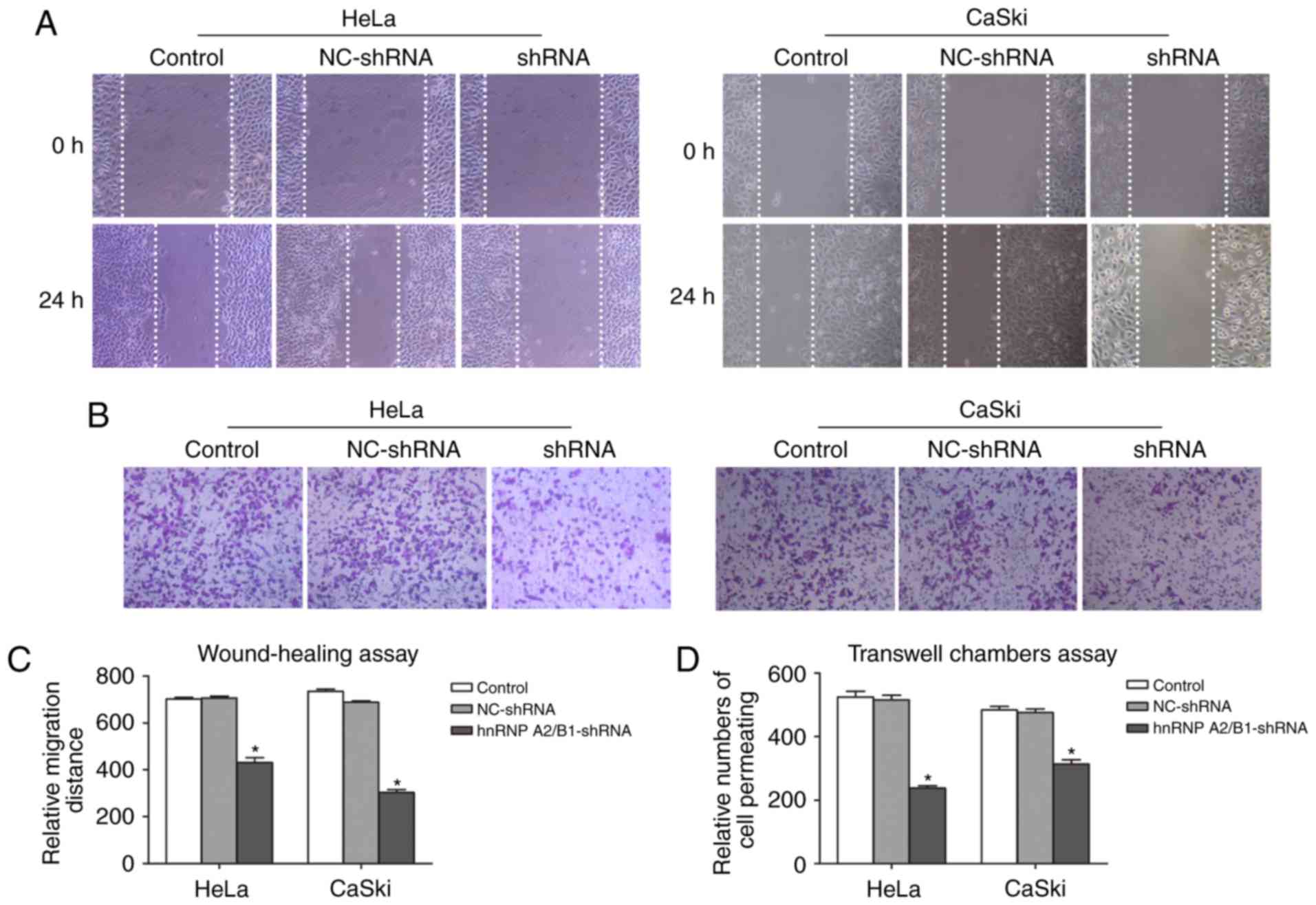
Inhibition of migration and invasion
in HeLa and CaSki cells after hnRNP A2/B1 knockdown
The migration defect mediated by knockdown hnRNP
A2/B1 in HeLa and CaSki cells were performed by wound-healing assay
(Fig. 4A and C). Cell motility
potential in hnRNP A2/B1-shRNA in HeLa and CaSki cells was
significantly decreased compared with NC-shRNA and blank control
group. For HeLa cells, the relative migration rate of hnRNA
A2/B1-shRNA cells was 431.33±20.03 as compared to 702.00±7.21 or
707.33±7.57 of blank control or NC-shRNA cells. For CaSki cells,
the relative migration rate was halved in hnRNA A2/B1 knockdown
cells as compared to blank control or NC-shRNA group (303.33±12.22
vs. 735.33±8.33 or 688.00±6.00).
Transwell chamber assay was used to exhibit invasion
ability (Fig. 4B and D). In both
cell lines, the number of cell permeating in hnRNA A2/B1 knockdown
cells dropped significantly from blank control or NC-shRNA cells
(237.67±7.51 vs. 524.67±17.5 or 515.33±15.01 in HeLa cells;
313.67±13.05 vs. 483.67±10.97 or 476.00±11.00 in CaSki cells) and
again we did not observe a significant difference between blank
control and NC-shRNA groups for either cell line.
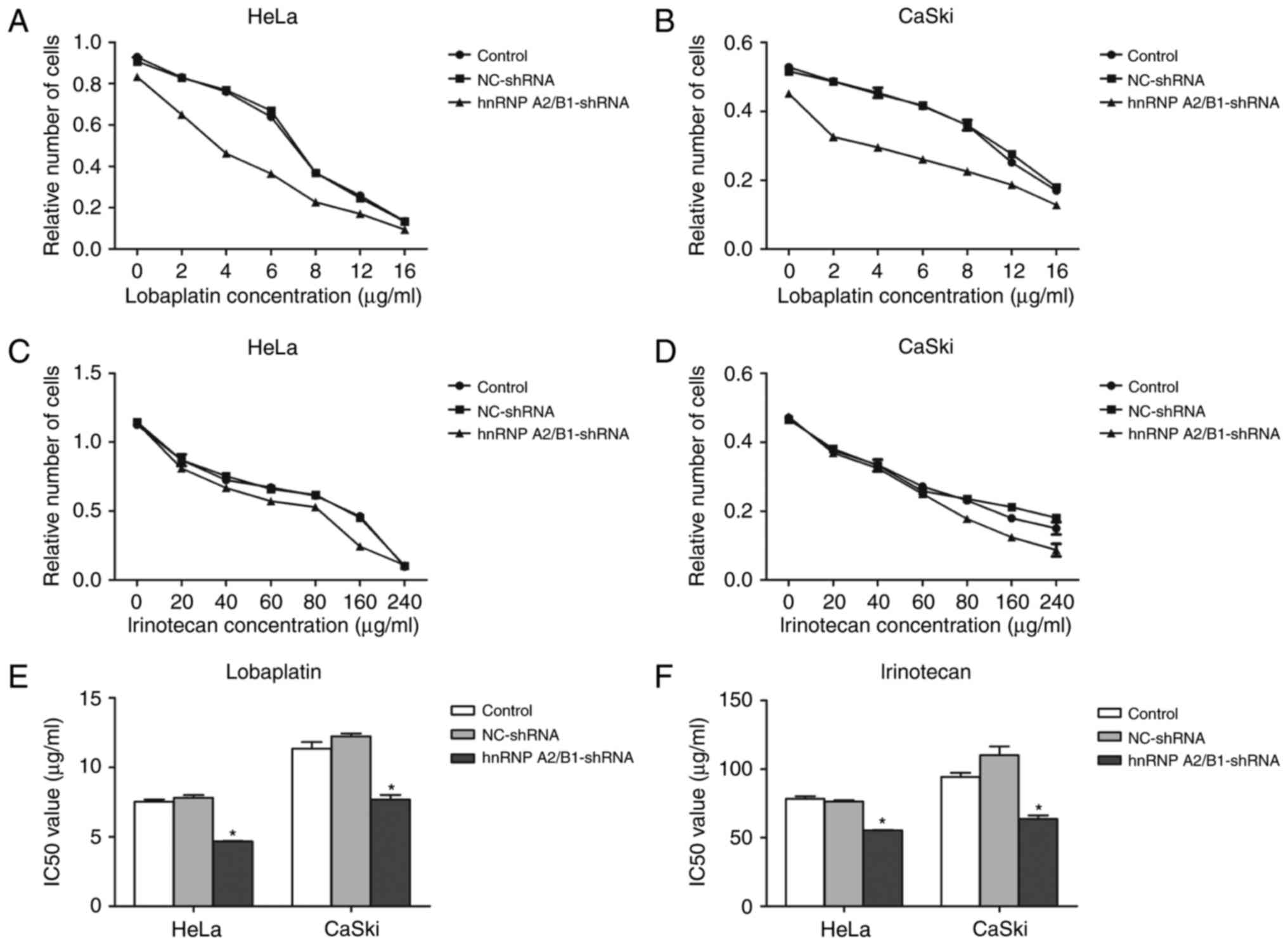
The enhancement of chemotherapy
sensitivity of lobaplatin or irinotecan by suppression of hnRNP
A2/B1 in HeLa and CaSki cells
The IC50 value of lobaplatin or
irinotecan in HeLa and CaSki cells was detected by MTT assay. The
IC50 of lobaplatin in HeLa blank control, HeLa NC-shRNA
and HeLa hnRNPA2/B1-shRNA was 7.526±0.17, 7.816±0.19, 4.669±0.03
µg/ml, respectively. The IC50 value of blank control
group, NC group, shRNA group was 11.340±0.49, 12.240±0.20 and
7.677±0.34 µg/ml in lobaplatin treated CaSki cells. The
IC50 of irinotecan in HeLa blank control group and HeLa
NC-shRNA group were 78.487±1.69 and 76.277±1.09 µg/ml, CaSki blank
control group and CaSki NC-shRNA group was 94.250±3.05 and
110.233±6.38 µg/ml, but the hnRNP A2/B1-shRNA group of HeLa and
CaSki cells were decreased significantly to 55.447±0.224 and
63.593±2.76 µg/ml. The IC50 value of lobaplatin and
irinotecan in HeLa and CaSki cells had no statistical significance
between NC-shRNA group and blank control group (Fig. 5A-F). The results indicate that
inhibition of hnRNP A2/B1 boosts the sensitivity of cervical cancer
lines towards lobaplatin and irinotecan.
PI3K/AKT signaling pathway plays an
important role in hnRNP A2/B1-regulated cell cycle and apoptosis in
cervical cancer cell lines
Flow cytometry was used to investigate the effect of
hnRNP A2/B1 knockdown on cell cycle distribution in HeLa and CaSki
cells. The proportion of G1 phase cells in hnRNP A2/B1 knockdown
HeLa and CaSki cells was significantly increased (Fig. 6A and Table III). No significant difference was
seen between NC-shRNA group and blank control group in HeLa or
CaSki cells.
 | Table III.The influence of hnRNPA2/B1 on the
cell cycle (n=3, x±s). |
Table III.
The influence of hnRNPA2/B1 on the
cell cycle (n=3, x±s).
| Group | G (%) | S (%) |
|---|
| CaSki |
57.56±0.75 |
42.52±0.22 |
| CaSki-shRNA |
60.57±0.14a |
38.35±0.36 |
| CaSki-NC |
57.36±0.55 |
42.53±0.16 |
| HeLa |
50.58±0.23 |
49.44±0.21 |
| HeLa-shRNA |
57.23±0.23a |
41.63±0.56 |
| HeLa-NC |
51.21±0.17 |
48.66±0.31 |
We also looked into the relationship between hnRNP
A2/B1 inhibition and cell apoptosis. In both HeLa and CaSki cell
lines, the apoptosis rate was increased after hnRNP knockdown as
compared to blank control or NC groups (25.53 vs. 11.83% or 14.01%
in HeLa cells; 46.20 vs. 12.40% or 11.97 in CaSki cells); (Fig. 6B). The apoptosis rate was similar in
the two control groups for both cell lines.
We further tested the change of PI3K/AKT pathway
related proteins by Western blot in hnRNP A2/B1 knockdown cervical
cancer cells (Fig. 6C). The hnRNP
A2/B1 knockdown group showed the upregulation of p21, p27 and
cleaved caspase-3 and downregulation of p-AKT (P<0.05). However,
there is no obvious change in the expression level of PI3K and AKT
in the knock-down cell lines.
Effects of hnRNPA2/B1 knockdown cells
treated with IGF-1 and LY294002, respectively, or not on the
activation or inhibition of PI3K/AKT pathway
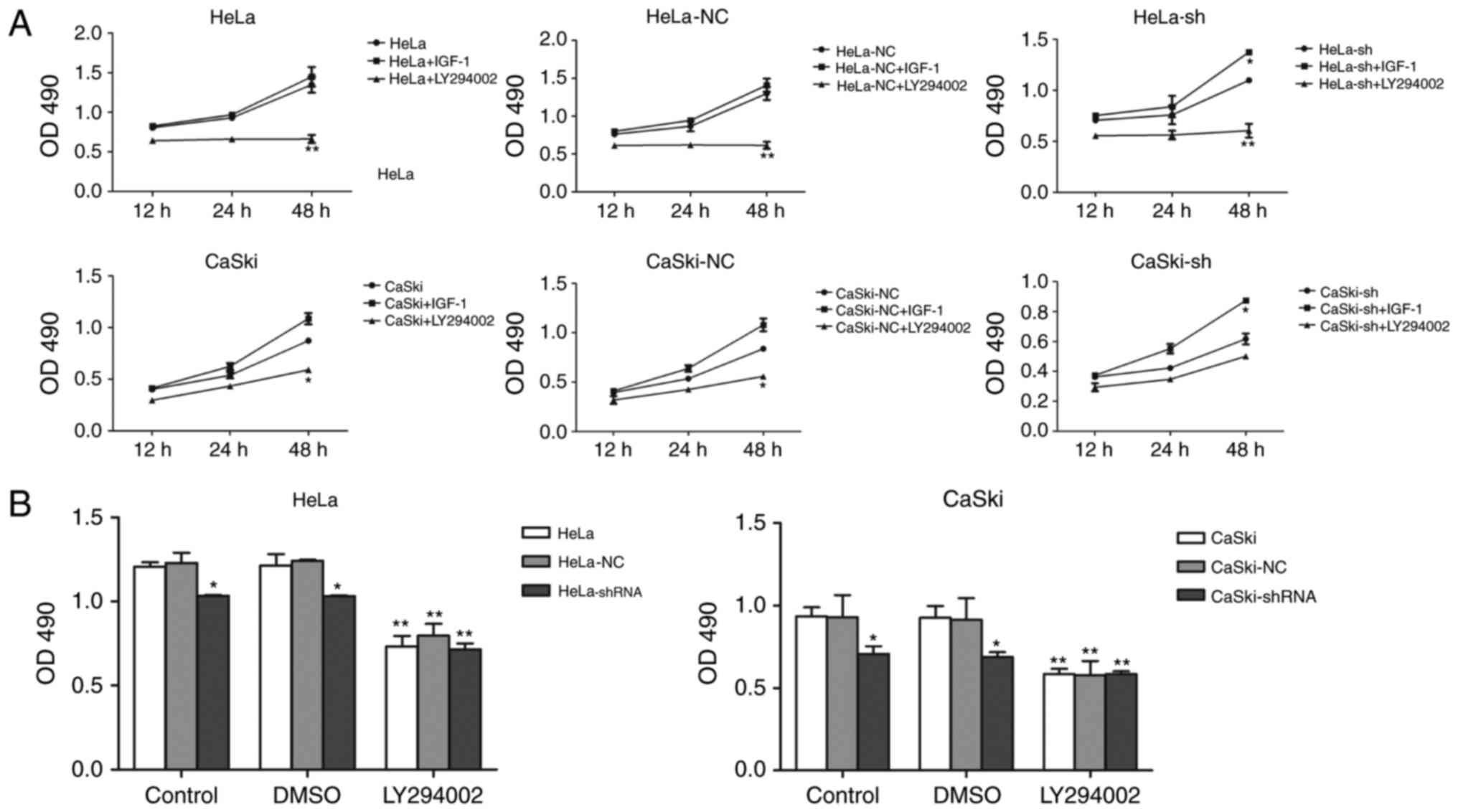
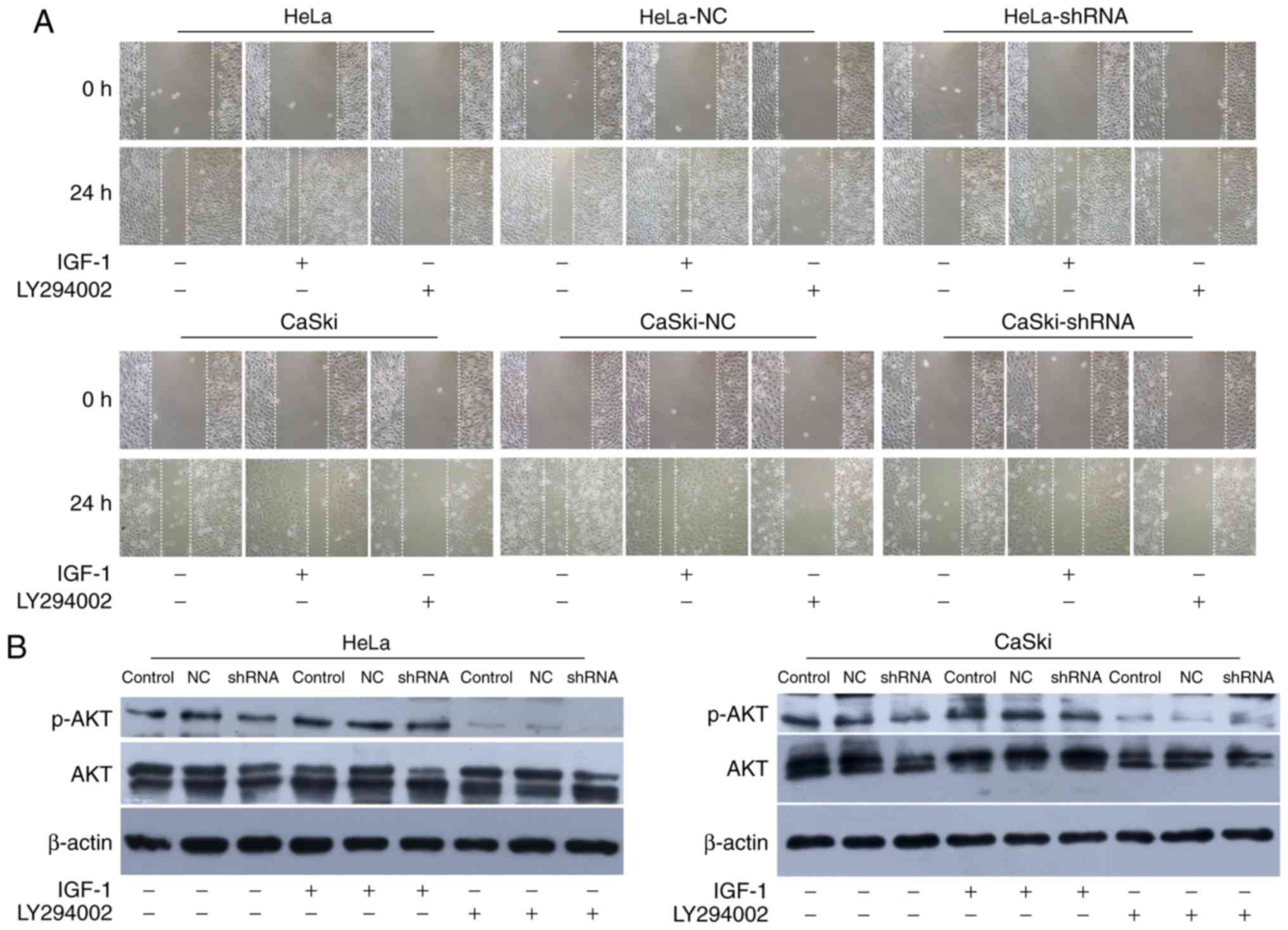
To further illustrate the relationship between hnRNP
A2/B1 and PI3K/AKT pathway, IGF-1 and LY294002 were used as agonist
and inhibitor of the PI3K/AKT pathway. MTT assay and wound healing
assay were used to investigate the proliferation and migration of
HeLa and CaSki cells after incubated with IGF-1 and LY294002
(Figs. 7A and 8A). The proliferation and migration
phenotype of hnRNP A2/B1 knockdown groups were rescued by IGF-1
treatment while worsened in LY294002 treated group. Validation of
activation or inhibition of PI3K/AKT pathway after exposed to IGF-1
or LY294002 was performed by western blotting (Fig. 8B). The expression of p-AKT was
significantly reduced after cultured in LY294002, while the
expression of p-AKT was increased both in HeLa and CaSki cells
after exposed in IGF-1 (P<0.01). The PI3K inhibitor LY294002 was
dissolved in DMSO solution, so the same amount of DMSO was added
into cells as the vehicle control group to avoid interference with
the experiment. The PI3K activator IGF-1 was reconstituted in
sterile 18M Omega-cm H2O according to instruction and so
no vehicle control group in this drug. The proliferation of HeLa
and CaSki cells at 48 h after cultured with DMSO was investigated
by MTT assay (Fig. 7B), the DMSO
group had no significant change compared to the non-medicated
group.
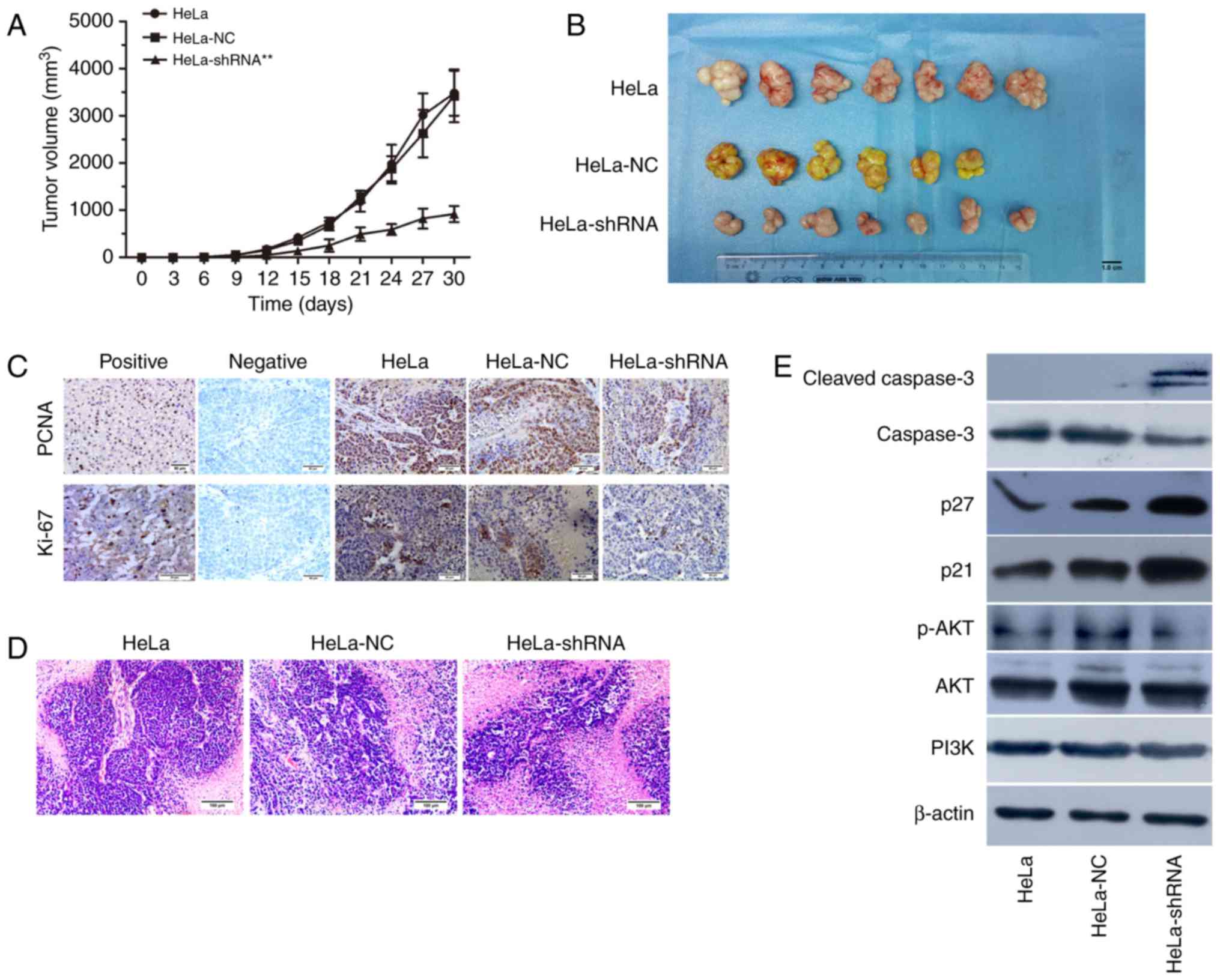
Effect of hnRNPA2/B1 silencing on nude
mouse xenograft in vivo
To determine the effect of hnRNP A2/B1 knockdown
in vivo, the transfected HeLa cells were injected into nude
mice to establish a tumor xenograft model. The nude mice were
randomly divied into 3 groups for injection with HeLa, HeLa-NC
shRNA and HeLa-hnRNP A2/B1 shRNA, respectively. Since a nude mouse
of NC group was dead before inoculating tumor cells, the survival
rate of nude mice injected with tumor cells was 100% and behavior
was normal throughout the experiment. The results showed that the
incidence of tumorigenesis in hnRNP A2/B1-shRNA transfected HeLa
cells injected group was significantly lower compared to the
control and NC-shRNA group (Fig. 9A and
B). To demonstrate whether the proliferation capacity was
consistent with the previous experiment results in vitro,
immunohistochemistry was used to confirm the expression of PCNA and
Ki-67 in vivo and the brown particles were labeled as
positive areas. In addition, H&E staining was used to observe
the morphological structure in tumor tissues. The results suggested
that the positive expression of PCNA (P<0.05) and Ki-67
(P<0.01) were significantly lower in hnRNP A2/B1 knockdown tumor
group compared to the other group (Fig.
9C and Table IV). As shown in
Fig. 9D, the characteristics of
xenograft tissues conformed to tumor cells and were as follows:
Acidophil hepatocytes with both nuclear and cytoplasmic
enlargement, nuclear pleomorphism and hyperchromasia, and frequent
multinucleation. In order to further demonstrate the relationship
between the PI3K/AKT signaling pathway and hnRNP A2/B1 in nude
mouse xenograft tissues, western blotting was used for
clarification. The xenograft tumor of hnRNP A2/B1-shRNA group could
suppress the expression of p-AKT protein, upregulating cleaved
caspase-3, p21 and p27 (Fig. 9E).
The results indicated that it was consistent with the earlier
apoptotic and cycle results in vitro from the protein level
of xenograft tumor tissues.
 | Table IV.The IOD values of PCNA and Ki-67
(x±s). |
Table IV.
The IOD values of PCNA and Ki-67
(x±s).
| Group | PCNA | Ki-67 |
|---|
| HeLa |
163256.50±38370.00 |
43485.35±26291.10 |
| HeLa-NC |
151597.80±33089.76 |
43102.09±12737.12 |
| HeLa-shRNA |
75461.77±22288.53a |
13669.97±4926.23b |
Discussion
hnRNP A2/B1 is a set of primer-mRNA binding proteins
involved in cell transcription and protein translation. Previous
studies suggested that uncontrolled expression of hnRNP A2/B1 is
one of the reasons for promoting tumor formation and thus highly
expressed in a variety of cancers (9,22–26).
Some recent studies suggested that hnRNP A2/B1 is a proto-oncogene,
especially in non-small cell lung cancer, the expression of hnRNP
A2/B1 may be used as the reference index for evaluating the status
and prognostic indicator of disease (27). The functional role of hnRNP A2/B1 in
cervical cancer is rarely reported. Following previous reports, we
used cervical cancer cell lines HeLa and CaSki cells with hnRNP
A2/B1 knockdown by shRNA as a model to study the role of hnRNP
A2/B1 in cervical cancer.
hnRNPA2/B1, as a new focus of cancer-associated
tumor antigen has gradually attracted scientists' attention. A
study by Sinha and colleague showed that hnRNP A2/B1 may be
combined with the telomere repeated sequence TTAGGG to protect the
telomere from destruction by ribozyme (28). To investigate how to interrupt these
factors, receptor and oncogene signaling pathway to inhibit cancer
cell proliferation, invasion and migration has become one of the
main strategies for the development of new anticancer drugs. As
before, hnRNP A2/B1 knockdown in this study suggested that the
proliferation of cervical cancer cell lines was markedly decreased
compared to the control group. The proliferation-related antigen
PCNA and Ki-67 were also significantly reduced after hnRNP A2/B1
knockdown in vivo. According to previous studies,
proliferation-related antigen Ki-67 and proliferating cell nuclear
antigen (PCNA) are proteins that are present in the cell
proliferative phase and are one of the markers of proliferating
cells (29). Similarly, our study
also showed that hnRNP A2/B1 knockdown could inhibit cell colony
formation. Upregulated proliferation of cancer cells is one of the
mechanism of tumor growth and is the basis of the occurrence and
development of cancer cells (30,31).
Previous data reported that hnRNP A2/B1 plays an
important role in the regulation of the migration, invasion and
drug resistance in partial cancer cells. In addition, the process
of therapy resistance during the development of pancreatic cancer
is related to the high expession of hnRNPA2/B1 (32–35).
This study showed that the inhibition of hnRNP A2/B1 in cervical
cancer cell lines could decrease the ability of migration and
invasion. After chemotherapy with lobaplatin and irinotecan
respectively, the IC50 value was significantly reduced
in hnRNP A2/B1 knockdown group, these results also confirmed
previous research conclusions. This suggests that hnRNP A2/B1 in
cervical cancer is associated with drug sensitivity and may be one
of the mechanism in enhancement of therapy sensitivity by hnRNP
A2/B1 knockdown.
Silencing hnRNP A2/B1 resulted in G1/S cell cycle
arrest and accumulation of G0/G1 phase cells (36). The restriction point of cell cycle
at G1/S transition is particularly important and determines the
conversion of cell cycle time, the number of S phase and G2/M phase
cell proportion can reflect the state of cell proliferation,
suggesting active cell growth. The upregulation of checkpoint in
cell cycle is closely related to the occurrence of tumors which
induce cell apoptosis (37), and
another study also suggested that hnRNP A2/B1 can regulate the
expression of p14 and p16 and activate cyclin-dependent kinase 4 to
assure the transition between G1 and S phase (38). The levels of S phase and G2/M phase
were decreased in hnRNP A2/B1 knockdown cervical cancer cells which
demonstrated that silencing hnRNP A2/B1 could block cervical cancer
cell cycle in G1 phase to prevent cell proliferation. Moreover, we
indicated that hnRNP A2/B1 knockdown can induce cell apoptosis. p21
and p27, as the inhibitor of cyclin-dependent kinases (CDKs), plays
an important part in regulation of cell cycle (39,40).
Just as our results, the expression of p21 and p27 were increased
in vitro and vivo at hnRNP A2/B1 downregulation group
and the result suggested that the hnRNP A2/B1 affected cell cycle
by regulated p21 and p27 in cervical cancer. Previous studies
showed that hnRNP A2/B1 can upregulate the proportion of
anti-apoptosis factors and proteins in cells to promote the
malignant growth of tumors (41),
our study also confirmed this argument. Caspase-3 may be involved
in cell apoptosis (42), our
results indicated that silencing hnRNP A2/B1 enhanced apoptosis in
cervical cancer via activation of caspase-3.
Aberrant activation of the PI3K/AKT pathway is
widespread in malignant tumors and is an important pathway to
mediate cell cycle, and apoptosis (43,44).
Licochalcone A induced autophagy by inactivation of PI3K/AKT/mTOR
pathway in cervical cancer cells (45). Activation of the PI3K/AKT pathway
could reflect phosphorylation levels of AKT proteins and after
phosphorylation, it could be further activated a variety of
downstream proteins, such as p21, p27 and caspase-3, which could
regulate the state of tumor cells. Our results demonstrated that
the expression of p-AKT was reduced in hnRNP A2/B1 knockdown group
both in vitro and in vivo and hnRNP A2/B1 was related
to PI3K/AKT pathway in promotion of cervical cancer. Previous
studies have reported that hnRNP A2/B1 regulates the self-renewal,
cell cycle and pluripotency in human embryonic stem cells is
related to PI3K/AKT pathway (46)
and this was similar to our results.
In conclusion, our findings demonstrate that
inhibiting hnRNP A2/B1 expression in cervical cancer can induce
apoptosis and cell cycle arrest and enhance the chemotherapy
sensitivity of cervical cancer cells to lobaplatin and irinotecan.
Analysis of cervical cancer cell lines HeLa and CaSki cells in
vitro shows that hnRNP A2/B1 knockdown can reduce the ability
of cell proliferation, invation and migration, indicating that
hnRNP A2/B1 may be one of the central regulators for cervical
cancer. The activation of PI3K/AKT pathway is one of the important
mechanisms for hnRNP A2/B1 to facilitate the development of
cervical cancer. Therefore, our study suggests that hnRNP A2/B1 may
be an important molecular target for cancer treatment of cervical
cancer and provide a new direction for clinical treatment of
cervical cancer.
Acknowledgements
This study was supported by National Natural Science
Foundation of China (2015–81560481) and The Joint Funds of Science
and Technology Department of Guizhou Province and Affiliated
Hospital of Guizhou Medical University (2015–7410).
Glossary
Abbreviations
Abbreviations:
|
hnRNP A2/B1
|
heterogeneous nuclear
ribonucleoprotein A2/B1
|
|
qRT-PCR
|
quantitative reverse transcription
polymerase chain reaction
|
|
shRNA
|
short hairpin RNA
|
|
RPMI-1640
|
Roswell Park Memorial
Institute-1640
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
|
|
PBS
|
Phosphate buffere saline
|
|
IC50
|
half maximal inhibitory
concentration
|
|
DMSO
|
dimethylsulphoxide
|
|
SDS
|
Sodium dodecyl sulfate
|
|
PI3K/AKT
|
phophatidylinositol 3-kinase/protein
kinase-B
|
|
CDK
|
cyclin dependent kinase
|
|
IGF-1
|
insulin-like growth factor 1
|
|
PCNA
|
proliferating cell nuclear antigen
|
|
H&E
|
hematoxylin and eosin
|
|
DAB
|
3,3′-diaminobenzidine
|
|
IOD
|
integrated option density
|
References
|
1
|
Yoysungnoen B, Bhattarakosol P, Changtam C
and Patumraj S: Effects of tetrahydrocurcumin on tumor growth and
cellular signaling in cervical cancer xenografts in nude mice.
Biomed Res Int. 2016:17812082016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin WC, Kuo KL, Shi CS, Wu JT, Hsieh JT,
Chang HC, Liao SM, Chou CT, Chiang CK, Chiu WS, et al: MLN4924, a
Novel NEDD8-activating enzyme inhibitor, exhibits antitumor
activity and enhances cisplatin-induced cytotoxicity in human
cervical carcinoma: In vitro and in vivo study. Am J Cancer Res.
5:3350–3362. 2015.PubMed/NCBI
|
|
4
|
Koh WJ, Greer BE, Abu-Rustum NR, Apte SM,
Campos SM, Chan J, Cho KR, Cohn D, Crispens MA, DuPont N, et al:
Cervical cancer. J Natl Compr Canc Netw. 11:320–343. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma X, Zhang J, Liu S, Huang Y, Chen B and
Wang D: Nrf2 knockdown by shRNA inhibits tumor growth and increases
efficacy of chemotherapy in cervical cancer. Cancer Chemother
Pharmacol. 69:485–494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dreyfuss G, Kim VN and Kataoka N:
Messenger-RNA-binding proteins and the messages they carry. Nat Rev
Mol Cell Biol. 3:195–205. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kozu T, Henrich B and Schäfer KP:
Structure and expression of the gene (HNRPA2B1) encoding the human
hnRNP protein A2/B1. Genomics. 25:365–371. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He Y, Brown MA, Rothnagel JA, Saunders NA
and Smith R: Roles of heterogeneous nuclear ribonucleoproteins A
and B in cell proliferation. J Cell Sci. 118:3173–3183. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Patry C, Bouchard L, Labrecque P, Gendron
D, Lemieux B, Toutant J, Lapointe E, Wellinger R and Chabot B:
Small interfering RNA-mediated reduction in heterogeneous nuclear
ribonucleoparticule A1/A2 proteins induces apoptosis in human
cancer cells but not in normal mortal cell lines. Cancer Res.
63:7679–7688. 2003.PubMed/NCBI
|
|
10
|
Izaurralde E, Jarmolowski A, Beisel C,
Mattaj IW, Dreyfuss G and Fischer U: A role for the M9 transport
signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 137:27–35.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He Y and Smith R: Nuclear functions of
heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci.
66:1239–1256. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kamma H, Horiguchi H, Wan L, Matsui M,
Fujiwara M, Fujimoto M, Yazawa T and Dreyfuss G: Molecular
characterization of the hnRNPA2/B1 proteins: Tissue-specific
expression and novel isoforms. Exp Cell Res. 246:399–411. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qu XH, Liu JL, Zhong XW, Li XI and Zhang
QG: Insights into the roles of hnRNP A2/B1 and AXL in non-small
cell lung cancer. Oncol Lett. 10:1677–1685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Joshi J, Fernandez-Marcos PJ, Galvez A,
Amanchy R, Linares JF, Duran A, Pathrose P, Leitges M, Cañamero M,
Collado M, et al: Par-4 inhibits Akt and suppresses Ras-induced
lung tumorigenesis. EMBO J. 27:2181–2193. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang M, Fang X, Liu H, Guo R, Wu X, Li B,
Zhu F, Ling Y, Griffith BN, Wang S and Yang D: Bioinformatics-based
discovery and characterization of an AKT-selective inhibitor
9-chloro-2-methylellipticinium acetate (CMEP) in breast cancer
cells. Cancer Lett. 252:244–258. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao N, Flynn DC, Zhang Z, Zhong XS, Walker
V, Liu KJ, Shi X and Jiang BH: G1 cell cycle progression and the
expression of G1 cyclins are regulated by PI3K/AKT/mTOR/P70S6KI
signaling in human ovarian cancer cells. Am J Physiol Cell Physiol.
287:C281–C291. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Polivka J Jr and Janku F: Molecular
targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol
Ther. 142:164–175. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Ran L, Fang W and Wang D: Lobaplatin
arrests cell cycle progression, induces apoptosis and alters the
proteome in human cervical cancer cell line CaSki. Biomed
Pharmacother. 68:291–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jang HJ, Hong EM and Lee J, Choi JE, Park
SW, Byun HW, Koh DH, Choi MH, Kae SH and Lee J: Synergistic effects
of simvastatin and Irinotecan against colon cancer cells with or
without Irinotecan resistance. Gastroenterol Res Pract.
2016:78913742016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kodawara T, Higashi T, Negoro Y, Kamitani
Y, Igarashi T, Watanabe K, Tsukamoto H, Yano R, Masada M, Iwasaki H
and Nakamura T: The inhibitory effect of Ciprofloxacin on the
β-Glucuronidase-mediated deconjugation of the Irinotecan metabolite
SN-38-G. Basic Clin Pharmacol Toxicol. 118:333–337. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ren C, Ren T, Yang K, Wang S, Bao X, Zhang
F and Guo W: Inhibition of SOX2 induces cell apoptosis and G1/S
arrest in Ewing's sarcoma through the PI3K/Akt pathway. J Exp Clin
Cancer Res. 35:442016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Golan-Gerstl R, Cohen M, Shilo A, Suh SS,
Bakàcs A, Coppola L and Karni R: Splicing factor hnRNPA2/B1
regulates tumor suppressor gene splicing and is an oncogenic driver
in glioblastoma. Cancer Res. 7:4464–4472. 2011. View Article : Google Scholar
|
|
23
|
Shilo A, Ben Hur V, Denichenko P, Stein I,
Pikarsky E, Rauch J, Kolch W, Zender L and Karni R: Splicing factor
hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf
splicing in hepatocellular carcinoma development. RNA. 20:505–515.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
David CJ, Chen M, Assanah M, Canoll P and
Manley JL: hnRNP proteins controlled by c-Myc deregulate pyruvate
kinase mRNA splicing in cancer. Nature. 463:364–368. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao CH, Li QF, Chen LY, Tang J, Song JY
and Xie Z: Expression and localization of hnRNP A2/B1 during
differentiation of human osteosarcoma MG-63 cells induced by HMBA.
Ai Zheng. 27:677–684. 2008.(In Chinese). PubMed/NCBI
|
|
26
|
Katsimpoula S, Patrinou-Georgoula M,
Makrilia N, Dimakou K, Guialis A, Orfanidou D and Syrigos KN:
Overexpression of hnRNPA2/B1 in bronchoscopic specimens: A
potential early detection marker in lung cancer. Anticancer Res.
29:1373–1382. 2009.PubMed/NCBI
|
|
27
|
Etcheverry GJ: 2006 Nobel prize in
physiology or medicine. The silence of genes. Medicina (B Aires).
67:92–95. 2007.PubMed/NCBI
|
|
28
|
Sinha P, Poland J, Kohl S, Schnölzer M,
Helmbach H, Hütter G, Lage H and Schadendorf D: Study of the
development of chemoresistance in melanoma cell line using proteome
analysis. Electrophoresis. 24:2386–2404. 2013. View Article : Google Scholar
|
|
29
|
Bologna-Molina R, Mosqueda-Taylor A,
Molina-Frechero N, Mori-Estevez AD and Sánchez-Acuña G: Comparison
of the value of PCNA and Ki67 as markers of cell proliferation in
ameloblastic tumors. Med Oral Patol Oral Cir Bucal. 18:e174–e179.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cotrim P, Martelli-Junior H, Graner E,
Sauk JJ and Coletta RD: Cyclosporin A induces proliferation in
human gingival fibroblasts via induction of transforming growth
factor-beta1. Periodontol. 74:1625–1633. 2003. View Article : Google Scholar
|
|
31
|
Stanley G, Harvey K, Slivova V, Jiang J
and Sliva D: Ganoderma lucidum suppresses angiogenesis through the
inhibition of secretion of VEGF and TGF-beta1 from prostate cancer
cells. Biochem Biophys Res Commun. 330:46–52. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Clower CV, Chatterjee D, Wang Z, Cantley
LC, Vander Heiden MG and Krainer AR: The alternative splicing
repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform
expression and cell metabolism. Proc Nati Acad Sci USA. 107:pp.
1894–1899. 2010; View Article : Google Scholar
|
|
33
|
Gu WJ and Liu HL: Induction of pancreatic
cancer cell apoptosis, invasion, migration, and enhancement of
chemotherapy sensitivity of gemcitabine, 5-FU, and oxaliplatin by
hnRNP A2/B1 siRNA. Anticancer Drugs. 24:566–576. 2013.PubMed/NCBI
|
|
34
|
Tauler J, Zudaire E, Liu H, Shih J and
Mulshine JL: hnRNP A2/B1 modulates epithelial-mesenchymal
transition in lung cancer cell lines. Cancer Res. 70:7137–7147.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Liu HL, Li Y and Yuan P: Proteomic
analysis of pancreatic intraepithelial neoplasia and pancreatic
carcinoma in rat models. World J Gastroenterol. 17:1434–1441. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hallett RM, Huang C, Motazedian A, Auf der
Mauer S, Pond GR, Hassell JA, Nordon RE and Draper JS:
Treatment-induced cell cycle kinetics dictate tumor response to
chemotherapy. Oncotarget. 6:7040–7052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Montague JW and Cidlowski JA: Cellular
catabolism in apoptosis: DNA degradation and end nuclease
activation. Experientia. 52:857–862. 1996. View Article : Google Scholar
|
|
38
|
Zhu D, Xu G, Ghandhi S and Hubbard K:
Modulation of the expression of p16INK4a and p14AKT by hnRNPA1 and
A2 RNA binding proteins: Implications for cellular senescence. J
Cell Physiol. 193:19–25. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chu I, Sun J, Arnaout A, Kahn H, Hanna W,
Narod S, Sun P, Tan CK, Hengst L and Slingerland J: p27
phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell.
128:281–294. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gartel AL and Radhakrishnan SK: Lost in
transcription: p21 repression, mechanisms and consequences. Cancer
Res. 65:3980–3985. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen ZY, Cai L, Zhu J, Chen M, Chen J, Li
ZH, Liu XD, Wang SG, Bie P, Jiang P, et al: Fyn requires hnRNPA2/B1
and Sam68 to synergistically regulate apoptosis in pancreatic
cancer. Carcinogenesis. 32:1419–1426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zimmermann KC, Bonzon C and Green DR: The
machinery of programmed cell death. Pharmacol Ther. 92:57–70. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Stegeman H, Span PN, Kaanders JH and
Bussink J: Improving chemoradiation efficacy by PI3-K/AKT
inhibition. Cancer Treat Rev. 40:1182–1191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Manning BD and Cantley LC: AKT/PKB
Signaling: Navigating downstream. Cell. 129:1261–1274. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tsai JP, Lee CH, Ying TH, Lin CL, Hsueh JT
and Hsieh YH: Licochalcone A induces autophagy through
PI3K/Akt/mTOR inactivation and autophagy suppression enhances
Licochalcone A-induced apoptosis of human cervical cancer cells.
Oncotarget. 6:28851–28866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Choi HS, Lee HM, Jang YJ, Kim CH and Ryu
CJ: Heterogeneous nuclear ribonucleoprotein A2/B1 regulates the
self-renewal and pluripotency of human embryonic stem cells via the
control of the G1/S transition. Stem Cells. 31:2647–2658. 2013.
View Article : Google Scholar : PubMed/NCBI
|