Introduction
Colorectal cancer (CRC), one of the most common
malignancies of the gastrointestinal tract, is a major cause of
tumor-associated morbidity and mortality worldwide (1). Its incidence rate continues to rise
(2). Over 1 million new cases of
CRC are diagnosed globally each year, resulting in about 0.5
million deaths annually (3,4). The poor prognosis for CRC is mainly
attributable to its insidious onset, atypical symptoms, and
aggressive malignancy. Most patients with CRC are diagnosed at an
advanced stage. Approximately 25% of patients with CRC present with
liver metastases at the time of the initial diagnosis (5). Despite overall advances in the
treatment of this disease, the overall cure rate of CRC has not
markedly improved, and the overall 5-year survival rate remains at
approximately 60% in Asia (6).
Nuclear factor κB (NF-κB) is a family of dimeric
transcription factors required to coordinate numerous physiological
and pathological processes, such as immunity, inflammation, and
tumorigenesis (7,8). Notably, constitutively activated NF-κB
signaling has been demonstrated to play vital roles in the
development and progression of a large array of malignancies,
including CRC (9,10). Stimulatory factors, such as tumor
necrosis factor-α (TNF-α), interleukin-1β (IL-1β), or
pathogen-derived components that include bacterial
lipopolysaccharides (LPS), bind to their respective receptors,
leading to the rapid recruitment of tumor necrosis factor receptor
type 1-associated DEATH domain (TRADD), cellular inhibitor of
apoptosis protein 1 (cIAP1), baculoviral IAP repeat-containing
protein 3 (cIAP2), and TNF receptor-associated factors (TRAFs),
including TRAF1, TRAF2, TRAF3, TRAF5, and TRAF6. As molecular
activators for the NF-κB signaling pathway, TRAFs function as E3
ubiquitin ligases that induce the K63 polyubiquitination of
receptor-interacting protein 1 (RIP1), resulting in activation of
the transforming growth factor-activated kinase-1 (TAK1)/TAB2/3/
TGF-β activated kinase 1 (MAP3K7) binding protein 1 (TAB1) complex.
During this process, TAB1, TAB2, and TAB3 form a complex with TAK1,
which phosphorylates and activates the inhibitor of the NF-κB
kinase (IKK)-α/β/γ kinase complex, leading to nuclear translocation
and activation of NF-κB (11–13).
Thus, further investigations into the mechanism of regulation of
the NF-κB pathway key components, such as TRAF2, TRAF5, TAK1, TAB1,
and TAB3, would increase our knowledge of the mechanisms
underpinning the constitutive activation of NF-κB in cancer.
MicroRNAs (miRNAs), a class of small non-coding
RNAs, function as negative regulators of gene expression by
interacting with the 3′ untranslated region (3′-UTR) of their
target mRNAs (14). These miRNAs,
which are approximately 20–25 nucleotides in length, play important
roles in a variety of physiological and pathological processes,
such as development, cell proliferation, differentiation, and
senescence (15). Previous research
has revealed that miRNAs are involved in carcinogenesis via
regulation of several key cellular processes, including cell
proliferation, apoptosis, migration, invasion, and angiogenesis
(16). Numerous miRNAs have been
reported to be upregulated or downregulated in various types of
cancer, demonstrating their potential roles as oncogenes or tumor
suppressors (17–20). The altered expression of miRNAs in
cancers suggests that they may serve as potential diagnostic or
prognostic biomarkers for cancer (21–23).
For instance, in glioma, ovarian cancer, and breast cancer, miR-873
is downregulated and may act as tumor suppressor (24–28).
However, miR-873 has been revealed to be upregulated in lung
adenocarcinoma, where it promotes tumor cell proliferation and
migration (29).
In the present study we revealed that miR-873 was
downregulated in CRC and this was correlated with a poor prognosis
for patients with CRC. Furthermore, we determined that
downregulation of miR-873 enhanced CRC cell proliferation by
directly targeting TRAF5 and TAB1, leading to activation of NF-κB
signaling. These results demonstrated that miR-873 served as a
tumor-suppressive miRNA in the development and progression of
CRC.
Materials and methods
Cells
The CRC cell lines (SW620, SW480, DLD1, HCT116,
LoVo, and HT-29) were purchased from the American Type Culture
Collection (Manassas, VA, USA). The normal human colon mucosal
epithelial cell line (NCM460) and the CRC cell line (KM12) were
purchased from the BeNa Culture Collection (Beijing, China). The
normal human colon mucosal epithelial cell line (NCM460) and seven
CRC cell lines (SW620, SW480, DLD1, HCT116, KM12, LoVo, and HT-29)
were grown in Dulbecco's modified Eagle's medium (Invitrogen;
Thermo Fisher Scientific, Carlsbad, CA, USA) supplemented with 10%
fetal bovine serum (Invitrogen; Thermo Fisher Scientific) and 100
units of penicillin-streptomycin.
Patient information and tissue
specimens
This study was conducted on a total of 125
paraffin-embedded and archived CRC samples, which were diagnosed
histopathologically at the Affiliated Shenzhen Sixth Hospital of
Guangdong Medical University from 2003 to 2012. Informed patient
consent and approval from the Institutional Research Ethics
Committee of the Affiliated Shenzhen Sixth Hospital of Guangdong
Medical University was obtained for use of these clinical materials
for research purposes. Clinical information regarding the samples
is summarized in Table I. Ten CRC
tissue samples and their matched adjacent non-cancerous colorectal
tissues were frozen and stored in liquid nitrogen until further
use.
 | Table I.Clinicopathological characteristics
of studied patients and expression of mir-873 in CRC. |
Table I.
Clinicopathological characteristics
of studied patients and expression of mir-873 in CRC.
| Factors | No. | (%) |
|---|
| Sex |
|
|
|
Male | 68 | 54.4 |
|
Female | 57 | 45.6 |
| Age (years) |
|
|
|
≤62 | 64 | 51.2 |
|
>62 | 61 | 48.8 |
| Tumor site |
|
|
|
Colon | 61 | 48.8 |
|
Rectal | 64 | 51.2 |
| Dukes' stage |
|
|
| A | 17 | 13.6 |
| B | 36 | 28.8 |
| C | 45 | 36.0 |
| D | 27 | 21.6 |
| Clinical stage |
|
|
| I | 17 | 13.6 |
| II | 37 | 29.6 |
|
III | 44 | 35.2 |
| IV | 27 | 21.6 |
| T
classification |
|
|
|
T1 | 2 |
1.6 |
|
T2 | 30 | 24.0 |
|
T3 | 42 | 33.6 |
|
T4 | 51 | 40.8 |
| N
classification |
|
|
|
N0 | 63 | 50.4 |
|
N1 | 39 | 31.2 |
|
N2 | 23 | 18.4 |
| M
classification |
|
|
|
M0 | 97 | 77.6 |
|
M1 | 28 | 22.4 |
| Histological
differentiation |
|
|
|
Well | 29 | 23.2 |
|
Moderate | 65 | 52.0 |
|
Poor | 31 | 24.8 |
| Vital status |
|
|
|
Alive | 49 | 39.2 |
|
Dead | 76 | 60.8 |
| Expression of
mir-873 |
|
|
|
Low | 62 | 49.6 |
|
High | 63 | 50.4 |
Plasmids and transfection
The 3′-UTR regions of human TRAF5 (from 1801
to 2211 nt, containing a predicted conserved miR-873 binding site)
and TAB1 (from 3288 to 3700 nt, containing a predicted
conserved miR-873 binding site) were generated by PCR and cloned
into the modified pGL3-control luciferase reporter plasmid (Promega
Corporation, Madison, WI, USA). Primer sequences were as follows:
TRAF5-3′UTR sense, 5′-ACTCCGCGGATCCCAGATGATTAAATT-3′ and antisense,
5′-CTAACTGCAGTTCCTTGTTCTGGGATCAC-3′; TAB1-3′UTR sense,
5′-ACTCCGCGGCGGAGGTCCTGGCCCTCAG-3′ and antisense,
5′-CTAACTGCAGCCCATGGAGGAAACAACAGGGAG-3′. Point mutations in the
putative miR-873-binding seed regions of the TRAF5-3′-UTR and
TAB1-3′-UTR constructs were created using a Stratagene QuikChange
Mutagenesis kit (Stratagene; Agilent Technologies Inc., La Jolla,
CA, USA). A miR-873 mimic, miR-873 inhibitor, and the negative
control (NC) were purchased from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The sequences were as follows: miR-873 mimic,
5′ GCAGGAACUUGUGAGUCU CCU-3′; miR-873-NC sense,
5′-UUUGUACUACACAAAAGUACUG-3′; miR-873 inhibitor,
5′-AGGAGACUCACAAGUUCCUGC-3′. The miR-873 mimic or miR-873 inhibitor
(the miR-873 inhibitor is a locked nucleic acid
(LNA)/O-Methyl oligo (OMe) modified antisense
oligonucleotide designed specifically to bind to and inhibit the
endogenous miR-873 molecule) was transfected into cells using the
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific)
according to the manufacturer's instructions.
RNA extraction, reverse transcription
(RT), and quantitative PCR (qPCR)
Total RNA from the indicated tissues or cells was
extracted using the TRIzol reagent (Life Technologies; Thermo
Fisher Scientific, Carlsbad, CA, USA), according to the
manufacturer's instructions. Complementary DNA (cDNA) was amplified
and quantified on an ABI PRISM 7500 Sequence Detection system
(Applied Biosystems; Thermo Fisher Scientific, Foster City, CA,
USA) using SYBR Green I (Roche Diagnostics, Grenzach-Wyhlen,
Germany). miRNA quantification was determined using Bulge-loop™
miRNA quantitative reverse transcription PCR (qRT-PCR) Primer Set
(one RT primer and a pair of qPCR primers for each set) specific
for U6 and miR-873 that were designed and synthesized by Guangzhou
RiboBio Co., Ltd. The catalog numbers of these primers were as
follows: miR-873 RT primer (ssD809230648), miR-873 forward primer
(ssD090525061), miR-873 reverse primer (ssD089261711), U6 RT primer
(ssD0904071008), U6 forward primer (ssD0904071006), and U6 reverse
primer (ss D0904071007). The quantitative PCR (qPCR) conditions
were as follows: incubation at 50°C for 2 min, 95°C for 10 min,
followed by 40 cycles at 95°C for 15 sec, and 60°C for 1 min. The
expression of miR-873 was defined based on the quantification cycle
(Cq), and the relative fold changes between normal human colon
mucosal epithelial cell line and the CRC cell lines, and between
CRC tissues and their tumor-adjacent tissues (TATs), were
calculated according to the formula 2−[(Cq of miR-873)
- (Cq of U6)] after normalization to the
expression of U6 small nuclear RNA as a reference (30). REST-MCS beta software version 2 was
used to further analyze the qPCR data.
MTT assays
Cells (2×103/well) were seeded into
96-well plates. Transfection was performed 12 h later. At the
indicated time-points, 100 µl of sterile
3-(4,5-Dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazoliumbromide
(MTT) dye (0.5 mg/ml; Sigma-Aldrich, St. Louis, MI, USA) was added
and incubated for 4 h at 37°C. The culture medium was subsequently
removed, and 150 µl of dimethyl sulfoxide (DMSO; Sigma-Aldrich) was
added. The absorbance was assessed at a wavelength of 490 nm. All
experiments were performed in triplicate.
Colony formation assay
Cells (8×102/well) were seeded into
6-well plates and cultured for 10 days. The colonies were stained
with 1% crystal violet for 30 sec after incubation with 10%
formaldehyde for 5 min. Colonies were counted only if they
contained more than 50 cells, according to the established criteria
for colony formation (31–33).
Western blot analysis
Cell lysates were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to polyvinylidene fluoride (PVDF) membranes (EMD
Millipore, Billerica, MA, USA). The membranes were probed with
antibodies against TRAF5 (anti-TRAF5 antibody; 1:1,000; mouse,
polyclonal; cat. no. SAB1409766) and TAB1 (anti-TAB1 antibody;
1:500; rabbit, polyclonal; cat. no. SAB4301002; both from
Sigma-Aldrich) overnight at 4°C, followed by incubation with
horseradish peroxidase-conjugated secondary antibodies (1:2,000;
goat; cat. no. 7074 and 1:2,000; horse; cat. no. 7076; Cell
Signaling Technology, Danvers, MA, USA) for 1 h at 20°C. The
membranes were stripped and re-probed with an anti-α-tubulin
antibody (Sigma-Aldrich) as a loading control.
Luciferase assay
Cells (2×104/well) were seeded in
triplicate in 48-well plates and allowed to settle for 24 h. Next,
100 ng of the luciferase reporter plasmids or the control plasmid,
both with 1 ng of pRL-TK Renilla plasmid (Promega
Corporation), were transfected into cells using the Lipofectamine
2000 reagent (Invitrogen; Thermo Fisher Scientific), according to
the manufacturer's recommendation. Luciferase and Renilla
signals were assessed 24 h after transfection using a Dual
Luciferase Reporter assay kit (Promega Corporation), according to
the manufacturer's instructions.
Immunohistochemistry (IHC) and
hematoxylin and eosin (H&E) staining
Histology was performed to quantify Ki67 expression
in 10 paraffin-embedded human CRC samples. IHC was performed on
sections using an anti-Ki67 antibody (1:1; mouse, monoclonal; cat.
no. IR62661-2; Dako; Agilent Technologies, Inc., Glostrup,
Denmark). H&E staining was performed using Mayer's hematoxylin
solution. Immunostaining of the sections was quantified and scored
independently by two observers based on both the proportion of
positively-stained tumor cells and the intensity of staining. The
proportion of tumor cells enriched for Ki67 was scored as follows:
0 (no positive tumor cells), 1 (<10% positive tumor cells), 2
(10–50% positive tumor cells), and 3 (>50% positive tumor
cells). The intensity of staining was scored according to the
following criteria: 0 (no staining), 1 (weak staining, light
yellow), 2 (moderate staining, yellow brown), and 3 (strong
staining, brown). The staining index (SI) was calculated as the
proportion of positive tumor cells × the staining intensity score.
Using this scoring method, we evaluated the expression of Ki67 by
determining the SI, which was scored as 0, 1, 2, 3, 4, 6 and 9.
Statistical analysis
All values are presented as the means ± standard
deviation (SD). Student's t-test was used to determine the
statistical differences. The Chi-square test was used to analyze
the relationship between miR-873 expression and clinicopathological
characteristics. Multivariate statistical analysis was performed
using a Cox regression model. Survival curves were plotted using
the Kaplan-Meier method and compared using the log-rank test.
Statistical analyses were performed using the SPSS 19.0 software
(SPSS, Chicago, IL, USA). P≤0.05 was considered to indicate a
statistically significant result.
Results
miR-873 is downregulated in CRC cell
lines and CRC tissues
To examine the expression levels of miR-873
expression in CRC, we conducted qPCR in one normal human colon
mucosal epithelial cell line, seven CRC cell lines, and ten pairs
of CRC tissues and their TATs. The results revealed that miR-873
was markedly decreased in all seven CRC cell lines (SW620, SW480,
DLD1, HCT116, KM12, LoVo, and HT-29) compared with the normal human
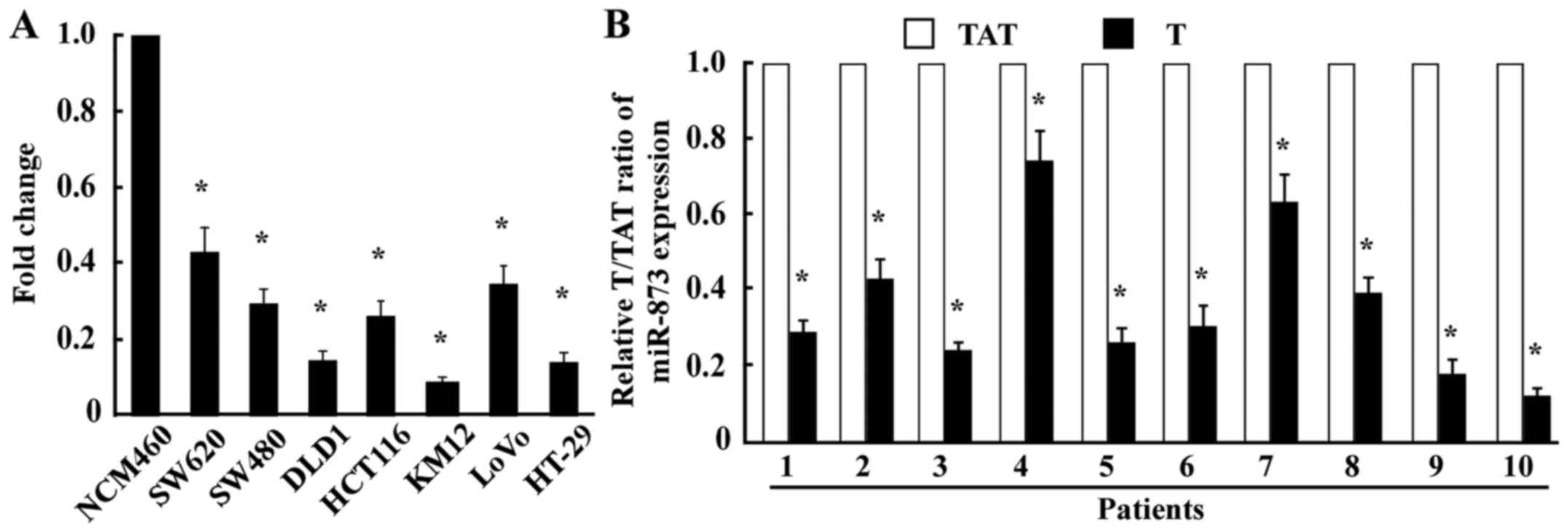
colon mucosal epithelial cell line NCM460 (Fig. 1A). Consistent with the results
obtained from the cell lines, miR-873 expression in the ten CRC
tissue samples was significantly lower compared with that in their
TATs (Fig. 1B), indicating that the
expression of miR-873 was downregulated in CRC.
miR-873 inhibits the proliferation of
CRC cells
To investigate the biological significance of
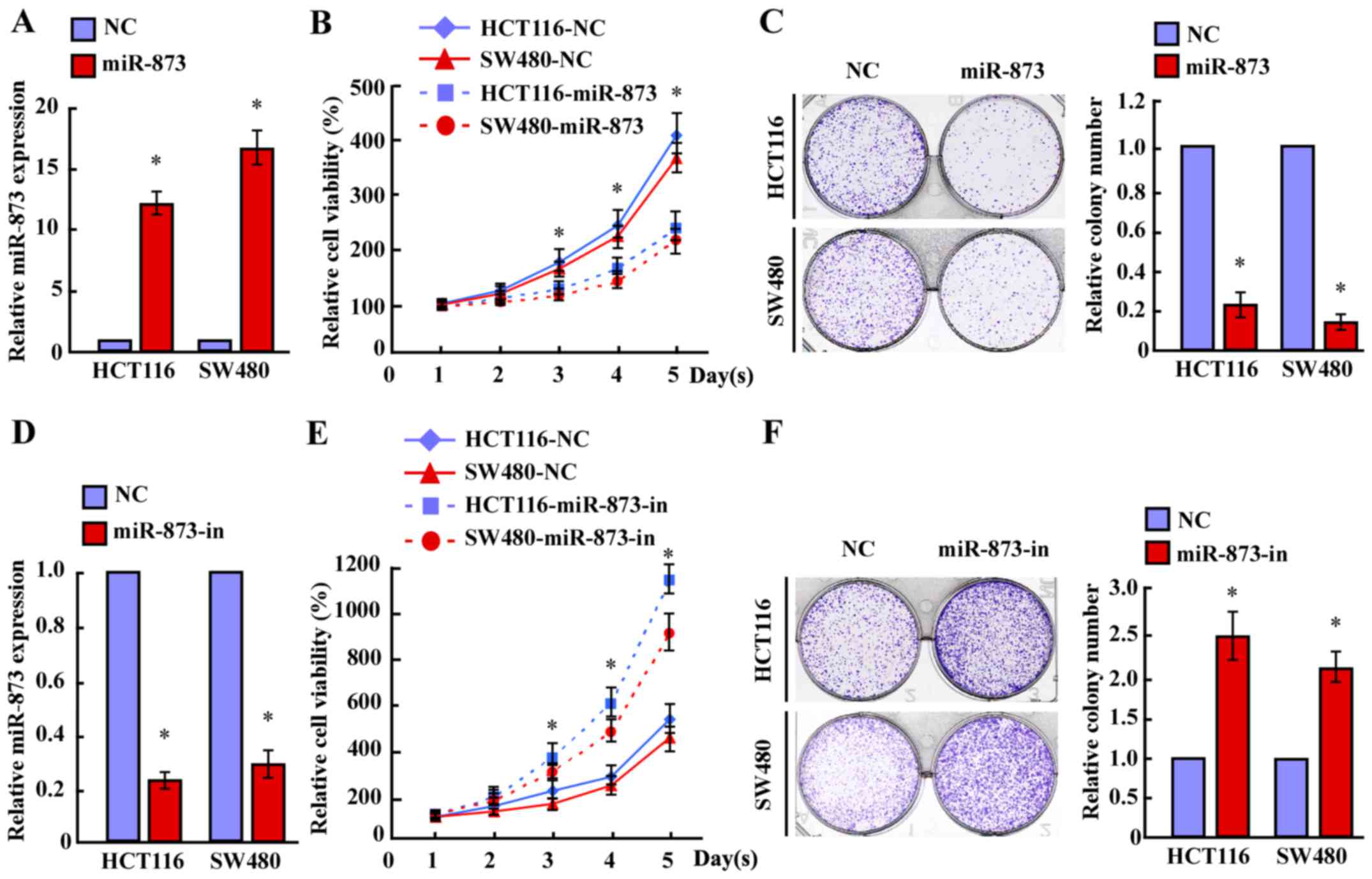
miR-873 downregulation during the progression of CRC, we
transfected the miR-873 mimic into HCT116 and SW480 cell lines. MTT
and colony formation assays revealed that overexpression of miR-873
markedly decreased the growth rates of both CRC lines compared with
the negative control (NC) (Fig.
2A-C). Furthermore, transfection of the miR-873 inhibitor
significantly increased the growth rates of both CRC lines compared
with that of the NC (Fig. 2D-F).
However, neither transfection of miR-873 nor the miR-873 inhibitor
altered the apoptotic rates of CRC cells (data not shown).
Therefore, these results revealed that miR-873 suppressed the
proliferation of CRC cells.
TRAF5 and TAB1 are direct targets of
miR-873 in CRC cells
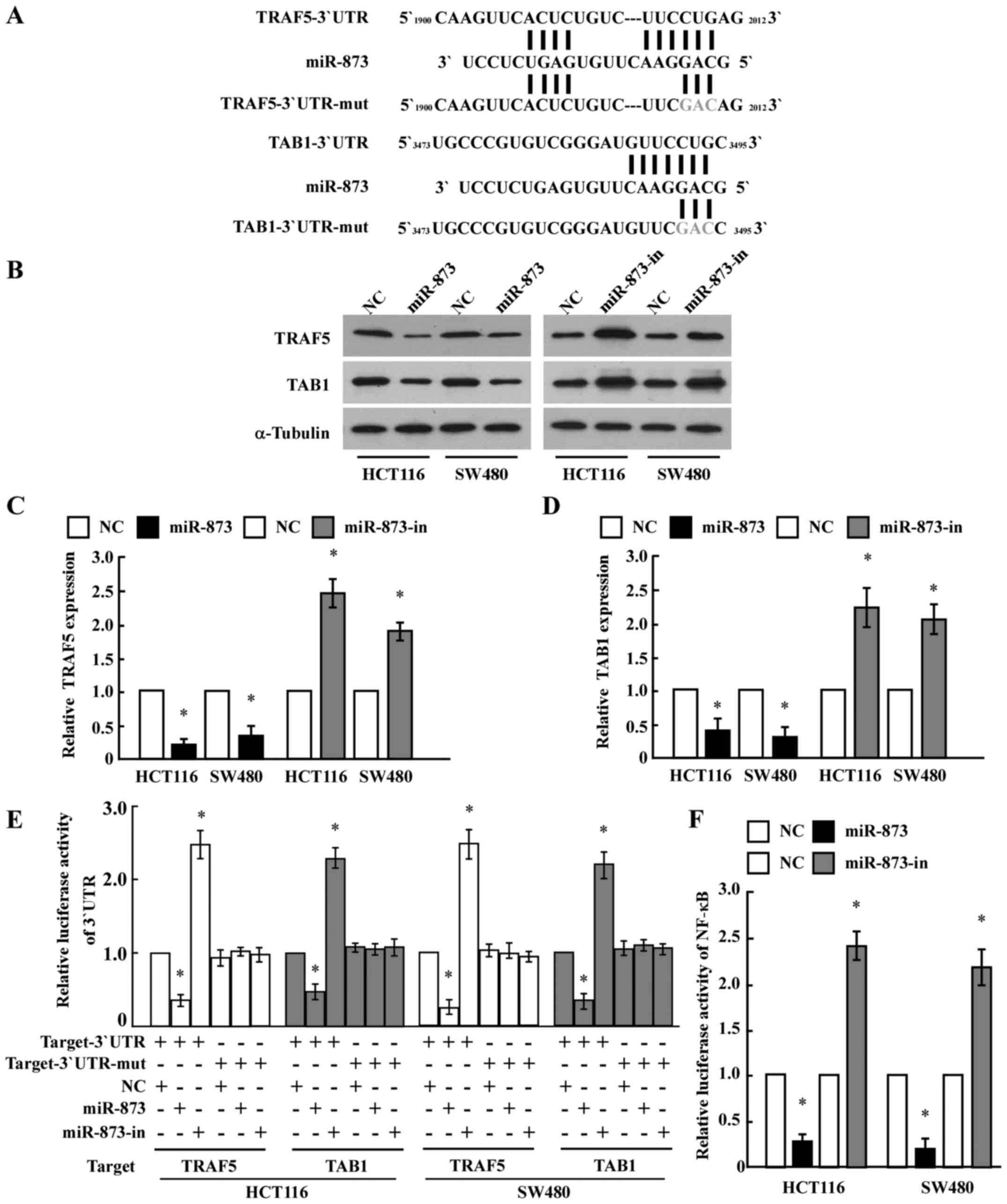
To identify the direct targets of miR-873
regulation, we searched publicly available databases (TargetScan,
Pictar, and miRANDA) and found that TRAF5 and TAB1,
which encode components of the NF-κB pathway, may be potential
targets (Fig. 3A). As predicted,
western blot analysis revealed that ectopic expression of miR-873
in HCT116 and SW480 cells decreased the levels of the TRAF5 and
TAB1 proteins, whereas ectopic expression of the miR-873 inhibitor
increased their levels (Fig. 3B-D).
However, we determined that the mRNA expression levels of
TRAF5 and TAB1 did not exhibit evident alterations in
the miR-873 dysregulated cells (data not shown), suggesting that
miR-873 negatively regulated the expression of these proteins at
the translation level. To further test this, we subcloned the
3′-UTRs of TRAF5 and TAB1 into the pGL3 luciferase
reporter. Transfection of miR-873 consistently attenuated the
luciferase activity of the TRAF5-3′-UTR and TAB1-3′-UTR luciferase
reporter in both HCT116 and SW480 cells, whereas transfection of
the miR-873 inhibitor rescued luciferase suppression. However,
dysregulation of miR-873 did not result in the alteration of the
reporter activities driven by the 3′-UTRs of TRAF5 and
TAB1 mutated within the miR-873-binding seed regions
(Fig. 3E). Collectively, these
results further supported the view that TRAF5 and
TAB1 are genuine targets of miR-873.
Furthermore, the luciferase assay revealed that
ectopic expression of miR-873 in HCT116 and SW480 cells
significantly reduced NF-κB luciferase activity, while ectopic
expression of the miR-873 inhibitor enhanced NF-κB luciferase
activity (Fig. 3F). This suggested
an important role for NF-κB signaling in the CRC cell proliferation
induced by miR-873 downregulation.
miR-873 expression is correlated with
clinical features and prognosis of patients with CRC
To further evaluate whether miR-873 downregulation
was associated with the clinical features or prognosis of CRC, we
examined the expression of miR-873 in a large cohort of clinical
CRC samples using qPCR and performed a correlation analysis between
the clinicopathological features and the expression of miR-873. The
data revealed that miR-873 expression was inversely correlated with
Dukes' stage (P=0.005), clinical stage (P=0.002),
tumor-node-metastasis (TNM) classification (T, P=0.036; N, P=0.001;
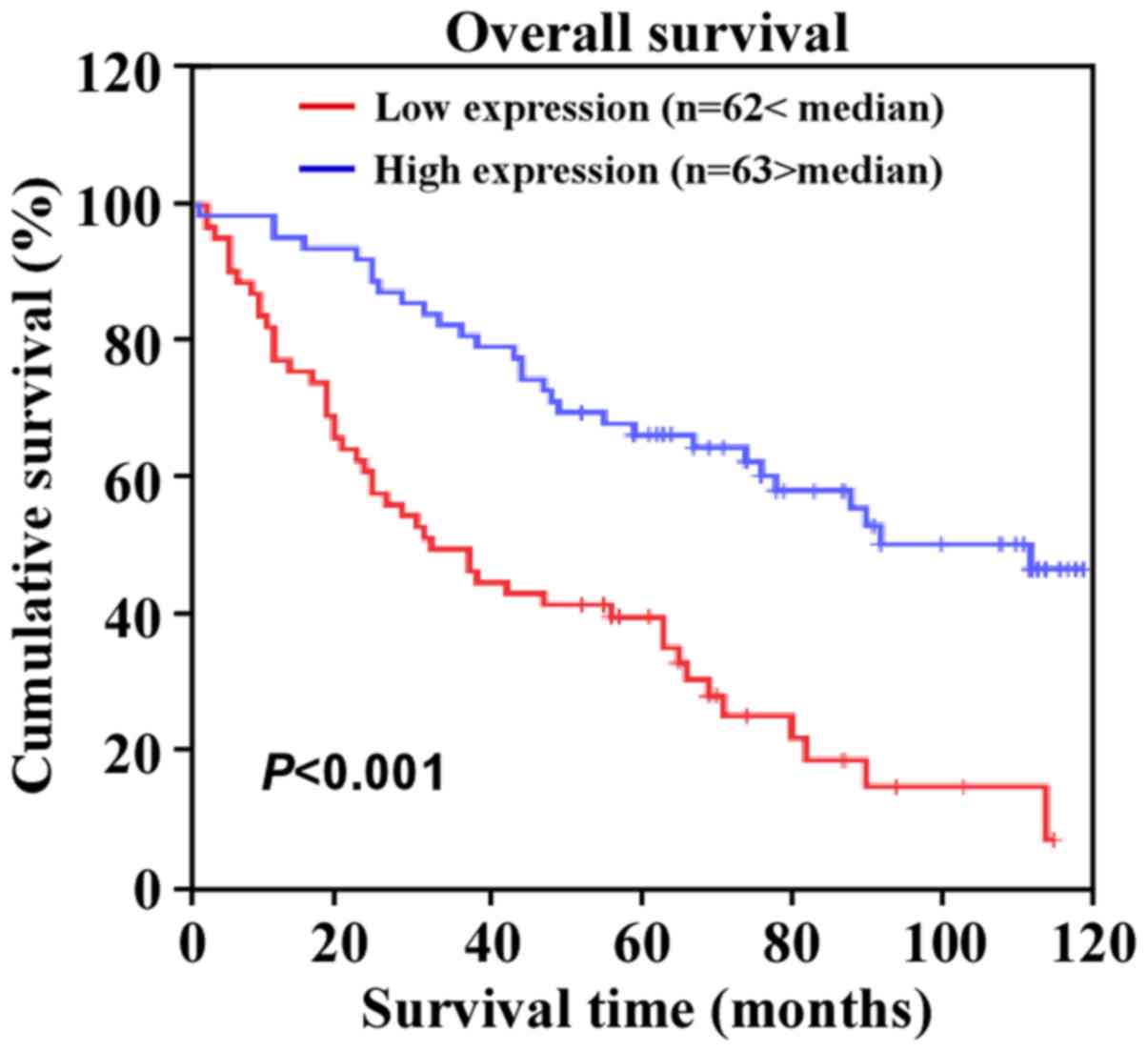
M, P=0.010), and histological differentiation (P=0.014) (Table II). Additionally, Kaplan-Meier
survival analysis revealed that patients with CRC with lower
miR-873 expression levels had shorter overall survival (Fig. 4). Moreover, univariate and
multivariate analysis indicated that miR-873 expression levels were
an independent prognostic factor for CRC (Table III). Collectively, these results
indicated a possible link between miR-873 downregulation and CRC
progression.
 | Table II.Correlation between
clinicopathological features of CRC patients and expression of
mir-873. |
Table II.
Correlation between
clinicopathological features of CRC patients and expression of
mir-873.
|
| mir-873
expression |
|
|---|
|
|
|
|
|---|
| Patient
characteristics | Low or none
(%) | High (%) | P-value |
|---|
| Sex |
|
|
|
|
Male | 31 (24.8) | 37 (29.6) | 0.372 |
|
Female | 31 (24.8) | 26 (20.8) |
|
| Age (years) |
|
|
|
|
≤62 | 31 (24.8) | 33 (26.4) | 0.859 |
|
>62 | 31 (24.8) | 30 (24.0) |
|
| Dukes' stage |
|
|
|
| A | 6 (4.8) | 11 (8.8) | 0.005 |
| B | 11 (8.8) | 25 (20.0) |
|
| C | 26 (20.8) | 19 (15.2) |
|
| D | 19 (15.2) | 8 (6.4) |
|
| Clinical stage |
|
|
|
| I | 6
(4.80) | 11 (8.80) | 0.002 |
| II | 11 (8.80) | 26 (20.8) |
|
|
III | 25 (20.0) | 19 (15.2) |
|
| IV | 20 (16.0) | 7 (5.6) |
|
| T
classification |
|
|
|
| T1 | 0
(0.0) | 2 (1.6) | 0.036 |
| T2 | 16 (12.8) | 14 (11.2) |
|
| T3 | 15 (12.0) | 27 (21.6) |
|
| T4 | 31 (24.8) | 20 (16.0) |
|
| N
classification |
|
|
|
|
N0 | 23 (18.4) | 40 (32.0) | 0.001 |
|
N1 | 20 (16.0) | 19 (15.2) |
|
|
N2 | 19 (15.2) | 4 (3.2) |
|
| M
classification |
|
|
|
|
M0 | 42 (33.6) | 55 (44.0) | 0.010 |
|
M1 | 20 (16.0) | 8 (6.4) |
|
| Histological
differentiation |
|
|
|
|
Well | 21 (16.8) | 8 (6.4) | 0.014 |
|
Moderate | 26 (20.8) | 39 (31.2) |
|
|
Poor | 15 (12.0) | 16 (12.8) |
|
| Vital status |
|
|
|
|
Alive | 15 (12.0) | 34 (27.2) | 0.001 |
|
Dead | 47 (37.6) | 29 (23.2) |
|
 | Table III.Univariate and multivariate analysis
of different prognostic parameters in patients with CRC by
Cox-regression analysis. |
Table III.
Univariate and multivariate analysis
of different prognostic parameters in patients with CRC by
Cox-regression analysis.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameters | No. of
patients | P-value | Regression
coefficient (SE) | P-value | Relative risk | 95% confidence
interval |
|---|
| N
classification |
| <0.001 | 0.150 | <0.001 | 1.746 | 1.306–2.336 |
|
N0 | 63 |
|
|
|
|
|
|
N1 | 39 |
|
|
|
|
|
|
N2 | 23 |
|
|
|
|
|
| M
classification |
| <0.001 | 0.260 |
0.001 | 2.405 | 1.430–4.044 |
|
M0 | 97 |
|
|
|
|
|
|
M1 | 28 |
|
|
|
|
|
| Expression of
mir-873 |
| <0.001 | 0.242 |
0.001 | 0.450 | 0.275–0.734 |
|
Low | 62 |
|
|
|
|
|
|
High | 63 |
|
|
|
|
|
Clinical relevance of miR-873
downregulation and cell proliferation in CRC
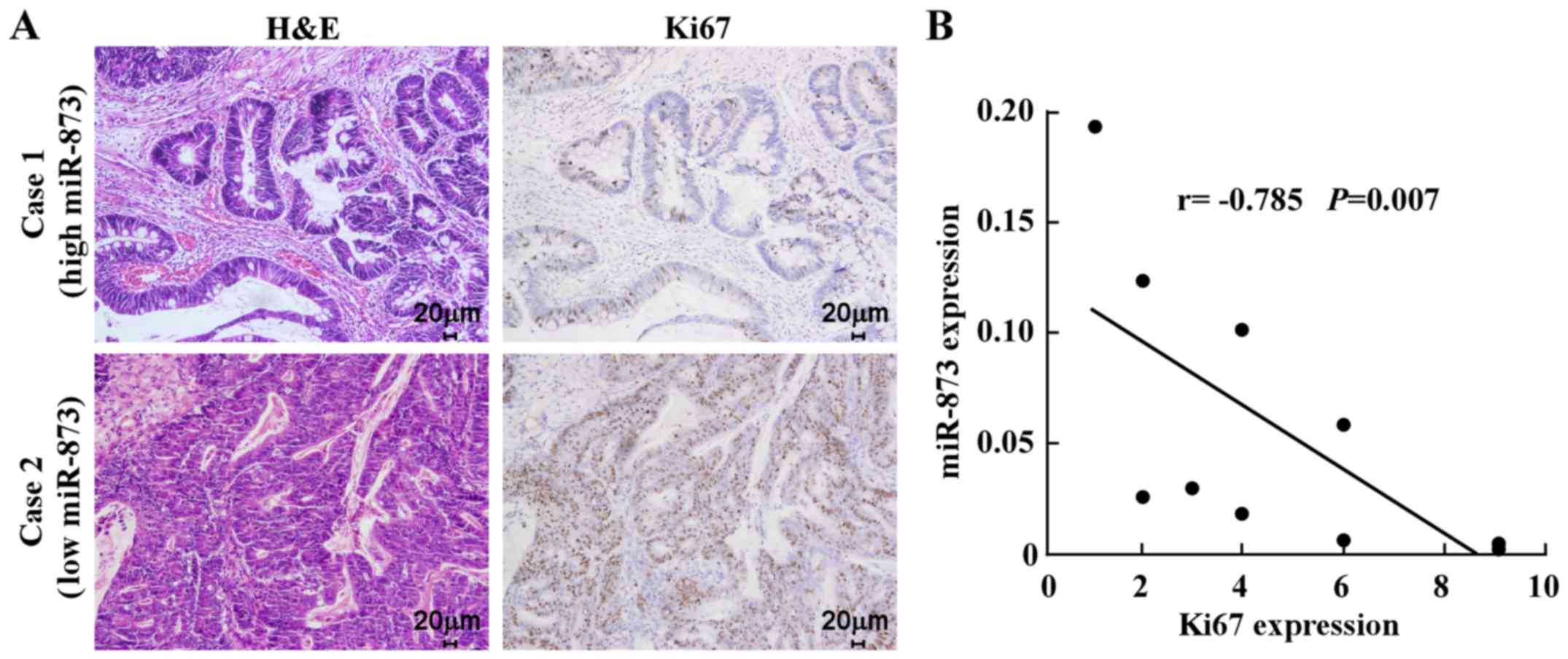
Finally, we examined whether the reduction in
miR-873 expression induced cell proliferation in CRC samples and
whether this was clinically relevant. IHC analysis of ten CRC
specimens revealed that low miR-873-expressed specimens had a
higher proportion of cells expressing the proliferation marker
Ki67. In contrast, high miR-873-expressed specimens displayed a
small proportion of Ki67-positive cells among the ten CRC specimens
(Fig. 5A). Correlation analysis
revealed that miR-873 expression levels were inversely correlated
with Ki67 in these CRC samples (Fig.
5B). Collectively, these results established that miR-873
suppressed CRC cell proliferation via inhibition of TRAF5 and TAB1,
which are key components of the NF-κB signaling pathway.
Discussion
The key finding of the current study is that miR-873
is a tumor-suppressive miRNA in CRC. Our data revealed that miR-873
was significantly downregulated in both CRC cell lines and primary
CRC specimens. Furthermore, downregulation of miR-873 expression
was associated with more advanced tumor stages and poor prognoses
for patients with CRC. Ectopic expression of miR-873 inhibited CRC
cell proliferation, whereas silencing of miR-873 promoted cell
proliferation. In addition, we demonstrated that miR-873 suppressed
the NF-κB pathway by directly targeting TRAF5 and
TAB1, which encode vital components of this pathway.
Collectively, our results demonstrated that miR-873 plays a
critical role in the tumorigenesis and progression of CRC and may
represent an important target for clinical intervention of CRC.
Previous research has revealed that miRNAs are
involved in tumor initiation, progression, metastasis, and response
to chemotherapy in CRC. In 2003, the first study describing the
roles of miRNAs in CRC was published, which revealed that miR-143
and miR-145 were specifically dysregulated in CRC, and multiple
other miRNAs were found to contribute to CRC by regulating critical
target mRNAs (34). For example,
several studies have revealed the dysregulation of a variety of
tissue-specific miRNAs, e.g., miR-21, miR-181b, miR-155, miR-92a,
and let-7, as well as some circulating miRNAs, e.g., miR-26a,
miR-21, miR-126, and miR-203 in CRC (35,36).
These miRNAs exert their effects by negatively regulating their
targets, such as p53, c-Met, K-Ras, COX-2, Rb, and the Bcl-2 family
(37). Accumulating evidence
indicates that miRNAs may serve as targets for miRNA-based
therapeutics of CRC. Inhibition of overexpressed oncogenic miRNAs
or introduction of tumor-suppressive miRNAs into cancer cells may
represent novel treatment strategies for CRC therapy in the future
(38).
Previously, it was shown that miR-873 is
dysregulated in a variety of malignancies. Notably, miR-873 was
downregulated in glioblastoma, and inhibited tumorigenesis and
metastasis by suppressing the expression of insulin-like growth
factor 2 mRNA-binding protein 1 (IGF2BP1) (24,25).
In addition, miR-873 enhanced the sensitivity of glioma cells to
cisplatin by targeting Bcl-2 (26).
It was also reported that overexpression of miR-873 increased the
sensitivity of ovarian cancer cells to cisplatin and paclitaxel by
targeting multidrug resistance protein 1 (MDR1) (27). However, there are also studies which
have reported that cisplatin is not a P-glycoprotein substrate and
multidrug resistance induced by cisplatin in ovarian carcinoma cell
lines was not due to overexpression of MDR1 and MDR3, both of which
are P-glycoproteins (39,40). Therefore, the controversial
mechanisms of multidrug resistance induced by cisplatin in ovarian
carcinoma warrant further investigation. In addition, miR-873 was
downregulated in tamoxifen-resistant breast cancer cell lines,
while overexpression of miR-873 reversed tamoxifen resistance by
targeting cyclin-dependent kinase 3 (CDK3) (28). By contrast, however, Gao et
al reported that miR-873 may act as an oncogene in lung
adenocarcinoma since it increased tumor cell proliferation and
migration via direct inhibition of SRCIN1 expression (29). Collectively, these findings
indicated that miR-873 can act as either a tumor-suppressive or
-promoting miRNA depending on the type of cancer. In this context,
we demonstrated that miR-873 inhibited CRC cell proliferation and
functioned as a tumor-suppressive miRNA in CRC. Meanwhile, the
inversed clinical relevance of miR-873 reduction with higher Ki67
signaling further supported the suppressive effect of miR-873 on
proliferation in CRC. However, the inhibitory effect of miR-873 on
proliferation in CRC warrants further investigation using an in
vivo mouse model. In addition, the expression and biological
function of miR-873 in other gastrointestinal tract cancers also
warrant further clarification.
NF-κB is a family of transcription factors that
controls the expression of a large number of genes related to
inflammation, immune responses, development, survival, and
proliferation (41). Since its
discovery nearly three decades ago (42), numerous studies have reported that
the NF-κB signaling pathway is frequently activated in a variety of
human cancers and it is associated with tumor initiation and
progression (6,8). The NF-κB signaling pathway plays
critical roles in the physiological and pathological processes of
CRC, and the relationship between CRC development and NF-κB
signaling is becoming clear (43).
Multiple research groups revealed that the constitutively activated
form of NF-κB was frequently expressed in CRC cells (44–46).
NF-κB may contribute to the progression of CRC by regulating the
expression of diverse target genes that are involved in cell
proliferation, angiogenesis, and metastasis (47). Therefore, the NF-κB pathway and its
upstream and downstream network constitute a potential druggable
target for therapeutic interventions (48). Although IKK complex-mediated NF-κB
activation has been studied in great detail, the regulatory
mechanism of the constitutive activation of NF-κB in CRC remains
largely unknown. Herein, we found that miR-873 significantly
inhibited the NF-κB pathway by directly targeting TRAF5 and TAB1,
key components of the NF-κB pathway. Thus, our results indicated
that miR-873 plays a regulatory role in NF-κB activation, and the
effect of miR-873-induced activation of NF-κB on invasion,
angiogenesis, or metastasis of CRC warrant further
investigation.
In conclusion, the present study reported that
miR-873 was downregulated in CRC and the expression level of
miR-873 was correlated with CRC progression and prognosis. We have
demonstrated, for the first time, that the upregulation of miR-873
markedly inhibited CRC cell proliferation by inhibiting the
expression of two key components (TRAF5 and TAB1) of the NF-κB
pathway. Therefore, our results demonstrated that miR-873 may play
an important role in the progression of CRC and could represent a
potential therapeutic target for CRC.
Acknowledgements
This study was supported by the Natural Science
Foundation of China (nos. 81402310, 81672957 and 91529301) and the
Science and Technology Innovation Committee of Shenzhen
Municipality (nos. JCYJ20140411093600199 and
JCYJ20160428180814307).
References
|
1
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weng W, Wei Q, Toden S, Yoshida K,
Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y and Goel A: Circular RNA
ciRS-7-a promising prognostic biomarker and a potential therapeutic
target in colorectal cancer. Clin Cancer Res. 23:3918–3928. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zeng J, Tang ZH, Liu S and Guo SS:
Clinicopathological significance of overexpression of interleukin-6
in colorectal cancer. World J Gastroenterol. 23:1780–1786. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deng J, Lei W, Fu JC, Zhang L, Li JH and
Xiong JP: Targeting miR-21 enhances the sensitivity of human colon
cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys
Res Commun. 443:789–795. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wieser M, Sauerland S, Arnold D, Schmiegel
W and Reinacher-Schick A: Peri-operative chemotherapy for the
treatment of resectable liver metastases from colorectal cancer: A
systematic review and meta-analysis of randomized trials. BMC
Cancer. 10:309–321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moghimi-Dehkordi B and Safaee A: An
overview of colorectal cancer survival rates and prognosis in Asia.
World J Gastrointest Oncol. 4:71–75. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karin M and Greten FR: NF-kappaB: Linking
inflammation and immunity to cancer development and progression.
Nat Rev Immunol. 5:749–759. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hayden MS and Ghosh S: Shared principles
in NF-kappaB signaling. Cell. 132:344–362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoesel B and Schmid JA: The complexity of
NF-κB signaling in inflammation and cancer. Mol Cancer. 12:862013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu D, Wu P, Zhao L, Huang L, Zhang Z, Zhao
S and Huang J: NF-κB expression and outcomes in solid tumors: A
systematic review and meta-analysis. Medicine (Baltimore).
94:e16872015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Z, Zhao L, Zhao F, Yang G and Wang J:
MicroRNA-26b regulates cancer proliferation migration and cell
cycle transition by suppressing TRAF5 in esophageal squamous cell
carcinoma. Am J Transl Res. 8:1957–1970. 2016.PubMed/NCBI
|
|
12
|
Jiang L, Yu L, Zhang X, Lei F, Wang L, Liu
X, Wu S, Zhu J, Wu G, Cao L, et al: miR-892b silencing activates
NF-κB and promotes aggressiveness in breast cancer. Cancer Res.
76:1101–1111. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harhaj EW and Dixit VM: Deubiquitinases in
the regulation of NF-κB signaling. Cell Res. 21:22–39. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khella HWZ, Daniel N, Youssef L, Scorilas
A, Nofech-Mozes R, Mirham L, Krylov SN, Liandeau E, Krizova A,
Finelli A, et al: miR-10b is a prognostic marker in clear cell
renal cell carcinoma. J Clin Pathol. 70:854–859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen J, Wang M, Guo M, Xie Y and Cong YS:
miR-127 regulates cell proliferation and senescence by targeting
BCL6. PLoS One. 8:e802662013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li M, Wang Y, Song Y, Bu R, Yin B, Fei X,
Guo Q and Wu B: MicroRNAs in renal cell carcinoma: A systematic
review of clinical implications (Review). Oncol Rep. 33:1571–1578.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao G, Cai C, Yang T, Qiu X, Liao B, Li
W, Ji Z, Zhao J, Zhao H, Guo M, et al: MicroRNA-221 induces cell
survival and cisplatin resistance through PI3K/Akt pathway in human
osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Go H, Jang JY, Kim PJ, Kim YG, Nam SJ,
Paik JH, Kim TM, Heo DS, Kim CW and Jeon YK: MicroRNA-21 plays an
oncogenic role by targeting FOXO1 and activating the PI3K/AKT
pathway in diffuse large B-cell lymphoma. Oncotarget.
6:15035–15049. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, VandenBoom TG II, Kong D, Wang Z,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-200 and let-7
by natural agents leads to the reversal of
epithelial-to-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Cancer Res. 69:6704–6712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng Y, Liu YM, Li LC, Wang LL and Wu XL:
microRNA-503 inhibits gastric cancer cell growth and
epithelial-to-mesenchymal transition. Oncol Lett. 7:1233–1238.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nofech-Mozes R, Khella HW, Scorilas A,
Youssef L, Krylov SN, Lianidou E, Sidiropoulos KG, Gabril M, Evans
A and Yousef GM: MicroRNA-194 is a marker for good prognosis in
clear cell renal cell carcinoma. Cancer Med. 5:656–664. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rapti SM, Kontos CK, Papadopoulos IN and
Scorilas A: High miR-96 levels in colorectal adenocarcinoma predict
poor prognosis, particularly in patients without distant metastasis
at the time of initial diagnosis. Tumour Biol. 37:11815–11824.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee TS, Jeon HW, Kim YB, Kim YA, Kim MA
and Kang SB: Aberrant microRNA expression in endometrial carcinoma
using formalin-fixed paraffin-embedded (FFPE) tissues. PLoS One.
8:e814212013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Skalsky RL and Cullen BR: Reduced
expression of brain-enriched microRNAs in glioblastomas permits
targeted regulation of a cell death gene. PLoS One. 6:e242482011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su
JL, Zhang MH and Liang HQ: MicroRNA-873 (miRNA-873) inhibits
glioblastoma tumorigenesis and metastasis by suppressing the
expression of IGF2BP1. J Biol Chem. 290:8938–8948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Zhang Y, Shi Y, Lian H, Tu H, Han
S, Peng B, Liu W and He X: miR-873 acts as a novel sensitizer of
glioma cells to cisplatin by targeting Bcl-2. Int J Oncol.
47:1603–1611. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu DD, Li XS, Meng XN, Yan J and Zong ZH:
MicroRNA-873 mediates multidrug resistance in ovarian cancer cells
by targeting ABCB1. Tumour Biol. 37:10499–10506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui J, Bi M, Overstreet AM, Yang Y, Li H,
Leng Y, Qian K, Huang Q, Zhang C, Lu Z, et al: miR-873 regulates
Erα transcriptional activity and tamoxifen resistance via targeting
CDK3 in breast cancer cells. Oncogene. 22:1–13. 2014.
|
|
29
|
Gao Y, Xue Q, Wang D, Du M, Zhang Y and
Gao S: miR-873 induces lung adenocarcinoma cell proliferation and
migration by targeting SRCIN1. Am J Transl Res. 7:2519–2526.
2015.PubMed/NCBI
|
|
30
|
Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY,
Cui YM, Qi L, Wu P, Jiao HL, Xie YJ, et al: MicroRNA-30b functions
as a tumour suppressor in human colorectal cancer by targeting
KRAS, PIK3CD and BCL2. J Pathol. 232:415–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miao Y, Li J, Qiu X, Li Y, Wang Z and Luan
Y: miR-27a regulates the self renewal of the H446 small cell lung
cancer cell line in vitro. Oncol Rep. 29:161–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li XX, Huang LY, Peng JJ, Liang L, Shi DB,
Zheng HT and Cai SJ: Klotho suppresses growth and invasion of colon
cancer cells through inhibition of IGF1R-mediated PI3K/AKT pathway.
Int J Oncol. 45:611–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu L, Deng B, Zeng Y and Cao Z: Decreased
expression of the Nkx2.8 gene correlates with tumor progression and
a poor prognosis in HCC cancer. Cancer Cell Int. 14:282014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Michael MZ, O' Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
35
|
Orang AV and Barzegari A: MicroRNAs in
colorectal cancer: From diagnosis to targeted therapy. Asian Pac J
Cancer Prev. 15:6989–6999. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kijima T, Hazama S, Tsunedomi R, Tanaka H,
Takenouchi H, Kanekiyo S, Inoue Y, Nakashima M, Iida M, Sakamoto K,
et al: MicroRNA-6826 and −6875 in plasma are valuable non-invasive
biomarkers that predict the efficacy of vaccine treatment against
metastatic colorectal cancer. Oncol Rep. 37:23–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Du Y, Liu X, Cho WC and Yang Y:
MicroRNAs as regulator of signaling networks in metastatic colon
cancer. Biomed Res Int. 2015:8236202015.PubMed/NCBI
|
|
38
|
Amirkhah R, Schmitz U, Linnebacher M,
Wolkenhauer O and Farazmand A: MicroRNA-mRNA interactions in
colorectal cancer and their role in tumor progression. Genes
Chromosomes Cancer. 54:129–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ren L, Xiao L, Hu J, Li Z and Wang Z: MDR1
and MDR3 genes and drug resistance to cisplatin of ovarian cancer
cells. J Huazhong Univ Sci Technolog Med Sci. 27:721–724. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stordal B, Hamon M, McEneaney V, Roche S,
Gillet JP, O'Leary JJ, Gottesman M and Clynes M: Resistance to
paclitaxel in a cisplatin-resistant ovarian cancer cell line is
mediated by P-glycoprotein. PLoS One. 7:e407172012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hayden MS and Ghosh S: Signaling to
NF-kappaB. Genes Dev. 18:2195–2224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sen R and Baltimore D: Multiple nuclear
factors interact with the immunoglobulin enhancer sequences. Cell.
46:705–716. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sakamoto K and Maeda S: Targeting
NF-kappaB for colorectal cancer. Expert Opin Ther Targets.
14:593–601. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sakamoto K, Maeda S, Hikiba Y, Nakagawa H,
Hayakawa Y, Shibata W, Yanai A, Ogura K and Omata M: Constitutive
NF-kappaB activation in colorectal carcinoma plays a key role in
angiogenesis, promoting tumor growth. Clin Cancer Res.
15:2248–2258. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Voboril R and Weberova-Voborilova J:
Constitutive NF-kappaB activity in colorectal cancer cells: Impact
on radiation-induced NF-kappaB activity, radiosensitivity, and
apoptosis. Neoplasma. 53:518–523. 2006.PubMed/NCBI
|
|
46
|
Lind DS, Hochwald SN, Malaty J, Rekkas S,
Hebig P, Mishra G, Moldawer LL, Copeland EM III and Mackay S:
Nuclear factor-κB is upregulated in colorectal cancer. Surgery.
130:363–369. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang S, Liu Z, Wang L and Zhang X:
NF-kappaB signaling pathway, inflammation and colorectal cancer.
Cell Mol Immunol. 6:327–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Vaiopoulos AG, Athanasoula KC and
Papavassiliou AG: NF-κB in colorectal cancer. J Mol Med (Berl).
91:1029–1037. 2013. View Article : Google Scholar : PubMed/NCBI
|