Introduction
Bladder cancer (BCa) is the most common malignant
tumor in the urinary system, and more than 300,000 new cases of the
disease are diagnosed each year worldwide (1,2). BCa
can be divided into the following two major types:
non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive
bladder cancer (MIBC). Patients with NMIBC have a good prognosis,
and the 5-year survival rate for such patients is over 88%
(3); however, ~70% of affected
patients will experience disease recurrence at the same position or
another bladder site (4). The
5-year survival rate for patients with MIBC is only ~60% (3,5).
Hence, studies investigating the potential mechanisms underlying
BCa occurrence and development are urgently needed. Recently, the
results of an increasing number of studies have suggested that
microRNAs (miRNAs) play an important role in BCa pathogenesis, and
may thus provide clinicians with new therapeutic targets and
strategies for treating the disease (6–9).
miRNAs are a class of conserved small non-coding
RNAs that exert their effects by binding to the 3′ untranslated
regions (3′UTRs) of their target messenger RNAs (mRNAs), leading to
mRNA degradation or translational inhibition and, ultimately, gene
expression regulation (10). The
results of previous studies indicate that miRNAs play important
roles in cancer occurrence and development as oncogenes or
tumor-suppressor genes (11).
miRNAs can affect cell proliferation, apoptosis, invasion,
metastasis and epithelial-mesenchymal transition (EMT) (12–14).
miR-130b, which is located at the 22q11 locus (15), has been shown to be aberrantly
expressed in some cancers, as it is overexpressed in renal cell
(16) and hepatocellular carcinoma
(17), and glioma (18), but is downregulated in papillary
thyroid (19) and endometrial
carcinoma (20), and prostate
cancer (21).
In a preliminary experiment, we found that miR-130b
was upregulated in BCa cells and tissues and that ectopic miR-130b
expression was closely related to BCa cell proliferation, migration
and invasion, indicating that miR-130b may function in BCa
progression.
Using bioinformatic analysis software, we identified
vestigial-like protein 4 (VGLL4) as the downstream target gene of
miR-130b. VGLL proteins are newly emerging TEAD-interacting
partners and transcriptional cofactors that participate in
tumorigenesis. Unlike other members of the VGLL family, VGLL4
contains an extra TDU domain, which is thought to be functionally
distinct from the other functional domains of the protein, and has
been identified as a transcriptional inhibitor that inhibits
YAP-induced tumor growth and development in Drosophila and
humans (22). The role of VGLL4 in
gastric, pancreatic and lung cancer, and esophageal squamous cell
carcinoma has recently attracted more attention. VGLL4 is
considered a new tumor-suppressor gene (23–28).
This study showed that the VGLL4 gene was a direct
target of miR-130b and that VGLL4 downregulation mediated by
miR-130b played a critical role in BCa cell proliferation,
migration and invasion.
Materials and methods
Cell cultures and clinical tissue
specimens
The indicated human BCa cell lines (T24, 5637 and
J82) and immortalized human bladder urothelial cell line (SV-HUC-1)
were obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China) and cultured in HyClone™ RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) (GE Healthcare Life
Sciences, HyClone Laboratories, Logan, UT, USA). All cells were
cultured in a humidified incubator at 37°C with 5% CO2
and 95% air. From January 2016 to September 2016, thirty pairs of
clinical tissue specimens (bladder tumor tissue specimens and
adjacent non-tumor bladder tissue specimens), including 19 men and
11 women with a median age of 63 years, were obtained from patients
undergoing transurethral bladder tumor resection (17 cases) or
radical cystectomy (13 cases) at the Department of Urology, The
First Hospital of China Medical University. All these cases were
staged by the 2002 UICC TNM classification. Of these 30 cases of
BCa, 17 cases were NMIBC (≤pT1), and the remaining cases were MIBC
(>pT1). The cases were also classified according to the WHO 2004
for grade, including 19-low grade papillary urothelial carcinoma,
and 11-high grade papillary urothelial carcinoma. No patients had
received radiotherapy, chemotherapy or adjuvant therapy before
surgery. All specimens were obtained after each patient provided
written informed consent to participate in this study, which was
approved by the Research Ethics Committee of The First Hospital of
China Medical University. Some specimens were fixed in formalin,
while others were immediately placed in liquid nitrogen and stored
at −80°C for further analysis.
RNA extraction and real-time PCR
(RT-PCR)
To detect VGLL4 mRNA, we extracted total RNA from
cultured cells and frozen tissues using Invitrogen™ TRIzol™ reagent
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to
the manufacturer's instructions. cDNA was synthesized with a
PrimeScript™ RT reagent kit (Takara, Shiga, Japan), and RT-PCR was
performed using a SYBR® Premix Ex Taq™ kit (Takara) in a
Roche LightCycler® 480 (Roche, Basel, Switzerland),
according to the manufacturers' instructions. To detect miR-130b,
we extracted total RNA (including microRNA) using a miRNeasy™ Mini
kit (Qiagen, Hilden, Germany), according to the manufacturer's
instructions. cDNA was synthesized using a miRCURY LNA™ Universal
cDNA Synthesis kit II (Exiqon A/S, Vedbaek, Denmark), and RT-PCR
was performed with miRCURY LNA™ ExiLENT SYBR®-Green
Master Mix (Exiqon A/S), according to the manufacturer's
instructions. miR-130b-3p and VGLL4 expression levels were
normalized to U6 and β-actin expression levels, respectively.
Relative expression levels were calculated using the 2-ΔCt method.
The primers for VGLL4 were: 5′-GCTGGTTTTCTTTGCTAGCCC-3′ (forward)
and 5′-CACCGGCAGGGTCTGTATTC-3′ (reverse), and the primers for
β-actin were: 5′-CTCACCATGGATGATGATATCGC-3′ (forward) and
5′-AGGAATCCTTCTGACCCATGC-3′ (reverse). The hsa-miR-130b-3p and U6
LNA™ PCR primer sets were provided by Exiqon.
Transient transfection
T24 and 5637 cells were transfected with
hsa-miR-130b-3p mimics, negative-control mimics, hsa-miR-130b-3p
inhibitors, negative-control inhibitors and VGLL4 siRNA
(GenePharma, Shanghai, China) using Invitrogen™ Lipofectamine™ 2000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The VGLL4 siRNA sense sequence was as follows:
5′-UCUGAACAAGACUGCCAAUTT-3′, and the VGLL4 siRNA antisense sequence
was as follows: 5′-AUUGGCAGUCUUGUUCAGATT-3′. The expression levels
of all transfected genes were confirmed with RT-PCR or western
blotting.
Luciferase assays
A total of 5637 cells were seeded in 24-well plates
and incubated for 24 h before transfection. miR-130b
NC/pmirGLO-VGLL4-3′UTR-WT, miR-130b mimic/pmirGLO-VGLL4-3′UTR-WT,
miR-130b NC/pmirGLO-VGLL4-3′UTR-MUT and miR-130b
mimic/pmirGLO-VGLL4-3′UTR-MUT (synthesized by GenePharma, Shanghai,
China) were transiently co-transfected into the cells using
Lipofectamine 2000. Cell lysates were gathered 48 h after
transfection and assessed using a Dual-Luciferase Reporter Assay
kit (Promega, Madison, WI, USA), according to the manufacturer's
protocol. All experiments were performed in triplicate.
Cell proliferation assay
Cell proliferation assay was conducted with the Cell
Counting Kit-8 (CCK-8) (Dojindo, Tokyo, Japan). The cells were
seeded in 96-well plates (3×103 cells/well) and
incubated for 24 h before transfection. Ten microliters of CCK-8
was added to every well at 1, 2, 3, 4 and 5 days after
transfection. Then, the cells were incubated at 37°C for 2 h, after
which the absorbance was measured at 450 nm using a microplate
reader. All experiments were performed in triplicate.
Colony formation assay
The cells (5×102 cells/well) were seeded
in 6-well plates in complete growth media and incubated for 2
weeks. The cell colonies were then fixed with methanol and stained
with 0.1% crystal violet before being imaged and counted (defined
as >50 cells/colony) using an AID iSpot Reader (Autoimmun
Diagnostika GmbH, Strassberg, Germany). The experiment was
performed three times for each cell line.
Wound healing assay
The cells were seeded in 6-well plates
(5×105 cells/well) and incubated until they reached 100%
confluence. Then, the cells were wounded by a 200-µl pipette tip,
and a cross-shape was generated in the monolayer. The cells were
subsequently washed with PBS for cellular debris removal and
cultured for 24 h. Then, the widths of wounds were measured under a
light microscope (Olympus Corp., Tokyo, Japan). The widths between
the cells which migrated the furthest on each side were measured to
calculate the relative wound closure area. The experiment was
performed three times for each cell line.
Invasion assay
Cell invasion assay was performed with Corning
Transwell insert chambers (Corning Incorporated, New York, NY, USA)
with an 8.0-µm pore size. Matrigel (Corning Incorporated) was used
to coat the top chamber. A total of 2×104 cells were
suspended in 200 µl of serum-free medium and seeded in the upper
chamber, and RPMI-1640 containing 10% FBS, which was used as a
chemoattractant, was added to the lower chamber. After incubating
for 24 h, the cells in the upper chamber were removed with cotton
swabs, and the cells that had passed through the membrane to invade
the lower chamber were fixed with methanol and stained with 0.1%
crystal violet before being counted under a light microscope. All
experiments were performed in triplicate.
Western blot analysis
The transfected cells and frozen tissues (including
the tumor and adjacent non-tumorous tissues) were lysed, and the
extracted proteins were quantified by a BCA Protein Assay kit
(Pierce Biotechnology, Rockford, IL, USA). Equal quantities of
protein were separated by 10% SDS-PAGE and transferred to
polyvinylidene fluoride (PVDF) membranes, which were blocked with
5% non-fat milk at room temperature for 2 h, and then incubated
with the appropriate primary antibody at 4°C overnight. Following 3
washes in Tris-buffered saline containing 0.1% Tween-20 (TBST), the
membranes were incubated with the appropriate secondary antibody
for 2 h at 37°C. Specific band signals were detected using a
chemiluminescence system (Bio-Rad, Philadelphia, PA, USA) and
analyzed using ImageJ Software (National Institutes of Health,
Bethesda, MD, USA) in accordance with the manufacturer's protocol.
The primary antibody rabbit polyclonal anti-VGLL4 (1:1,000; cat.
no. V2890; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and a
secondary antibody to mouse IgG (1:5,000; cat. no. ab193651; Abcam,
Cambridge, MA, USA) or rabbit IgG (1:5,000; cat. no. ab191866;
Abcam) were used for the experiments. The protein levels were
normalized to those of GAPDH (1:1,000; cat. no. ab8245; Abcam).
Statistical analysis
All data were processed with SPSS 13.0 (SPSS, Inc.,
Chicago, IL, USA) and presented as the mean ± standard deviation
(SD) of three independent experiments. Differences were evaluated
and analyzed with the Student's t-test. P-values <0.05 were
considered statistically significant.
Results
miR-130b expression is upregulated in
BCa
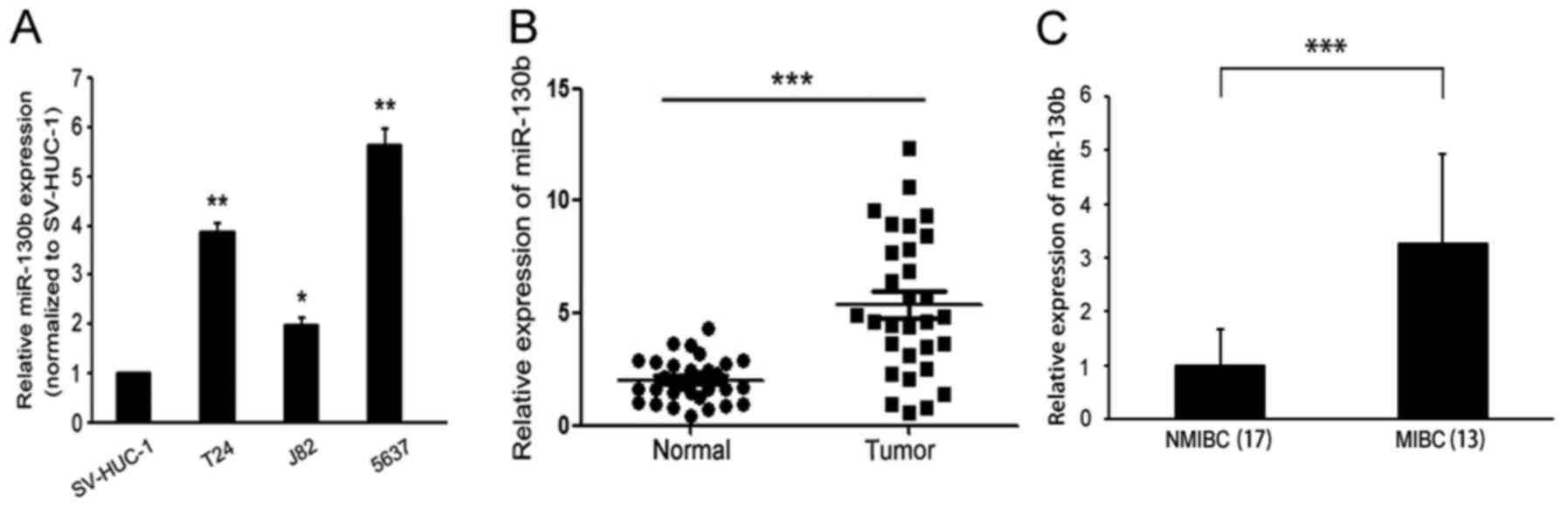
RT-PCR was used to detect miR-130b expression levels
in BCa cell lines and tissues. miR-130b was found to be
significantly elevated in the indicated BCa cell lines (T24, J82
and 5637) compared with the immortalized human bladder epithelial
cell line (SV-HUC-1) (Fig. 1A). We
also examined miR-130b expression levels in 30 BCa clinical tissue
samples and matched adjacent non-tumor tissue samples. The results
of the analysis showed that miR-130b expression levels in BCa
tissues were significantly higher than those in matched adjacent
non-tumor tissues (Fig. 1B). In
addition, miR-130b expression levels in the MIBC tissues were
significantly higher than those in the NMIBC tissues (Fig. 1C). These results indicated that
miR-130b expression levels were upregulated in BCa, suggesting that
miR-130b may play a role in promoting tumor progression in BCa.
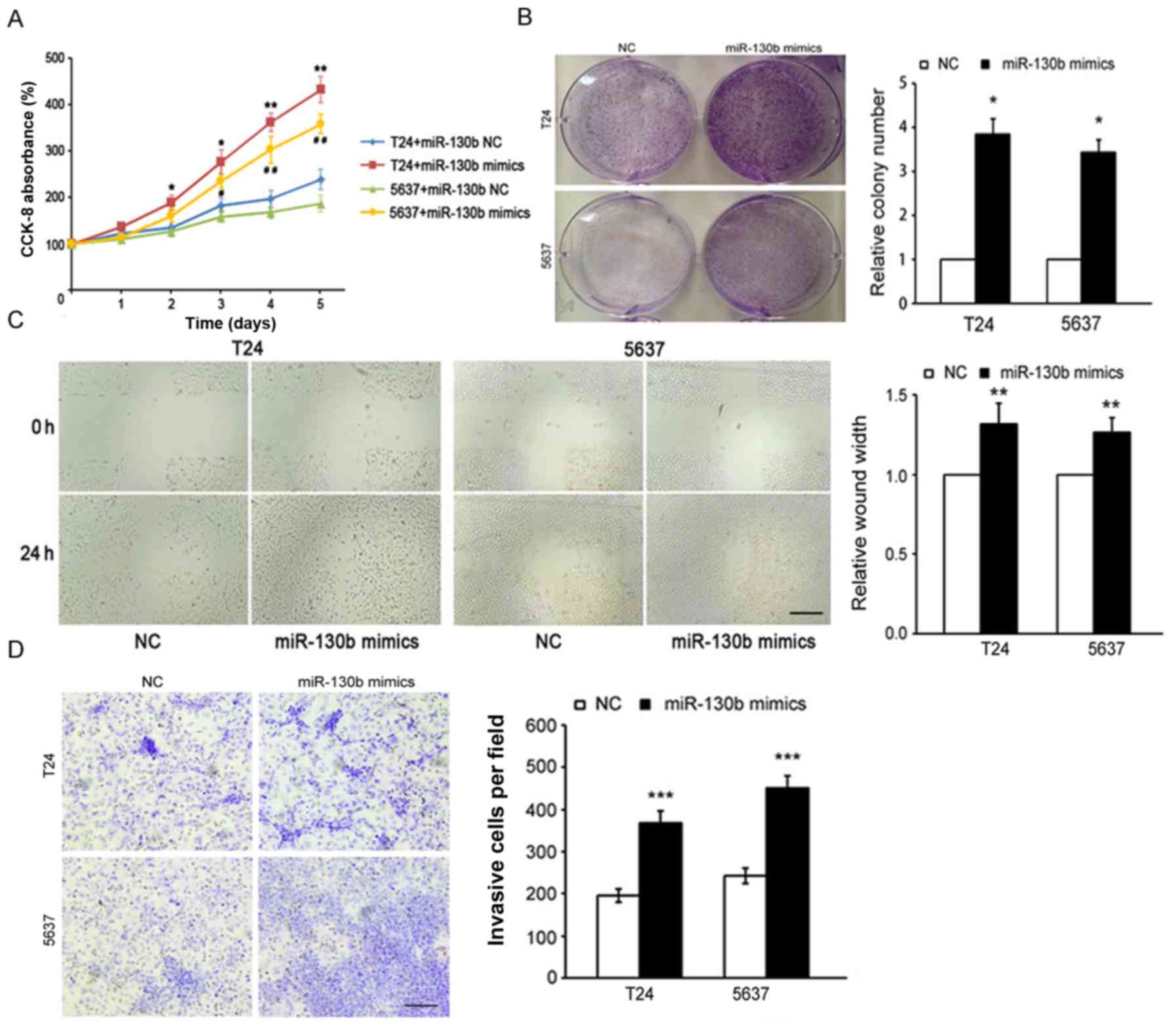
miR-130b promotes cancer cell
proliferation, migration and invasion
To further investigate the potential role of
miR-130b in the promotion of tumor progression in BCa, we
transfected miR-130b-3p mimics and miR-130b-3p mimic controls (as
control) into the T24 and 5637 cells and confirmed miR-130b-3p
overexpression by RT-PCR (data not shown). We used CCK-8 and colony
formation assays to assess the effects of miR-130b on cell
proliferation. The results of the CCK-8 assay indicated that
miR-130b overexpression significantly increased BCa cell growth
rates compared with the negative control (NC) cell growth rates
(Fig. 2A). The results of the
colony formation assay showed that BCa cells overexpressing
miR-130b formed a higher number of colonies than the control NC
cells (Fig. 2B). Additionally, the
results of the wound healing assay showed that the migration
ability of BCa cells overexpressing miR-130b was significantly
higher than that of the control NC cells (Fig. 2C). Furthermore, the results of the
Transwell assay indicated that miR-130b overexpression caused
significant increases in the invasive capabilities of BCa cells
compared with that of the control NC cells (Fig. 2D). All of these results indicated
that miR-130b overexpression promoted BCa cell proliferation,
migration and invasion.
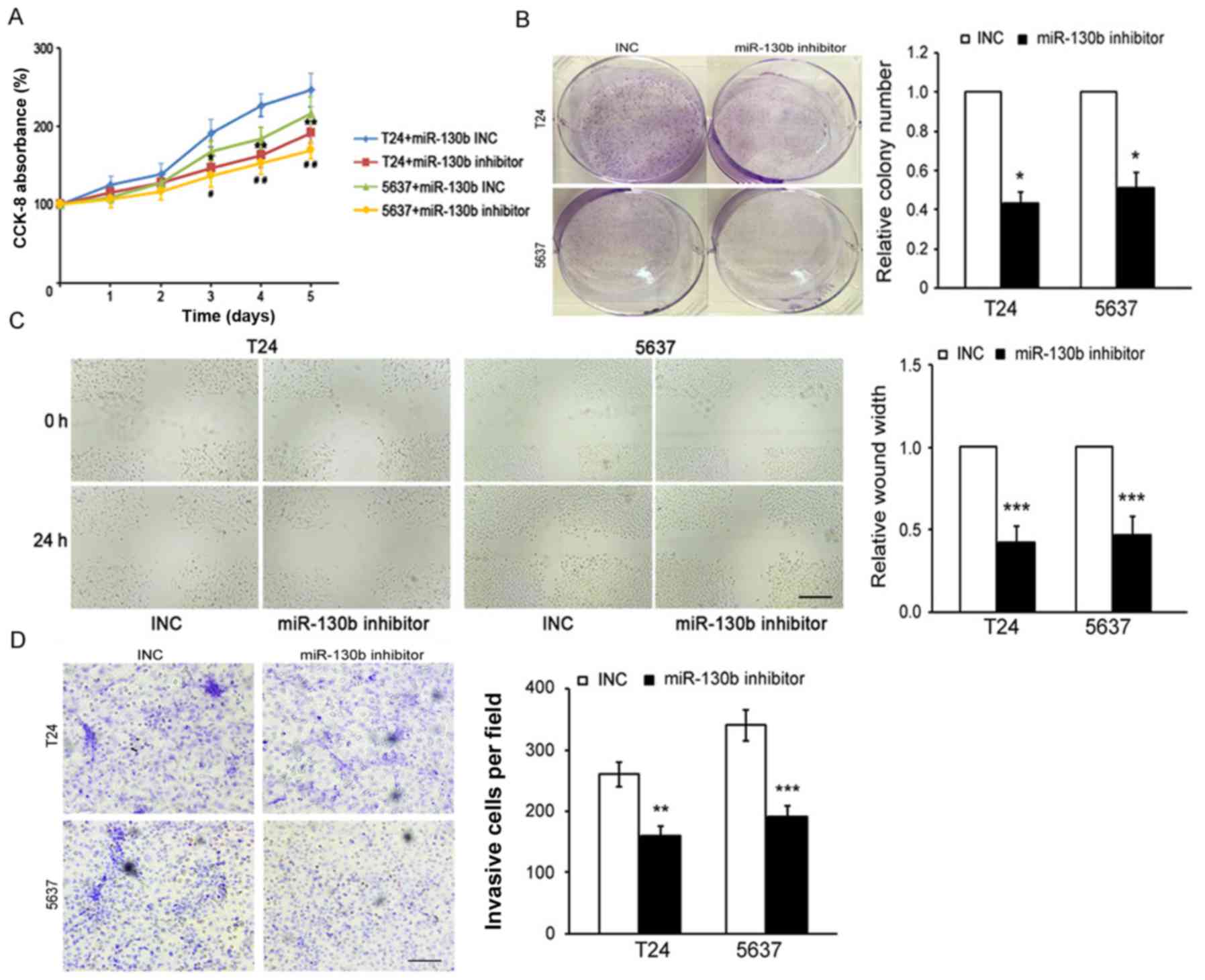
miR-130b inhibition suppresses BCa
proliferation, migration and invasion
To confirm the role of miR-130b in BCa
proliferation, migration and invasion, we performed
loss-of-function studies by transfecting miR-130b-3p inhibitors and
miR-130b-3p inhibitor controls [as control (INC)] into T24 and 5637
cells. The results of the CCK-8 and colony formation assays showed
that miR-130b suppression significantly decreased the proliferative
capacity of the BCa cells compared with that of the INC cells
(Fig. 3A and B). Additionally, the
results of the wound healing and Transwell assays indicated that
miR-130b inhibition caused a significant decrease in the migratory
and invasive capabilities of the BCa cells compared to those of the
INC cells (Fig. 3C and D). These
results indicated that miR-130b knockdown inhibited BCa cell
proliferation, migration and invasion.
VGLL4 is a direct target gene of
miR-130b in BCa cells
To further explore the molecular mechanisms
underlying the effects of miR-130b in BCa, we used bioinformatic
analysis software (TargetScan, miRBase) to predict the target genes
of miR-130b. VGLL4 was identified as one of the potential target
genes of miR-130b in BCa cells (Fig.
4F). Regarding the clinical specimens, VGLL4 protein expression
levels in BCa tissues were significantly decreased compared with
those in the adjacent non-tumor bladder tissues, and VGLL4 protein
expression levels were lower in MIBC tissues than in NMIBC tissues
(Fig. 4C). In BCa cells, VGLL4
protein expression levels were decreased in the
miR-130b-overexpressing cells, while VGLL4 protein expression
levels were significantly increased in the miR-130b-silenced cells
(Fig. 4A and B). However, there was
no significant difference in VGLL4 mRNA expression levels between
the two cell types (data not shown). miR-130b expression levels
were negatively correlated with those of VGLL4 in the BCa tissue
specimens (r=−0.7757, P<0.0001) (Fig. 4D). To determine whether VGLL4 is
regulated by the direct binding of miR-130b to its 3′UTR, we
performed luciferase assays. PmirGLO-VGLL4-3′UTR-WT and miR-130b
mimics were co-transfected into 5637 cells and caused a significant
decrease in luciferase activity in those cells compared with
control cells. In contrast, pmirGLO-VGLL4-3′UTR-MUT and miR-130b
mimics were co-transfected into 5637 cells and caused no
significant changes in luciferase activity in the treated group
compared with the NC group (Fig.
4E). These results suggested that the VGLL4 gene was a direct
target of miR-130b in BCa cells.
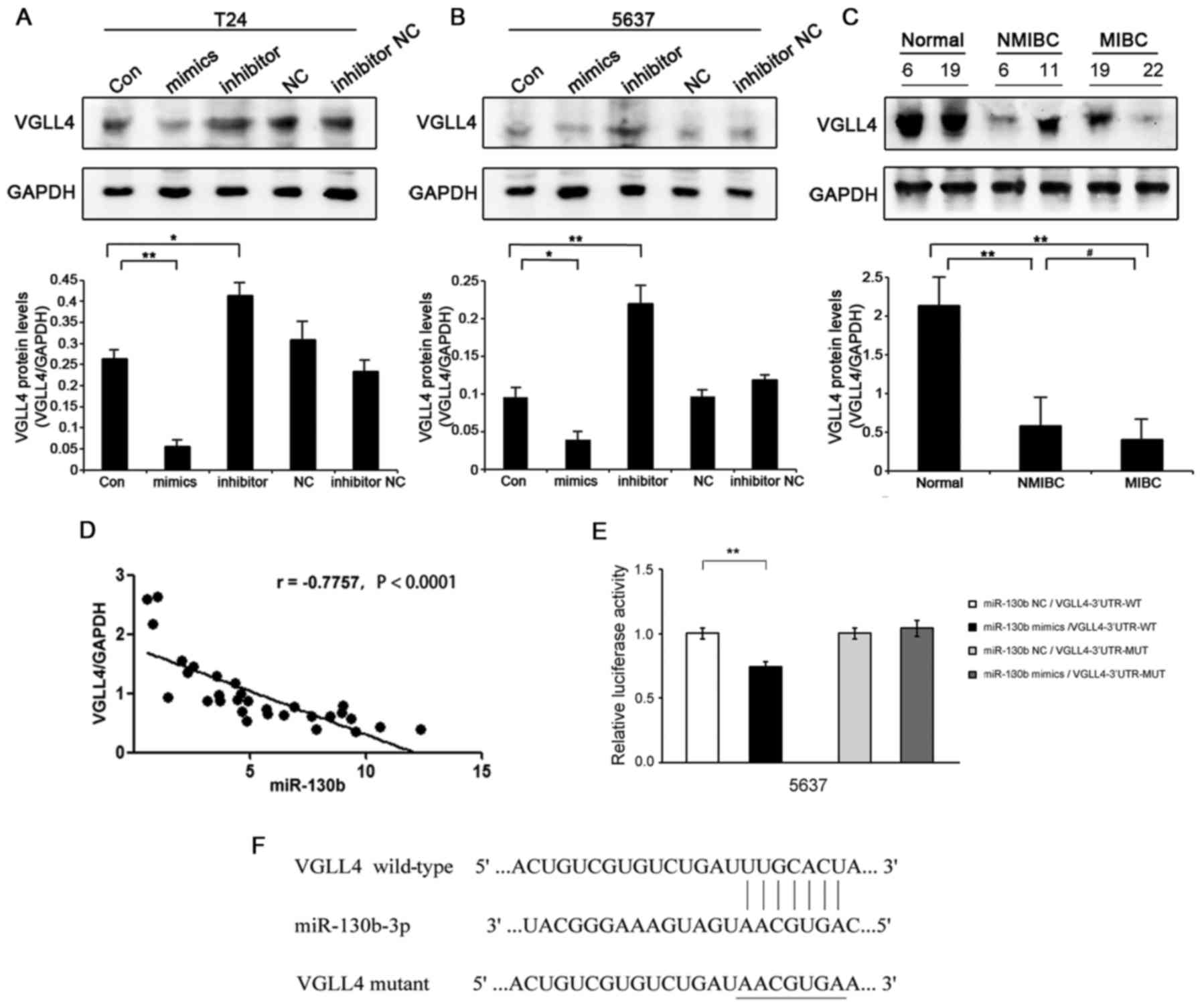 | Figure 4.miR-130b directly targets VGLL4 in
bladder cancer (BCa) cells. (A) VGLL4 protein expression levels in
T24 cells transfected with miR-130b mimics and inhibitors compared
with those in the negative control (NC) cells, as demonstrated by
western blotting. (B) VGLL4 protein expression levels in 5637 cells
transfected with miR-130b mimics and inhibitors compared with those
in the NC cells, as analyzed by western blotting. (C) Western blot
analysis of VGLL4 expression in BCa tissues, non-muscle-invasive
(NMIBC) tissues (T6, T11) and muscle-invasive (MIBC) tissues (T19,
T22) compared with those in adjacent non-tumor bladder tissues (N6,
N19). (D) Correlation between miR-130b and VGLL4 expression in BCa
tissues. (E) Luciferase activity levels were measured by luciferase
assay in the indicated cells. (F) Putative sequence of miR-130b,
with binding sites in the VGLL4 3′UTR. Data represent the mean ± SD
of three independent experiments; *P<0.05, **P<0.01,
#P<0.05. UTR, untranslated region. |
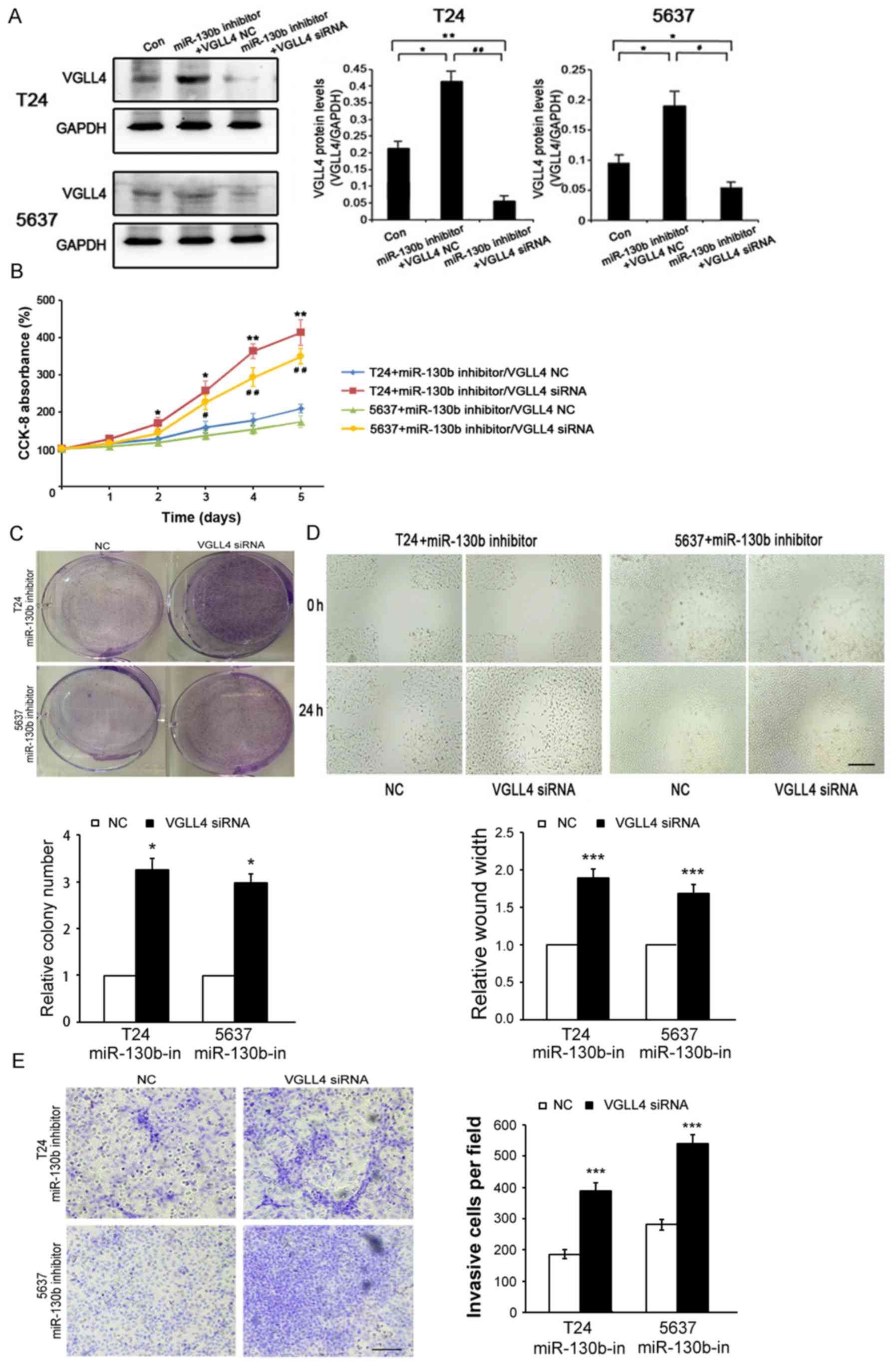
VGLL4 suppression is crucial for
miR-130b-induced BCa cell proliferation, migration and
invasion
To determine whether miR-130b-mediated BCa cell
proliferation, migration and invasion is attributable to VGLL4, we
transfected a miR-130b inhibitor/VGLL4-siRNA and miR-130b
inhibitor/control vector into T24 and 5637 cells (Fig. 5A). The results of the
above-mentioned assays showed that VGLL4 inhibition resulted in
significantly increased BCa cell proliferation (Fig. 5B and C), migration (Fig. 5D) and invasion (Fig. 5E) ability in the cells transfected
with the miR-130b inhibitor. These results showed that the
inhibitory effects of the miR-130b inhibitor on BCa cell
proliferation, migration and invasion were significantly reversed
by VGLL4 suppression. Hence, we confirmed that miR-130b-induced
cell proliferation, migration and invasion were facilitated by
VGLL4 suppression.
Discussion
More and more studies have shown that aberrant miRNA
expression plays an oncogenic or tumor suppressive role. The
oncogenic effects are exerted by tumor suppressor gene repression,
and the tumor suppressive effects are exerted by oncogene
inhibition (29,30). The important roles of miRNAs in
cancer occurrence and development have also been experimentally
demonstrated in animal models (31). Bladder cancer (BCa) is the most
common malignant tumor in the urinary system, hence, increasing
numbers of studies have focused on BCa biological characteristics,
occurrence and development and have elucidated the roles of miRNAs
in BCa. For example, a study reported that miR-200c functions as an
oncogene in BCa and that miR-200c downregulation significantly
inhibits BCa cell migration and invasion (32). Another study reported that miRNA-206
acted as a tumor suppressor in BCa by targeting YRDC; inhibiting
BCa cell proliferation, colony formation, migration and invasion;
and inducing cell cycle arrest at G0/G1 phase (33). Moreover, the expression levels of
some miRNAs, including miR-125b, miR-30b, miR-204, miR-99a and
miR-532-3p, were significantly downregulated in patient urine
supernatants. These miRNAs could be used as promising diagnostic
markers for the non-invasive diagnosis of BCa (34).
In the present study, the role of miR-130b in BCa
was studied in further detail. The results of this study showed
that miR-130b expression levels in surgically resected tumor
specimens were significantly higher than those in adjacent
non-tumor bladder tissue specimens. Moreover, higher miR-130b
expression levels were observed in MIBC tissues than in NMIBC
tissues. We also studied the effects of miR-130b
gain/loss-of-function on various aspects of T24 and 5637 cell
behaviors. We found that miR-130b overexpression or knockdown
promoted or inhibited BCa cell proliferation, migration and
invasion. Overall, we concluded that miR-130b may be an oncogene
and plays an important role in the pathogenesis of BCa.
To study the detailed mechanisms underlying the
function of miR-130b in BCa, we identified the downstream target
gene of miR-130b using bioinformatics analysis software (miRBase,
TargetScan). Bioinformatics analysis demonstrated that VGLL4 may be
a direct target of miR-130b in BCa. VGLL4 is a member of the VGLL
family, and VGLL proteins have recently emerged as a new
TEAD-interacting partners and participate in cancer development.
VGLL proteins are transcriptional cofactors. Recently, more
attention has been devoted to the role of VGLL4 in cancer. VGLL4
was believed to act as a tumor suppressor gene in many types of
cancer (24–26,35).
The results of a previous study indicated that VGLL4 directly
competes with YAP to bind TEADs and that VGLL4-mimicking peptides
potently suppressed gastric cancer tumor growth in vitro and
in vivo (23). Previous
study demonstrated that YAP was upregulated in some BCa cell (5637)
and overexpressed YAP promoted BCa cell growth and migration
(36). Furthermore, the inhibition
of YAP expressions significantly increased cytotoxic drug
sensitivity and reduced the migration of chemoresistant BCa cells
(37). In the present study, VGLL4
protein expression levels in BCa tissues were significantly
decreased compared with those in adjacent non-tumor bladder
tissues, and the inhibitory effects of the miR-130b inhibitor on
BCa cell proliferation, migration and invasion were significantly
reversed by VGLL4 suppression. Furthermore, we found that the VGLL4
gene is a direct target of miR-130b and that miR-130b downregulated
VGLL4 expression. miR-130b overexpression and knockdown in BCa
cells decreased and increased VGLL4 protein levels, respectively,
but did not significantly affect VGLL4 mRNA levels. These findings
indicated that miR-130b played an important role in regulating
VGLL4 translation rather than VGLL4 mRNA degradation. Therefore,
what is the specific role of VGLL4 in BCa is to be further
confirmed in our next studies.
In summary, our results indicate that miR-130b, a
potential oncogene, decreased VGLL4 expression by directly binding
to the 3′UTR of VGLL4 mRNA and promoted BCa cell proliferation,
migration and invasion. Thus, miR-130b/VGLL4 may be a new target
for the diagnosis and treatment of BCa.
Acknowledgements
The language of our manuscript had been polished by
the Nature Publishing Group Language Editing (NPG Language
Editing).
References
|
1
|
Acloque H, Thiery JP and Nieto MA: The
physiology and pathology of the EMT. Meeting on the
epithelial-mesenchymal transition. EMBO Rep. 9:322–326. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luke C, Tracey E, Stapleton A and Roder D:
Exploring contrary trends in bladder cancer incidence, mortality
and survival: Implications for research and cancer control. Intern
Med J. 40:357–362. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Rhijn BW, Burger M, Lotan Y, Solsona
E, Stief CG, Sylvester RJ, Witjes JA and Zlotta AR: Recurrence and
progression of disease in non-muscle-invasive bladder cancer: From
epidemiology to treatment strategy. Eur Urol. 56:430–442. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan YG, Eu E, Kam Lau On W and Huang HH:
Pretreatment neutrophil-to-lymphocyte ratio predicts worse survival
outcomes and advanced tumor staging in patients undergoing radical
cystectomy for bladder cancer. Asian J Urol. 4:239–246. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Urquidi V, Netherton M, Gomes-Giacoia E,
Serie DJ, Eckel-Passow J, Rosser CJ and Goodison S: A microRNA
biomarker panel for the non-invasive detection of bladder cancer.
Oncotarget. 7:86290–86299. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Falzone L, Candido S, Salemi R, Basile MS,
Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M and
Libra M: Computational identification of microRNAs associated to
both epithelial to mesenchymal transition and NGAL/MMP-9 pathways
in bladder cancer. Oncotarget. 7:72758–72766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Amir S and Mabjeesh NJ: microRNA
expression profiles as decision-making biomarkers in the management
of bladder cancer. Histol Histopathol. 32:107–119. 2017.PubMed/NCBI
|
|
9
|
Shin SS, Park SS, Hwang B, Moon B, Kim WT,
Kim WJ and Moon SK: MicroRNA-892b influences proliferation,
migration and invasion of bladder cancer cells by mediating the
p19ARF/cyclin D1/CDK6 and Sp-1/MMP-9 pathways. Oncol
Rep. 36:2313–2320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang D, Qiu C, Zhang H, Wang J, Cui Q and
Yin Y: Human microRNA oncogenes and tumor suppressors show
significantly different biological patterns: From functions to
targets. PLoS One. 5:52010.
|
|
12
|
Tang J, Ahmad A and Sarkar FH: The role of
microRNAs in breast cancer migration, invasion and metastasis. Int
J Mol Sci. 13:13414–13437. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Y, Zhang J, Wang H, Zhao J, Xu C, Du
Y, Luo X, Zheng F, Liu R, Zhang H, et al: miRNA-135a promotes
breast cancer cell migration and invasion by targeting
HOXA10. BMC Cancer. 12:1112012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ochoa AE, Choi W, Su X, Siefker-Radtke A,
Czerniak B, Dinney C and McConkey DJ: Specific micro-RNA expression
patterns distinguish the basal and luminal subtypes of
muscle-invasive bladder cancer. Oncotarget. 7:80164–80174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Burmistrova OA, Goltsov AY, Abramova LI,
Kaleda VG, Orlova VA and Rogaev EI: MicroRNA in schizophrenia:
Genetic and expression analysis of miR-130b (22q11). Biochemistry.
72:578–582. 2007.PubMed/NCBI
|
|
16
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang RM, Xu JF, Fang F, Yang H and Yang
LY: MicroRNA-130b promotes proliferation and EMT-induced metastasis
via PTEN/p-AKT/HIF-1α signaling. Tumour Biol. 37:10609–10619. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yip L, Kelly L, Shuai Y, Armstrong MJ,
Nikiforov YE, Carty SE and Nikiforova MN: MicroRNA signature
distinguishes the degree of aggressiveness of papillary thyroid
carcinoma. Ann Surg Oncol. 18:2035–2041. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dong P, Karaayvaz M, Jia N, Kaneuchi M,
Hamada J, Watari H, Sudo S, Ju J and Sakuragi N: Mutant p53
gain-of-function induces epithelial-mesenchymal transition through
modulation of the miR-130b-ZEB1 axis. Oncogene. 32:3286–3295. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Q, Zhao X, Zhang H, Yuan H, Zhu M,
Sun Q, Lai X, Wang Y, Huang J, Yan J, et al: MiR-130b suppresses
prostate cancer metastasis through down-regulation of MMP2. Mol
Carcinog. 54:1292–1300. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gabriel BM, Hamilton DL, Tremblay AM and
Wackerhage H: The Hippo signal transduction network for exercise
physiologists. J Appl Physiol 1985. 120:1105–1117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiao S, Wang H, Shi Z, Dong A, Zhang W,
Song X, He F, Wang Y, Zhang Z, Wang W, et al: A peptide mimicking
VGLL4 function acts as a YAP antagonist therapy against gastric
cancer. Cancer Cell. 25:166–180. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mann KM, Ward JM, Yew CC, Kovochich A,
Dawson DW, Black MA, Brett BT, Sheetz TE and Dupuy AJ: Australian
Pancreatic Cancer Genome Initiative, Chang DK, Biankin AV, Waddell
N, Kassahn KS, Grimmond SM, Rust AG, Adams DJ, Jenkins NA and
Copeland NG: Sleeping Beauty mutagenesis reveals cooperating
mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad
Sci USA. 109:5934–5941. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang W, Gao Y, Li P, Shi Z, Guo T, Li F,
Han X, Feng Y, Zheng C, Wang Z, et al: VGLL4 functions as a new
tumor suppressor in lung cancer by negatively regulating the
YAP-TEAD transcriptional complex. Cell Res. 24:331–343. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang W, Yao F and He J, Lv B, Fang W, Zhu
W, He G, Chen J and He J: Downregulation of VGLL4 in the
progression of esophageal squamous cell carcinoma. Tumour Biol.
36:1289–1297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Wang Z, Zhang W, Qian K, Liao G, Xu
W and Zhang S: VGLL4 inhibits EMT in part through suppressing
Wnt/β-catenin signaling pathway in gastric cancer. Med Oncol.
32:832015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li N, Yu N, Wang J, Xi H, Lu W, Xu H, Deng
M, Zheng G and Liu H: miR-222/VGLL4/YAP-TEAD1 regulatory loop
promotes proliferation and invasion of gastric cancer cells. Am J
Cancer Res. 5:1158–1168. 2015.PubMed/NCBI
|
|
29
|
Yoshino H, Seki N, Itesako T, Chiyomaru T,
Nakagawa M and Enokida H: Aberrant expression of microRNAs in
bladder cancer. Nat Rev Urol. 10:396–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bibi F, Naseer MI, Alvi SA, Yasir M,
Jiman-Fatani AA, Sawan A, Abuzenadah AM, Al-Qahtani MH and Azhar
EI: microRNA analysis of gastric cancer patients from Saudi Arabian
population. BMC Genomics. 17 Suppl 9:S7512016. View Article : Google Scholar
|
|
31
|
Li M, Li J, Ding X, He M and Cheng SY:
microRNA and cancer. AAPS J. 12:309–317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng Y, Zhang X and Li P, Yang C, Tang J,
Deng X, Yang X, Tao J, Lu Q and Li P: MiR-200c promotes bladder
cancer cell migration and invasion by directly targeting RECK. Onco
Targets Ther. 9:5091–5099. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang B, Zhai W, Hu G, Huang C, Xie T,
Zhang J and Xu Y: MicroRNA-206 acts as a tumor suppressor in
bladder cancer via targeting YRDC. Am J Transl Res. 8:4705–4715.
2016.PubMed/NCBI
|
|
34
|
Pospisilova S, Pazourkova E, Horinek A,
Brisuda A, Svobodova I, Soukup V, Hrbacek J, Capoun O, Hanus T,
Mares J, et al: MicroRNAs in urine supernatant as potential
non-invasive markers for bladder cancer detection. Neoplasma.
63:799–808. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koontz LM, Liu-Chittenden Y, Yin F, Zheng
Y, Yu J, Huang B, Chen Q, Wu S and Pan D: The Hippo effector Yorkie
controls normal tissue growth by antagonizing scalloped-mediated
default repression. Dev Cell. 25:388–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dong L, Lin F, Wu W, Huang W and Cai Z:
Transcriptional cofactor Mask2 is required for YAP-induced cell
growth and migration in bladder cancer cell. J Cancer. 7:2132–2138.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ciamporcero E, Daga M, Pizzimenti S,
Roetto A, Dianzani C, Compagnone A, Palmieri A, Ullio C, Cangemi L,
Pili R, et al: Crosstalk between Nrf2 and YAP contributes to
maintaining the antioxidant potential and chemoresistance in
bladder cancer. Free Radic Biol Med. 115:447–457. 2018. View Article : Google Scholar : PubMed/NCBI
|