Introduction
Colorectal cancer (CRC), the world's fourth leading
cause of cancer-associated mortality, is the third most common type
of malignancy in females and the third most common type in males
(1–3). At present, >1 million patients
suffer from CRC, and >600,000/year succumb to it worldwide.
Although CRC may occur at any age, the risk is highest for
individuals aged >50 years (4).
At present, the standard therapies for CRC are
surgical resection, radiotherapy, chemotherapy and targeted therapy
(5,6). Metastasis is one of the most critical
factors impacting the prognosis of CRC patients and the efficiency
of their treatment. Even though the 5-year survival rate for CRC
patients without metastasis is high, ~10% of them gradually develop
metastasis, which eventually leads to death, rendering the above
therapeutic therapies inefficient (7). Furthermore, the majority of patients
are diagnosed with CRC at stages when the cancer cells have further
metastasized to other tissues and organs in the body (8,9).
CRC patients tend to develop lymph node metastasis
and distant metastasis at late stages. Previous investigations have
corroborated that changes in various processes, including increased
cell proliferation, altered cell metabolism, decreased apoptosis,
activated protease systems take part in the development of
adenoma-carcinoma metastasis (10).
However, the underlying molecular mechanisms and key genes involved
in CRC progression or metastasis have remained to be fully
elucidated. Consequently, the exposure of these molecular
mechanisms and key genes is the most critical step towards
preventing metastasis. In recent years, bioinformatics analysis,
including high-throughput sequencing technology and protein-protein
interactions (PPIs), has provided a deeper understanding of the
aberrant genetic pathways involved in cancers (11,12).
In the present study, a bioinformatics analysis was performed to
identify the underlying molecular mechanisms and key genes involved
in the metastasis of CRC.
Initially, differentially expressed genes (DEGs)
between primary CRC tissues and metastatic CRC were identified from
datasets downloaded from the Gene Expression Omnibus (GEO) online
database. Subsequently, function and pathway enrichment analysis
were performed on the DEGs. The ten leading hub genes which may be
involved in the CRC progression of metastasis were selected from
the PPI network. Finally, tumor tissues from four patients with
metastatic CRC were subjected to reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assays to verify these five
leading potential metastasis-associated DEGs.
Materials and methods
Microarray data processing
The GEO dataset GSE2509, including the mRNA
expression profile of 3 SW480 and 3 SW620 cell samples, derived
from sequencing and subsequent data analysis by Provenzani et
al (13), was downloaded from
the National Center of Biotechnology Information GEO database
(http://www.ncbi.nlm.nih.gov/geo/). The
data had been generated using the GPL96 (HG-U133A) platform
(Affymetrix Human Genome U133A Array; Affymetrix; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The SW480 cell line originated
from a primary rectal gland carcinoma sample and the SW620 cell
line was derived one year later from a lymph node metastasis sample
of the same patient.
Screening of DEGs
The raw data were uploaded to the Gene-Cloud of
Biotechnology Information online laboratory for further analysis
(https://www.gcbi.com.cn/gclib/html/index). DEGs were
identified between SW480 and SW620. The threshold for the DEGs was
defined with a corrected P-value of <0.05 and a fold change (FC)
of >1.2.
Function and pathway enrichment
analysis of DEGs
Gene Ontology (GO) analysis is a useful
Bioinformatics tool used to annotate information about gene product
function and identify characteristic biological attributes by
analyzing high-throughput genome data (14,15).
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a tool for
the systematic analysis of gene function, performed by linking
series of genes with networks of interacting cellular molecules,
including complex pathways (16).
To facilitate the function and pathway analysis, the DEGs were
copied into the Database for Annotation, Visualization and
Integrated Discovery (DAVID), to perform the GO and KEGG enrichment
analyses. The human genome was chosen as the background list.
P<0.05 was considered to indicate a statistically significant
difference.
PPI network and module analysis
The PPI is described through an undirected diagram
with nodes symbolizing the genes and edges symbolizing the mutual
interactions of proteins encoded by the corresponding genes
(17). In the present study, all of
the DEGs were imported into the Search Tool for the Retrieval of
Interacting Genes (STRING; http://www.string-db.org) for analysis and only
interactions with a combined score of >0.7 were pasted into the
Cytoscape plugin to create the network visualization. Subsequently,
the PPI network was subjected to module analysis by using the
Plugin MCODE with the default parameters (Degree cutoff ≥2, Node
score cutoff ≥2, K-core ≥2 and Max depth =100). Finally, function
and pathway enrichment analysis of the DEGs in the most prominent
three modules were performed via DAVID.
Verification of the leading five
potential metastasis-associated DEGs in clinical tumor tissue
samples
In the present study, RT-qPCR assays were used to
validate the expression levels of the top five potential
metastasis-associated genes with the highest degree of interaction
and significance in the PPI network. Total RNA was extracted from
metastatic and primary CRC samples from four patients who had been
diagnosed with metastatic CRC by the Pathology Department of Sir
Run Run Shaw Hospital (Hangzhou, China) from October 2016 to March
2017.
First, the mRNA was extracted from the patients'
samples using TRIzol reagent (Thermo Fisher Scientific, Inc.) and
then stored at −80°C. A Reverse Transcription System
(GoTaq® Real-Time PCR System; Promega Corp., Madison,
WI, USA) was employed for the synthesis of complementary DNA
according to the manufacturer's instructions. The mRNA expression
levels of the key genes were measured by quantitative real-time-PCR
using the ABI PRISM 7500 Sequence Detector System (Applied
Biosystems; Thermo Fisher Scientific, Inc.), and GAPDH was used as
an internal standard. The primers used for qPCR were used as
follows: Epidermal growth factor receptor (EGFR) forward,
5′-AGGCACGAGTAACAAGCTCAC-3′ and reverse,
5′-ATGAGGACATAACCAGCCACC-3′; cyclin-dependent kinase inhibitor 1A
(CDKN1a) forward, 5′-TGTCCGTCAGAACCCATGC-3′ and reverse,
5′-AAAGTCGAAGTTCCATCGCTC-3′; Wnt family member 5A (Wnt5a) forward,
5′-TTCTTGGTGGTCGCTAGGT-3′ and reverse, 5′-TTCTTTGATGCCTGTCTTCG-3′;
Has proto-oncogene GTPase (HRas) forward, 5′-TTTGCCATCAACAACACCA-3′
and reverse, 5′-TCCTGAGCCTGCCGAGAT-3′; serine/threonine kinase 1
(Akt1) forward, 5′-TCCTCCTCAAGAATGATGGCA-3′ and reverse,
5′-GTGCGTTCGATGACAGTGGT-3′; GAPDH forward,
5′-AGACAGCCGCATCTTCTTGT-3′ and reverse, 5′-TGATGGCAACAATGTCCACT-3′.
The reaction protocol included heating for 3 min at 95°C, followed
by 40 cycles of amplification (5 sec at 95°C and 20 sec at 60°C).
qPCR reactions were performed in triplicate. The final results were
analyzed via the 2−∆∆Cq method with normalization to
GAPDH (18).
Statistical analysis
PCR results are presented as the mean ± standard
error of the mean (n=3). Differences between the PCR results for
the primary and metastatic tumors were determined by the two-tailed
unpaired Student's t-test. Prism 4.0 statistical software (GraphPad
Inc., La Jolla, CA, USA) was used for statistical analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of DEGs between the
SW480 and SW620 CRC cell lines
With the aim of exploring the mechanisms underlying
CRC metastasis, publicly accessible microarray datasets were
retrieved from the GEO database. In total, 3 cell lines each of
SW480 and SW620 pertaining to primary and secondary CRC,
respectively, were screened for DEGs. The specified criteria,
including FC >1.2 and P<0.05, were met for 7,384 genes,
including 3,949 upregulated genes and 3,435 downregulated genes.
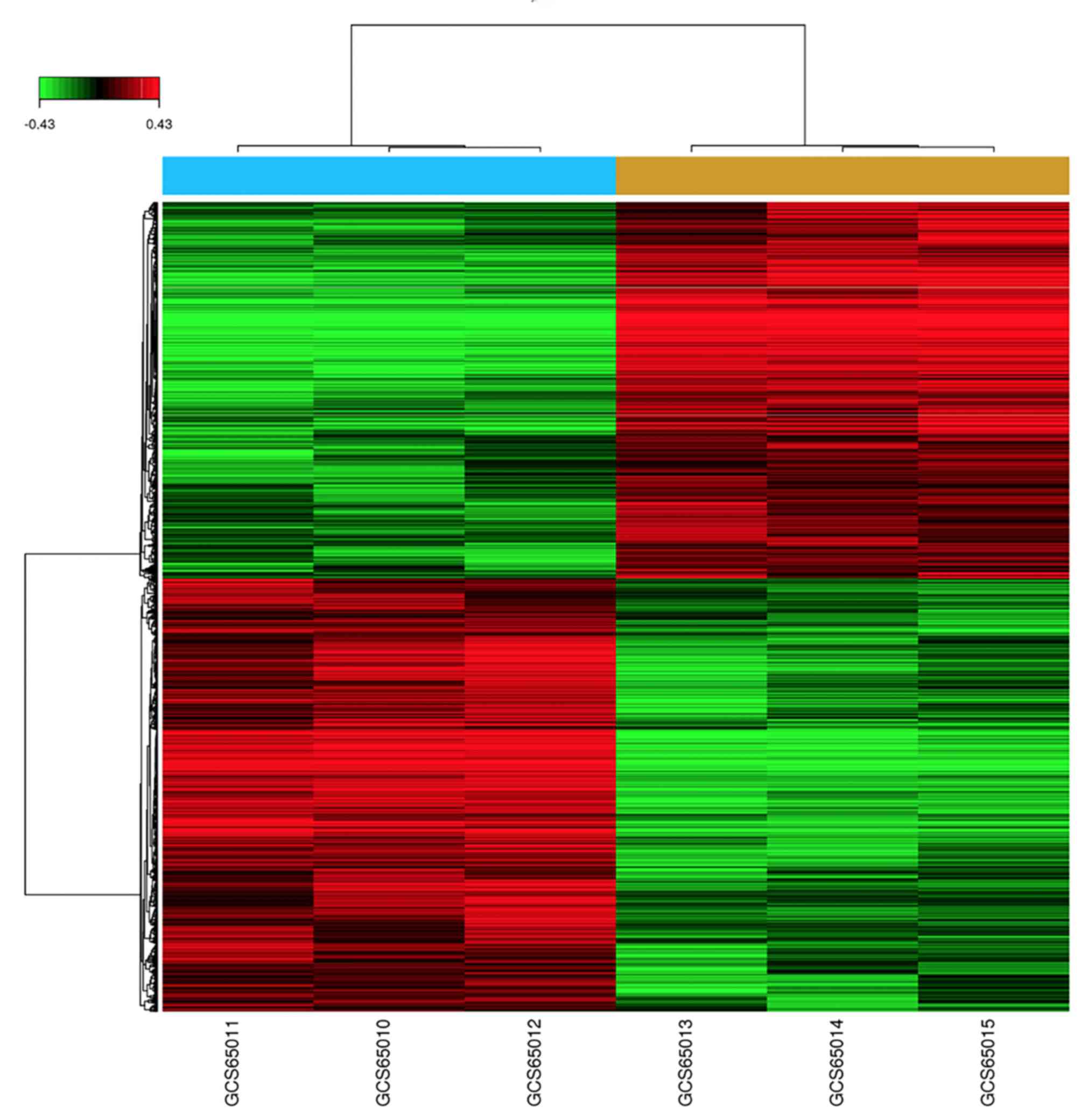
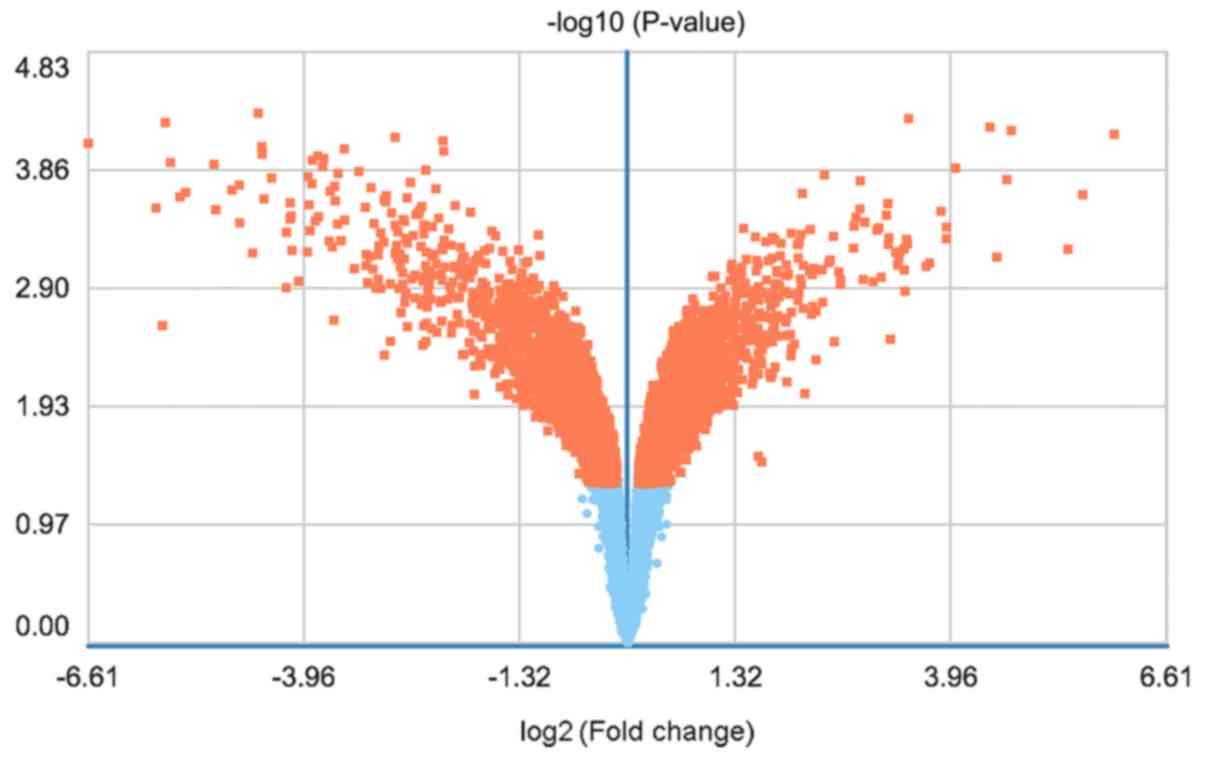
The DEGs were displayed in a heat map (Fig. 1) and a volcano plot (Fig. 2).
GO and KEGG pathway enrichment
analysis
The online biological classification tool DAVID was
then employed to analyze the functions and pathways of the 7,384
DEGs. GO analysis suggested that the DEGs were fully involved in
873 biological processes with the premise of false discovery rate
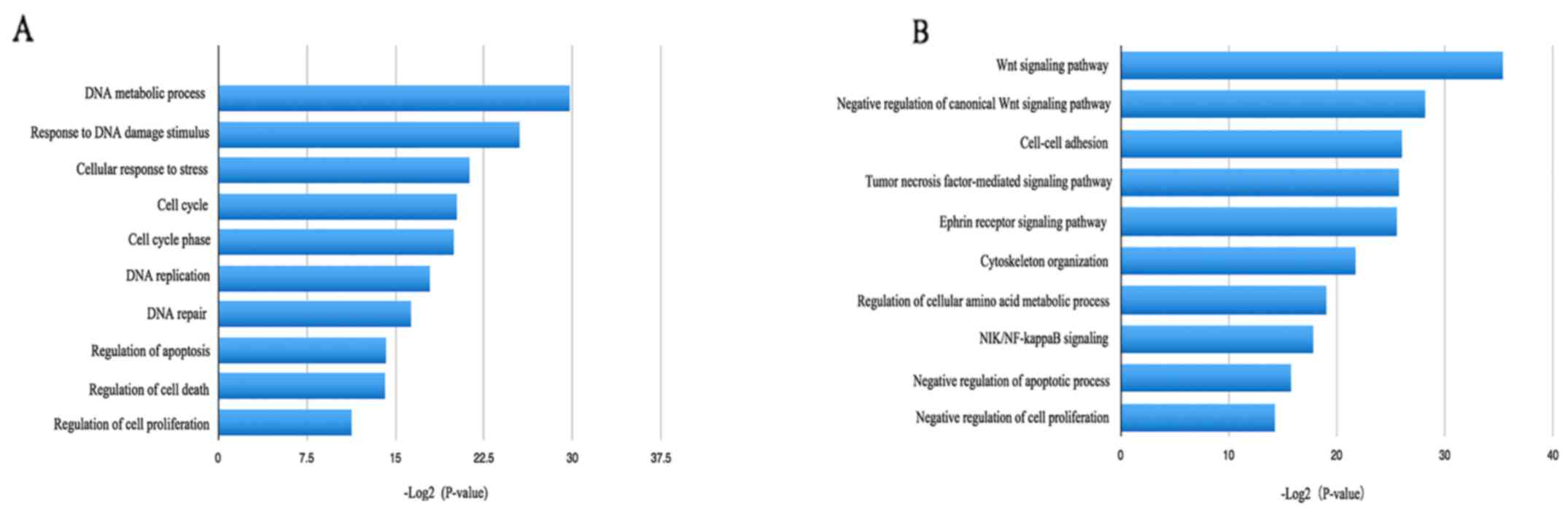
<0.05 and P<0.05. The upregulated DEGs were mainly involved
in DNA metabolic processes, responses to DNA damage stimuli,
cellular responses to stress and the cell cycle (Fig. 3), while the downregulated DEGs were
significantly involved in the Wnt signaling pathway, negative
regulation of the canonical Wnt signaling pathway, cell-cell
adhesion and negative regulation of apoptotic processes (Fig. 3).
The biological functions of cells are complex
processes mediated by numerous molecules and genes. Through KEGG
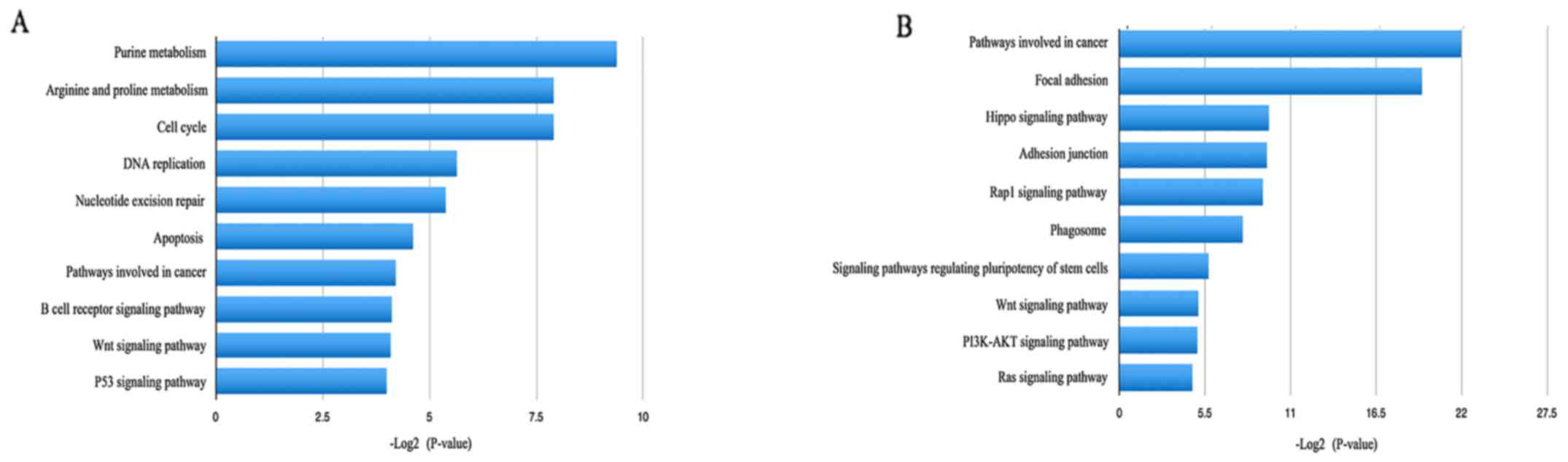
pathway enrichment analysis, the upregulated DEGs were identified
to be significantly enriched in purine metabolism, arginine and
proline metabolism, the cell cycle and DNA replication, which may
be associated with tumor proliferation and migration (Fig. 4). As to the downregulated DEGs,
pathways involved in cancer, focal adhesions, adhesion junctions
and the Hippo signaling pathway were most significant (Fig. 4).
PPI network and module analysis
To investigate the interactions and acquire the hub
genes for potential metastasis-associated DEGs in CRC, all of the
DEGs were analysed by using STRING. Subsequently, the genes whose
combined score were >0.7 revealed close association between
genes and were imported into Cytoscape for further analysis. The
PPI network contained 954 nodes and 3,326 interactions (data not
shown). The ten leading genes defined as the hub genes were as
follows: EGFR, HRas, Akt1, Wnt5a, CDKN1a, early growth response 1,
Cd44, Ras homolog family member A, cyclin D1 and Ras-related C3
botulinum toxin substrate 1 (data not shown). Next, the Plugin
MCODE was employed to recognize the module genes from the PPI
analysis (Fig. 5). Function
annotation and pathway analysis of the three most significant
modules, performed by DAVID, revealed that the module genes were
mainly associated with the cell cycle, the forkhead box (FoxO)
signaling pathway, the Wnt signaling pathway, ubiquitin-mediated
proteolysis, the ERBB2 signaling pathway and the vascular
endothelial growth factor (VEGF) signaling pathway (Tables I–III).
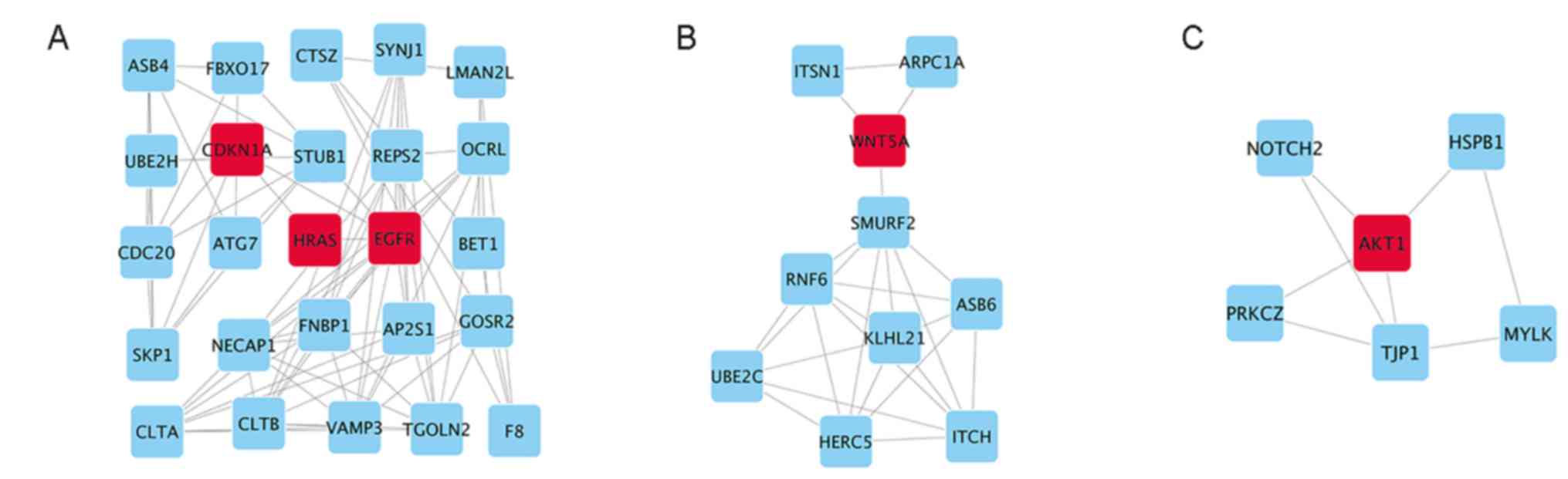 | Figure 5.Top 3 modules from analysis of the
PPI network. (A) Module 1, (B) module 2 and (C) module 3. EGFR,
epidermal growth factor receptor; HRas, Has proto-oncogene GTPase;
Akt1, serine/threonine kinase 1; Wnt5a, Wnt family member 5A;
CDKN1a, cyclin-dependent kinase inhibitor 1A; ASB4, ankyrin repeat
and SOCS box containing 4; FBXO17, F-box protein 17; CTSZ,
cathepsin Z; SYNJ1, synaptojanin 1; LMAN2L, lectin, mannose binding
2 like; UBE2H, ubiquitin conjugating enzyme E2 H; STUB1, STIP1
homology and U-box containing protein 1; REPS2, RALBP1 associated
Eps domain containing 2; OCRL, inositol
polyphosphate-5-phosphatase; ATG7, autophagy related 7; BET1, Bet1
golgi vesicular membrane trafficking protein; SKP1, S-phase kinase
associated protein 1; NECAP1, NECAP endocytosis associated 1;
FNBP1, formin binding protein 1; AP2S1, adaptor related protein
complex 2 sigma 1 subunit; GOSR2, golgi SNAP receptor complex
member 2; CLTA, clathrin light chain A; CLTB, clathrin light chain
B; VAMP3, vesicle associated membrane protein 3; TGOLN2,
trans-golgi network protein 2; F8, coagulation factor VIII; ITSN1,
intersectin 1; ARPC1A, actin related protein 2/3 complex subunit
1A; SMURF2, SMAD specific E3 ubiquitin protein ligase 2; RNF6, ring
finger protein 6; KLHL21, kelch like family member 21; ASB6,
ankyrin repeat and SOCS box containing 6; UBE2C, ubiquitin
conjugating enzyme E2 C; HERC5, HECT and RLD domain containing E3
ubiquitin protein ligase 5; ITCH, itchy E3 ubiquitin protein
ligase; NOTCH2, notch 2; HSPB1, heat shock protein family B (small)
member 1; PRKCZ, protein kinase C ζ; TJP1, tight junction protein
1; MYLK, myosin light chain kinase; PPI, protein-protein
interaction. |
 | Table I.Functional annotation of the
significant module 1. |
Table I.
Functional annotation of the
significant module 1.
| Analysis | Term | Genes | P-value |
|---|
|
GOTERM_BP_DIRECT | Negative regulation
of epidermal growth factor receptor signaling pathway | AP2S1, CLTA,
EGFR |
1.1×10−3 |
|
| ERBB2 signaling
pathway | HRas, STUB1,
EGFR | 1.2
×10−3 |
|
| Positive regulation
of MAPK activity | HRas, EGFR |
7.8×10−2 |
| KEGG PATHWAY | Cell cycle | SKP1, CDC20,
CDKN1a | 3.6
×10−2 |
|
| FoxO signaling
pathway | HRas, CDKN1a,
EGFR | 4.2
×10−2 |
 | Table III.Functional annotation of the
significant module 3. |
Table III.
Functional annotation of the
significant module 3.
| Analysis | Term | Genes | P-value |
|---|
|
GOTERM_BP_DIRECT | Regulation of
apoptotic process | Akt1, HSPB1,
NOTCH2, PRKCZ |
1.9×10−4 |
|
| Negative regulation
of oxidative stress-induced intrinsic apoptotic signaling
pathway | Akt1, HSPB1 |
4.2×10−3 |
|
| Positive regulation
of blood vessel endothelial cell migration | Akt1, HSPB1 |
5.6×10−3 |
| KEGG PATHWAY | Tight junction | Akt1, PPKCZ,
TJP1 |
3.8×10−3 |
|
| VEGF signaling
pathway | Akt1, HSPB1 |
4.3×10−2 |
Verification of the five most
prominent potential metastasis-associated DEGs through analysis of
clinical tumor tissue samples
Every DEG analysed with Cytoscape has an
interactional degree with others. The five hub genes that had the
highest interactional degree of all DEGs were selected as the most
prominent potential metastasis-associated DEGs. RT-qPCR analysis
was performed to corroborate the expression levels of five
potential metastasis-associated genes (EGFR, HRas, Wnt5a, Akt1 and
CDKN1a), identified by the above analyses, in four metastatic and
four primary CRC samples. EGFR, HRas and Akt1 were upregulated DEGs
and their expression levels were identified to be increased, while
Wnt5a and CDKN1a were downregulated DEGs and their expression
levels were decreased when compared with those in the primary
controls. The results of the RT-qPCR analysis are presented in
Fig. 6.
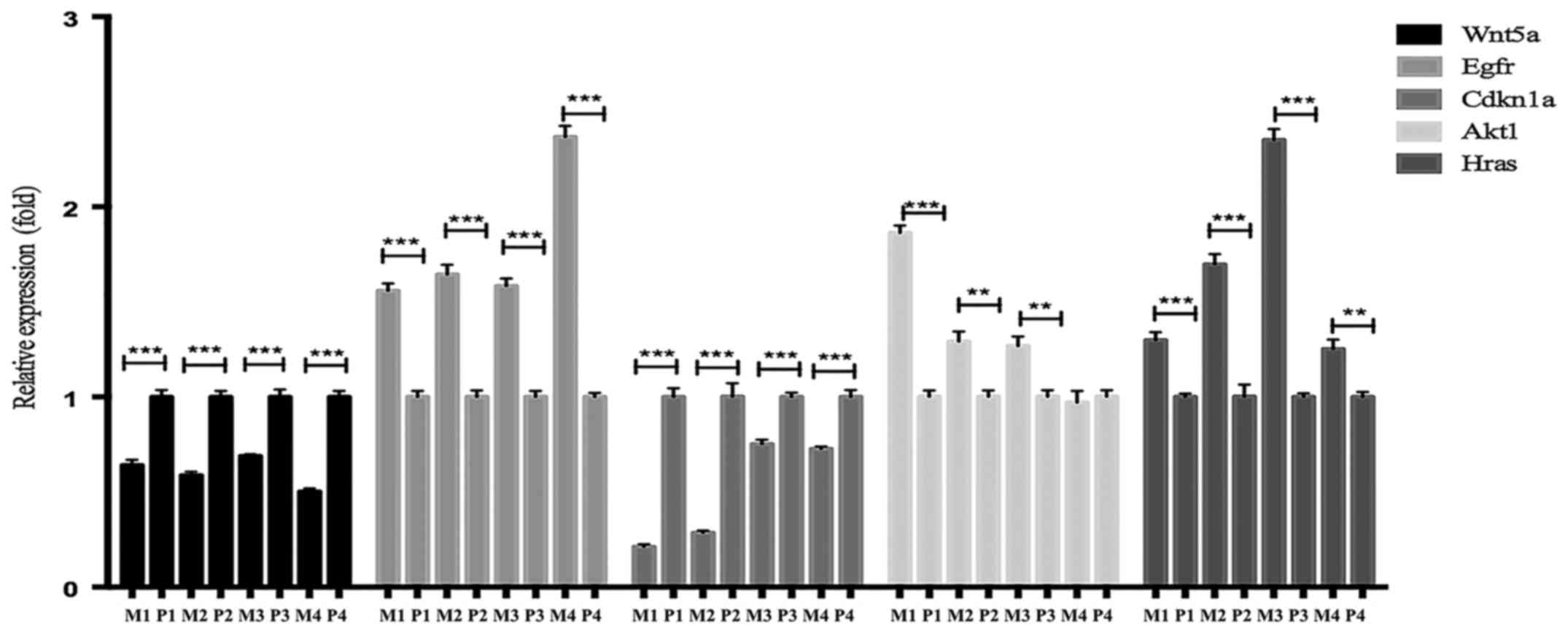 | Figure 6.Analysis of the expression levels of
hub genes in primary and metastatic clinical CRC samples selected
by the interactional degree with other DEGs closely associated with
metastasis via Cytoscape. Relative mRNA expression levels were
measured by RT-qPCR analysis. **P<0.01; ***P<0.001. P,
primary tumor tissues; M, metastatic tumor tissues; EGFR, epidermal
growth factor receptor; HRas, Has proto-oncogene GTPase; Akt1,
serine/threonine kinase 1; Wnt5a, Wnt family member 5A; CDKN1a,
cyclin-dependent kinase inhibitor 1A; CRC, colorectal cancer; DEGs,
differentially expressed genes; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
Discussion
CRC is considered to be a histological heterogeneous
disease, and genetic aberrations have a conspicuous role in its
occurrence, progression and metastasis (19,20).
However, the underlying molecular mechanisms and key genes involved
in the progression or metastasis of CRC have remained to be fully
elucidated. Consequently, exposure of these molecular mechanisms
and key genes is the most critical step in preventing
metastasis.
In the present study, 7,384 DEGs, including 3,949
upregulated and 3,435 downregulated genes, were identified from
available gene expression profiles. GO and KEGG pathway enrichment
analyses subsequently revealed that the DEGs were mainly involved
in the cell cycle, regulation of apoptosis, purine metabolism, DNA
metabolic process, as well as the Hippo and Wnt signaling
pathways.
It is in accord with the current knowledge that the
abnormal regulation of apoptosis and the cell cycle are the causes
for cancer initiation and progression (21,22).
Uric acid is the final breakdown product in the
cycle of nucleotide metabolism. Xanthine oxidoreductase (XOR) acts
as a catalyst when uric acid is produced from xanthine (23). Several prior studies revealed that
the absence of XOR in tumor cells is significantly associated with
poor clinical prognosis and reduced survival rates in breast,
stomach, ovarian, colorectal and non-small cell lung cancer
(24–26). A recent study, independent of all
these variables, revealed that stage IIIA/IIIB CRC patients with
high serum uric acid levels may develop early metastasis (27). The present study indicated that DEGs
were enriched in purine metabolism, which may correlate with CRC
metastasis.
The Hippo pathway participates in the mediation of
cell proliferation, differentiation, growth and apoptosis (28). Its negative regulation is often
discovered in various types of malignant human tumor, demonstrating
that the Hippo signaling pathway is associated with tumor
initiation and progression (29,30).
In the intestine, the deregulation of the Hippo pathway may lead to
neoplastic growth and migration. Furthermore, it has been
demonstrated that interactions among the Hippo, Wnt and EGFR
signaling pathways have an effect on the development of CRC
(31).
In the present study, GO and KEGG pathway enrichment
analysis revealed several possible biological processes and
pathways, which may be involved in the initiation and development
of CRC associated with metastasis.
The results of the module analysis indicated an
association of CRC progression with cell cycle activity, negative
regulation of the apoptotic process, as well as the Wnt, EGFR, VEGF
and FoxO signaling pathways, which is consistent with the above
analyses. In addition, the five leading potential
metastasis-associated DEGs (EGFR, HRas, WNT5a, Akt1 and CDKN1a)
were involved in the three leading modules.
Wnt5a, located at 3p14.2-p21.1, is known as a
non-canonical signaling pathway member of the Wnt family (the Wnt
signaling pathway can be classified as canonical Wnt signaling and
non-canonical Wnt signaling) (32).
Prior studies have observed an elevated expression of Wnt5a in
numerous types of cancer and particularly in aggressive tumor cells
with greater invasive ability than that of the non-aggressive tumor
cells. This delineates the important role of Wnt5a in promoting the
progression of tumors through the non-canonical Wnt signaling
pathway (33). High expression
levels of Wnt5a have been reported to be associated with cancer
progression and metastasis in gastric cancer, osteosarcoma and
glioblastoma (34–36). Of note, the various functions of
Wnt5a rely on disparate cellular contexts in diverse cancer types
(33). Wnt5a also inhibits the
canonical Wnt signaling pathway (37). In breast cancer, Wnt5a has been
reported to inhibit metastasis in vivo, as well as cell
migration and invasion in vitro (38,39).
In addition, downregulation of Wnt5a is significantly linked with
the malignant characteristics of primary invasive breast cancer
(40). However, Wnt5a was reported
to act as a tumor suppressor by obstructing canonical Wnt signaling
in CRC (41). Studies focusing on
the roles of Wnt5a in the metastatic progression of CRC,
determining whether a lower expression of Wnt5a is correlated with
a poorer clinical outcome, have been rarely conducted. In the
present study, it was discovered that Wnt5a was a downregulated DEG
and the expression levels of Wnt5a in primary versus metastatic CRC
were varied. GO and KEGG analysis indicated that Wnt5a was
significantly enriched in Wnt signaling, which may be the mechanism
underlying the higher metastatic rate in CRC patients with
decreased expression levels of Wnt5a. Consequently, targeting Wnt5a
is likely to be a novel therapeutic strategy for metastatic
CRC.
HRas is a member of the Ras superfamily, which
comprises KRas, HRas and NRas (42–44).
Since HRas mutations do not appear frequently as do mutations in
KRas, few studies have been performed with this regard. Several
previous studies have reported that HRas directly activates the
Ras/BRAF/mitogen-activated protein kinase (MAPK) kinase (MEK)/MAPK
and phosphoinositide-3 kinase (PI3K)/Akt/mammalian target of
rapamycin (mTOR) pathway, bypassing EGFR-initiated signaling
(45–47). The activation of HRas is likewise
associated with galectin-1. The inhibitors of galectin-1, including
OTX008, and of mTOR, including rapamycin, may nearly completely
suppress Hras-mutant cancers (48).
A recent study has revealed that HRas mutations have the tendency
to affect patients' outcomes (hazard ratio =0.545, 95% confidence
interval =0.277–1.073, P=0.079) (49). Furthermore, non-mutated HRas has
been identified to be upregulated in CRC cells when compared with
normal adjacent tissues. A positive association has been noted
between HRas expression levels and the up and downstream signaling
factors of EGFR, MEK and extracellular signal-regulated kinase,
indicating that HRas overexpression contributes to the
carcinogenesis of CRC (50).
Despite these studies, HRas expression levels have rarely been
assessed with regard to the metastatic progression of CRC, and the
connection between HRas and metastasis has remained elusive. In the
present study, PPI and module analysis disclosed that HRas is a
significant DEG participating in the metastatic progression of CRC.
KEGG enrichment analysis revealed that the RAS/BRAF/MEK/MAPK
pathway is likely to be an important pathway involved in
metastasis. Subsequent RT-qPCR assays performed on tissues of CRC
patients further confirmed that high expression levels of HRas were
positively associated with CRC metastasis. Although HRas mutations
are rare, the upregulation of HRas implies CRC progression.
Therefore, the prognostic value of this indication calls for
further study.
Proteins of the Akt family, including Akt1, 2 and 3,
possess high homology but are expressed by different genes. Akt1,
known as an essential molecule in the PI3K/Akt signaling pathway,
has a vital role in multiple tumor-associated events, including the
promotion of cancer cell proliferation, metabolism and survival
(51,52). It is implicated in
epithelial-mesenchymal transition and metastasis (53,54).
Häggblad Sahlberg et al (53) have confirmed that knockout of Akt1
attenuates metastasis and tumor cell growth by upregulating
apoptosis and certain metastasis-inhibitory genes in CRC. In
addition, Akt1 directly activates focal adhesion kinase via serine
phosphorylation, facilitating the metastasis of pressure-induced
CRC (55). In the present study,
the PPI network revealed that Akt1 exhibited a high degree of
connectivity. In addition, the present KEGG enrichment analysis
demonstrated that Akt1 participates in regulating the VEGF
signaling pathway, while the latter promotes tumor angiogenesis,
development and metastasis (56,57).
Of note, previous studies have confirmed that inhibition of Akt1
reduces the secretion of VEGF (58). Furthermore, the upregulated
expression levels of Akt1 in certain metastatic versus paired
primary CRC samples were confirmed in the present study by RT-qPCR
assays, which was consistent with the above analyses. In summary,
Akt1 is associated with increased cell invasion and motility in
CRC. For CRC patients, a high level of Akt1 may herald advanced
colorectal tumor progression and unfavorable outcomes.
EGFR may provide cancer cells with self-sufficient
growth ability. EGFR has been reported to be overexpressed in
>80% of all CRC patients (59),
and to be correlated with a higher metastatic risk (60). Simultaneously, an increased
expression of EGFR is associated with tumor progression and a
lessened survival time of patients with metastatic CRC (61). In addition, the RAS/BRAF/MEK/MAPK
and the PI3K/Akt/mTOR pathway, the two major signaling cascades of
the EGFR signaling pathway, are activated by phosphorylated EGFR
and have an indispensable role in the progression of CRC (10). EGFR-targeting drugs are the
first-line therapy and a powerful strategy for treating CRC
patients without Ras mutations based on their molecular profiles
(62). In the present study,
RT-qPCR assays revealed that the expression levels of EGFR were
significantly increased in metastatic tissues, implying the
possible involvement of EGFR in CRC metastasis.
The CDKN1a gene, which encodes p21, a protein
regulating anti-growth signals, is located on chromosome 6 (6p21.2)
(63). The expression levels of
CDKN1a, a key intermediary agent of the p53 response, are mediated
by p53 in response to innumerable stress-stimulating factors. It
then binds to and blocks several CDKs, restraining cell cycle
progression at the G1- and S-phase (64–66).
Suppression of CDKN1a leads to the transformation of colorectal
adenoma into a malignant tumor and likewise accelerates the
proliferation and metastasis of CRC cells (67). Furthermore, downregulation of CDKN1a
has been confirmed to be associated with venous involvement, as
well as lymph node and liver metastasis (68,69).
The present study, consistent with the abovementioned studies,
implied that CDKN1a may be a key DEG in the metastasis of CRC.
Targeting this gene may be a novel therapeutic strategy.
In conclusion, the present study identified several
hub genes and key pathways participating in the progression and
metastasis of CRC. These efforts may contribute towards an improved
understanding of the molecular mechanisms underlying the
progression of CRC, and provide potential biomarkers for clinical
surveillance and therapy. However, as the total number of cases
analyzed in the present study was insufficient, there may be some
bias; further molecular biology experiments are required to further
confirm the specific functions of these identified genes involved
in CRC metastasis in the future.
Acknowledgements
Not applicable.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
DEGs
|
differentially expressed genes
|
|
GEO
|
Gene Expression Omnibus
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
STRING
|
Search Tool the Retrieval of
Interacting Genes
|
|
DAVID
|
Database for Annotation Visualization
and Integrated Discovery
|
|
GO
|
Gene Ontology
|
|
PPI
|
protein-protein interaction
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
EGFR
|
epidermal growth factor receptor
|
|
HRas
|
Has proto-oncogene GTPase
|
|
Akt1
|
serine/threonine kinase 1
|
|
Wnt5a
|
Wnt family member 5A
|
|
CDKN1a
|
cyclin-dependent kinase inhibitor
1A
|
|
XOR
|
xanthine oxidoreductase
|
References
|
1
|
Choi Y, Sateia HF, Peairs KS and Stewart
RW: Screening for colorectal cancer. Semin Oncol. 44:34–44. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Manku G, Hueso A, Brimo F, Chan P,
Gonzalez-Peramato P, Jabado N, Gayden T, Bourgey M, Riazalhosseini
Y and Culty M: Changes in the expression profiles of claudins
during gonocyte differentiation and in seminomas. Andrology.
4:95–110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Basu A, Seth S, Arora K and Verma M:
Evaluating estradiol levels in male patients with colorectal
carcinoma. J Clin Diagn Res. 9:BC08–BC10. 2015.PubMed/NCBI
|
|
5
|
Ciombor KK, Wu C and Goldberg RM: Recent
therapeutic advances in the treatment of colorectal cancer. Annu
Rev Med. 66:83–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao
X, Jia W and Huang J: Decreased expression of miR-218 is associated
with poor prognosis in patients with colorectal cancer. Int J Clin
Exp Pathol. 6:2904–2911. 2013.PubMed/NCBI
|
|
7
|
Zhou C, Cui F, Li J, Wang D, Wei Y, Wu Y,
Wang J, Zhu H and Wang S: miR-650 represses high-risk
non-metastatic colorectal cancer progression via inhibition of
AKT2/GSK3β/E-cadherin pathway. Oncotarget. 8:49534–49547.
2017.PubMed/NCBI
|
|
8
|
De Rosa M, Pace U, Rega D, Costabile V,
Duraturo F, Izzo P and Delrio P: Genetics, diagnosis and management
of colorectal cancer (Review). Oncol Rep. 34:1087–1096. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamasaki M, Takemasa I, Komori T, Watanabe
S, Sekimoto M, Doki Y, Matsubara K and Monden M: The gene
expression profile represents the molecular nature of liver
metastasis in colorectal cancer. Int J Oncol. 30:129–138.
2007.PubMed/NCBI
|
|
10
|
Hagland HR, Berg M, Jolma IW, Carlsen A
and Søreide K: Molecular pathways and cellular metabolism in
colorectal cancer. Dig Surg. 30:12–25. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao M, Zhong A, Patel N, Alur C and Vyas
D: High throughput RNA sequencing utility for diagnosis and
prognosis in colon diseases. World J Gastroenterol. 23:2819–2825.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Serratì S, De Summa S, Pilato B, Petriella
D, Lacalamita R, Tommasi S and Pinto R: Next-generation sequencing:
Advances and applications in cancer diagnosis. Onco Targets Ther.
9:7355–7365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Provenzani A, Fronza R, Loreni F, Pascale
A, Amadio M and Quattrone A: Global alterations in mRNA polysomal
recruitment in a cell model of colorectal cancer progression to
metastasis. Carcinogenesis. 27:1323–1333. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gene Ontology Consortium, . The Gene
Ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D326.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gene Ontology Consortium, . Gene Ontology
Consortium: Going forward. Nucleic Acids Res. 43(D1): D1049–D1056.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28(1): 27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Köhler S, Bauer S, Horn D and Robinson PN:
Walking the interactome for prioritization of candidate disease
genes. Am J Hum Genet. 82:949–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aran V, Victorino AP, Thuler LC and
Ferreira CG: Colorectal Cancer: Epidemiology, disease mechanisms
and interventions to reduce onset and mortality. Clin Colorectal
Cancer. 15:195–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taylor-Weiner A, Zack T, O'Donnell E,
Guerriero JL, Bernard B, Reddy A, Han GC, AlDubayan S, Amin-Mansour
A, Schumacher SE, et al: Genomic evolution and chemoresistance in
germ-cell tumours. Nature. 540:114–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang YT, Tseng HC, Huang CC, Chen YP,
Chiang HC and Chou FP: Relative down-regulation of apoptosis and
autophagy genes in colorectal cancer. Eur J Clin Invest. 41:84–92.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perez R, Wu N, Klipfel AA and Beart RW Jr:
A better cell cycle target for gene therapy of colorectal cancer:
Cyclin G. J Gastrointest Surg. 7:884–889. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fini MA, Elias A, Johnson RJ and Wright
RM: Contribution of uric acid to cancer risk, recurrence, and
mortality. Clin Transl Med. 1:162012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ikegami T, Natsumeda Y and Weber G:
Decreased concentration of xanthine dehydrogenase (EC 1.1.1.204) in
rat hepatomas. Cancer Res. 46:3838–3841. 1986.PubMed/NCBI
|
|
25
|
Sun AS and Cederbaum AI: Oxidoreductase
activities in normal rat liver, tumor-bearing rat liver, and
hepatoma HC-252. Cancer Res. 40:4677–4681. 1980.PubMed/NCBI
|
|
26
|
Tanriverdi O, Cokmert S, Oktay E, Pilanci
KN, Menekse S, Kocar M, Sen CA, Avci N, Akman T, Ordu C, et al:
Prognostic significance of the baseline serum uric acid level in
non-small cell lung cancer patients treated with first-line
chemotherapy: A study of the Turkish Descriptive Oncological
Researches Group. Med Oncol. 31:2172014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cetin AO, Omar M, Calp S, Tunca H, Yimaz
N, Ozseker B and Tanriverdi O: Hyperuricemia at the time of
diagnosis is a factor for poor prognosis in patients with stage II
and III colorectal cancer (uric acid and colorectal cancer). Asian
Pac J Cancer Prev. 18:485–490. 2017.PubMed/NCBI
|
|
28
|
Zhao B, Li L, Lei Q and Guan KL: The
Hippo-YAP pathway in organ size control and tumorigenesis: An
updated version. Genes Dev. 24:862–874. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harvey KF, Zhang X and Thomas DM: The
Hippo pathway and human cancer. Nat Rev Cancer. 13:246–257. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu H, Jiang D, Chi F and Zhao B: The
Hippo pathway regulates stem cell proliferation, self-renewal, and
differentiation. Protein Cell. 3:291–304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wierzbicki PM and Rybarczyk A: The Hippo
pathway in colorectal cancer. Folia Histochem Cytobiol. 53:105–119.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Serra R, Easter SL, Jiang W and Baxley SE:
Wnt5a as an effector of TGFβ in mammary development and cancer. J
Mammary Gland Biol Neoplasia. 16:157–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Endo M, Nishita M, Fujii M and Minami Y:
Insight into the role of Wnt5a-induced signaling in normal and
cancer cells. Int Rev Cell Mol Biol. 314:117–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu B, Wang Q, Wang YA, Hua S, Sauvé CG,
Ong D, Lan ZD, Chang Q, Ho YW, Monasterio MM, et al: Epigenetic
activation of WNT5A drives glioblastoma stem cell differentiation
and invasive growth. Cell. 167:1281–1295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kurayoshi M, Oue N, Yamamoto H, Kishida M,
Inoue A, Asahara T, Yasui W and Kikuchi A: Expression of Wnt-5a is
correlated with aggressiveness of gastric cancer by stimulating
cell migration and invasion. Cancer Res. 66:10439–10448. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yong BC, Lu JC, Xie XB, Su Q, Tan PX, Tang
QL, Wang J, Huang G, Han J, Xu HW, et al: LDOC1 regulates Wnt5a
expression and osteosarcoma cell metastasis and is correlated with
the survival of osteosarcoma patients. Tumour Biol.
39:10104283176911882017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mikels AJ and Nusse R: Purified Wnt5a
protein activates or inhibits beta-catenin-TCF signaling depending
on receptor context. PLoS Biol. 4:e1152006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Prasad CP, Chaurasiya SK, Axelsson L and
Andersson T: WNT-5A triggers Cdc42 activation leading to an ERK1/2
dependent decrease in MMP9 activity and invasive migration of
breast cancer cells. Mol Oncol. 7:870–883. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Säfholm A, Tuomela J, Rosenkvist J, Dejmek
J, Härkönen P and Andersson T: The Wnt-5a-derived hexapeptide
Foxy-5 inhibits breast cancer metastasis in vivo by targeting cell
motility. Clin Cancer Res. 14:6556–6563. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jönsson M, Dejmek J, Bendahl PO and
Andersson T: Loss of Wnt-5a protein is associated with early
relapse in invasive ductal breast carcinomas. Cancer Res.
62:409–416. 2002.PubMed/NCBI
|
|
41
|
Ying J, Li H, Yu J, Ng KM, Poon FF, Wong
SC, Chan AT, Sung JJ and Tao Q: WNT5A exhibits tumor-suppressive
activity through antagonizing the Wnt/β-catenin signaling, and is
frequently methylated in colorectal cancer. Clin Cancer Res.
14:55–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Elbjeirami WM and Sughayer MA: KRAS
mutations and subtyping in colorectal cancer in Jordanian patients.
Oncol Lett. 4:705–710. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nakano H, Yamamoto F, Neville C, Evans D,
Mizuno T and Perucho M: Isolation of transforming sequences of two
human lung carcinomas: Structural and functional analysis of the
activated c-K-ras oncogenes. Proc Natl Acad Sci USA. 81:71–75.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Santos E, Martin-Zanca D, Reddy EP,
Pierotti MA, Della Porta G and Barbacid M: Malignant activation of
a K-ras oncogene in lung carcinoma but not in normal tissue of the
same patient. Science. 223:661–664. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Espada J, Pérez-Moreno M, Braga VM,
Rodriguez-Viciana P and Cano A: H-Ras activation promotes
cytoplasmic accumulation and phosphoinositide 3-OH kinase
association of beta-catenin in epidermal keratinocytes. J Cell
Biol. 146:967–980. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mendoza MC, Er EE and Blenis J: The
Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends
Biochem Sci. 36:320–328. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rodriguez-Viciana P, Warne PH,
Vanhaesebroeck B, Waterfield MD and Downward J: Activation of
phosphoinositide 3-kinase by interaction with Ras and by point
mutation. EMBO J. 15:2442–2451. 1996.PubMed/NCBI
|
|
48
|
Michael JV, Wurtzel JG and Goldfinger LE:
Inhibition of galectin-1 sensitizes HRAS-driven tumor growth to
rapamycin treatment. Anticancer Res. 36:5053–5061. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chang YY, Lin PC, Lin HH, Lin JK, Chen WS,
Jiang JK, Yang SH, Liang WY and Chang SC: Mutation spectra of RAS
gene family in colorectal cancer. Am J Surg. 212:537–544. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Feng J, Hua F, Shuo R, Chongfeng G,
Huimian X, Nakajima T, Subao W and Tsuchida N: Upregulation of
non-mutated H-ras and its upstream and downstream signaling
proteins in colorectal cancer. Oncol Rep. 8:1409–1413.
2001.PubMed/NCBI
|
|
51
|
Riggio M, Perrone MC, Polo ML, Rodriguez
MJ, May M, Abba M, Lanari C and Novaro V: AKT1 and AKT2 isoforms
play distinct roles during breast cancer progression through the
regulation of specific downstream proteins. Sci Rep. 7:442442017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hofbauer SW, Krenn PW, Hofbauer Piñόn J,
Pucher S, Asslaber D, Egle A, Hartmann TN and Greil R: The AKT1
isoform plays a dominant role in the survival and chemoresistance
of chronic lymphocytic leukaemia cells. Br J Haematol. 172:815–819.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sahlberg Häggblad S, Mortensen AC, Haglöf
J, Engskog MK, Arvidsson T, Pettersson C, Glimelius B, Stenerlöw B
and Nestor M: Different functions of AKT1 and AKT2 in molecular
pathways, cell migration and metabolism in colon cancer cells. Int
J Oncol. 50:5–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Irie HY, Pearline RV, Grueneberg D, Hsia
M, Ravichandran P, Kothari N, Natesan S and Brugge JS: Distinct
roles of Akt1 and Akt2 in regulating cell migration and
epithelial-mesenchymal transition. J Cell Biol. 171:1023–1034.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang S and Basson MD: Akt directly
regulates focal adhesion kinase through association and serine
phosphorylation: Implication for pressure-induced colon cancer
metastasis. Am J Physiol Cell Physiol. 300:C657–C670. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Papadimitriou K, Rolfo C, Dewaele E, VanDe
Wiel M, Van den Brande J, Altintas S, Huizing M, Specenier P and
Peeters M: Incorporating anti-VEGF pathway therapy as a continuum
of care in metastatic colorectal cancer. Curr Treat Options Oncol.
16:182015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xu WW, Li B, Lam AK, Tsao SW, Law SY, Chan
KW, Yuan QJ and Cheung AL: Targeting VEGFR1- and VEGFR2-expressing
non-tumor cells is essential for esophageal cancer therapy.
Oncotarget. 6:1790–1805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang H, Zhang C, Ning Z, Xu L, Zhu X and
Meng Z: Bufalin enhances anti-angiogenic effect of sorafenib via
AKT/VEGF signaling. Int J Oncol. 48:1229–1241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Salomon DS, Brandt R, Ciardiello F and
Normanno N: Epidermal growth factor-related peptides and their
receptors in human malignancies. Crit Rev Oncol Hematol.
19:183–232. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mayer A, Takimoto M, Fritz E, Schellander
G, Kofler K and Ludwig H: The prognostic significance of
proliferating cell nuclear antigen, epidermal growth factor
receptor, and mdr gene expression in colorectal cancer. Cancer.
71:2454–2460. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Srivatsa S, Paul MC, Cardone C, Holcmann
M, Amberg N, Pathria P, Diamanti MA, Linder M, Timelthaler G,
Dienes HP, et al: EGFR in tumor-associated myeloid cells promotes
development of colorectal cancer in mice and associates with
outcomes of patients. Gastroenterology. 153:178–190. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bardelli A and Siena S: Molecular
mechanisms of resistance to cetuximab and panitumumab in colorectal
cancer. J Clin Oncol. 28:1254–1261. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Georgakilas AG, Martin OA and Bonner WM:
p21: A two-faced genome guardian. Trends Mol Med. 23:310–319. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
el-Deiry WS, Harper JW, O'Connor PM,
Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill
DE and Wang Y: WAF1/CIP1 is induced in p53-mediated G1 arrest and
apoptosis. Cancer Res. 54:1169–1174. 1994.PubMed/NCBI
|
|
65
|
el-Deiry WS, Tokino T, Velculescu VE, Levy
DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW and
Vogelstein B: WAF1, a potential mediator of p53 tumor suppression.
Cell. 75:817–825. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Harper JW, Adami GR, Wei N, Keyomarsi K
and Elledge SJ: The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yasui W, Akama Y, Yokozaki H, Semba S,
Kudo Y, Shimamoto F and Tahara E: Expression of p21WAF1/CIP1 in
colorectal adenomas and adenocarcinomas and its correlation with
p53 protein expression. Pathol Int. 47:470–477. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Matsushita K, Kobayashi S, Kato M, Itoh Y,
Okuyama K, Sakiyama S and Isono K: Reduced messenger RNA expression
level of p21 CIP1 in human colorectal carcinoma tissues and its
association with p53 gene mutation. Int J Cancer. 69:259–264. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Schwandner O, Bruch HP and Broll R:
Prognostic significance of p21 and p27 protein, apoptosis, clinical
and histologic factors in rectal cancer without lymph node
metastases. Eur Surg Res. 34:389–396. 2002. View Article : Google Scholar : PubMed/NCBI
|