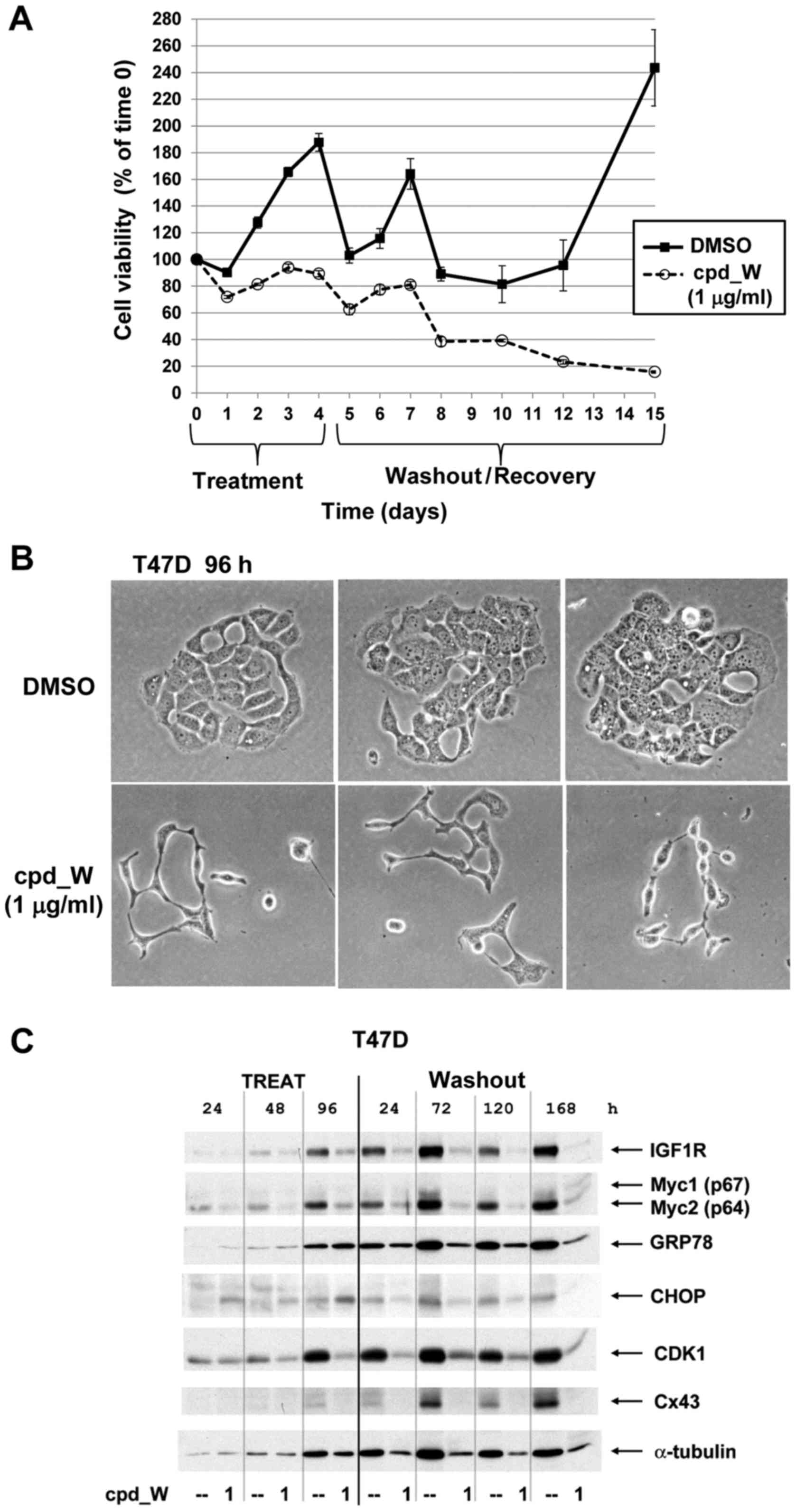
Introduction
Internal ribosome entry sites (IRESs) are
translation-regulatory features found in association with the mRNAs
encoding many cancer-relevant proteins (1–2).
IRES-mediated translation is a specialized mode of protein
synthesis which is independently regulated and remains operational
even when the general protein synthesis mediated by ribosome
scanning from the beginning of the mRNA is shut down (Fig. 1A). Malignant cells are particularly
dependent on IRES-mediated translation and exploit this mechanism
to synthesize oncogenic proteins to promote their own survival,
particularly under adverse microenvironmental conditions such as
those to which the tumor is exposed in vivo (3–7).
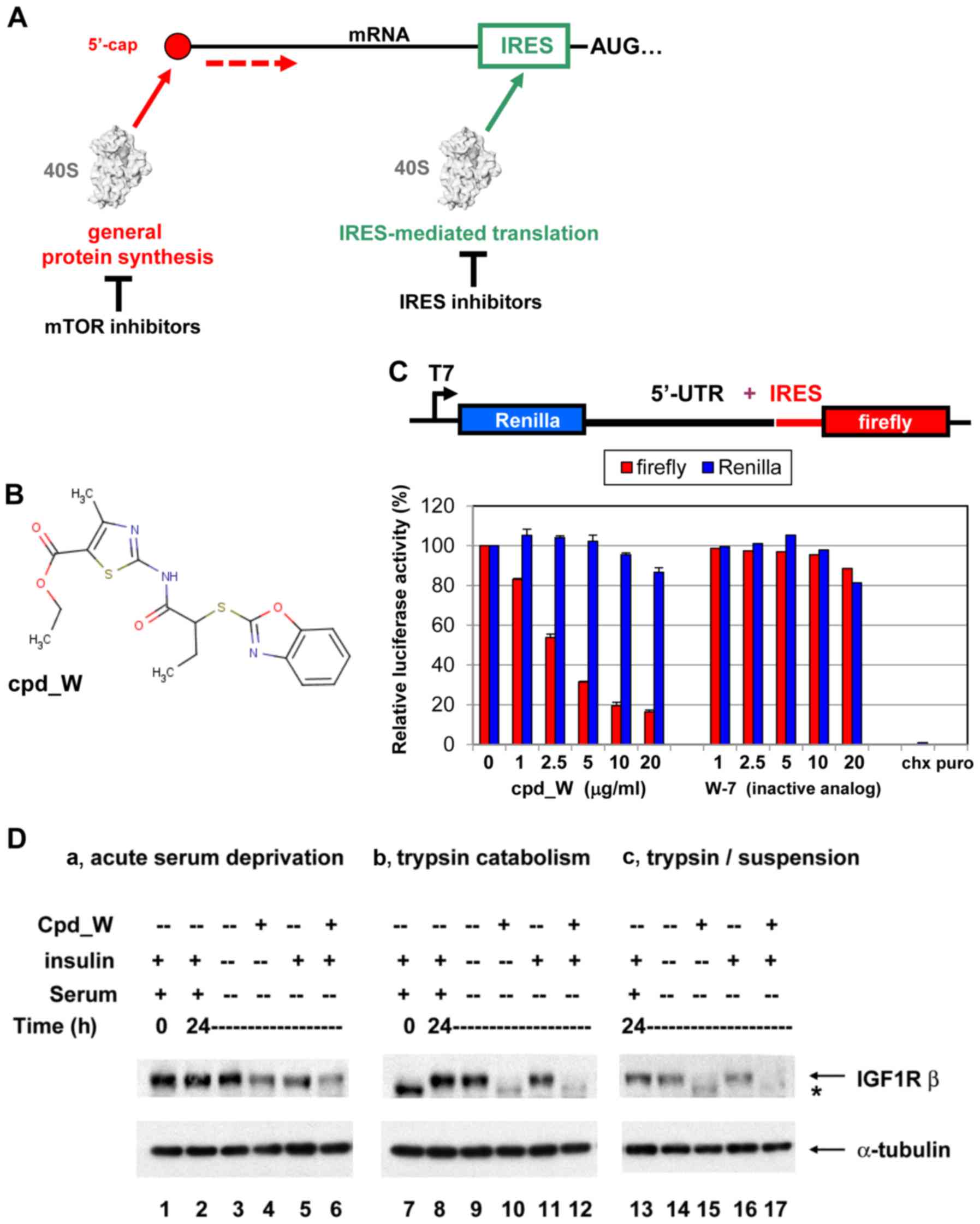 | Figure 1.IRES-mediated translation and IRES
inhibition in a cell-free system and in cells. (A) Diagrammatic
comparison of general protein synthesis to IRES-mediated
translation. General protein synthesis is mediated by cap-dependent
ribosomal scanning from the 5′-end of the mRNA and may be modulated
by mTOR inhibitors. Internal ribosome entry sites (IRESs) allow the
40S ribosome to engage the mRNA at a position much closer (in many
cases immediately adjacent to) the AUG initiation codon.
IRES-mediated translation is independently regulated and serves as
a fail-safe mechanism ensuring the synthesis of proteins most
critical for cell survival. (B) Structure of IRES inhibitor lead
compound W (cpd_W): Ethyl
2-{[2-(1,3-benzoxazol-2-ylthio)butanoyl]amino}-4-methyl-1,3-thiazole-5-carboxylate,
MW 405. (C) In vitro translation assays: Rabbit reticulocyte
lysate was programmed with a bicistronic reporter RNA in which
translation of the second cistron (firefly luciferase coding
sequence) is mediated by the IGF1R IRES, while translation of the
first cistron (Renilla luciferase coding sequence) is
mediated by ribosomal scanning. IRES inhibitor cpd_W (or vehicle
control) was included in the reaction in increasing concentrations
as indicated. The result is indicative of selective inhibition of
IRES-mediated translation. A structural analog of cpd_W (W-7) in
which a single atom has been modified (converting the benzoxazole
to a benzimidazole) was completely inactive in this assay,
indicative of the chemical specificity of IRES inhibition.
Cycloheximide (5 µg/ml, chx) and puromycin (250 µg/ml, puro) were
included as reference standards for non-specific translational
inhibition (far right). (D) IRES inhibitor cpd_W completely blocked
de novo synthesis of IGF1R in breast tumor cells under
adverse conditions (serum-deprivation, loss of adhesion) relevant
to the microenvironment of the tumor. T47D breast tumor cells were
seeded in 6-well plates and allowed 48 h to recover and resume
proliferation, then incubated in the presence of IRES inhibitor
cpd_W (10 µg/ml) or vehicle control (0.1% DMSO) as indicated. The
cells were simultaneously subjected to acute serum deprivation
(0.5% fetal calf serum, no added insulin) to increase dependence on
IRES-mediated translation. After 24 h, the cells were harvested and
whole cell lysates prepared, equivalent aliquots separated by
SDS-PAGE and immunoblotted for IGF1R-β and α-tubulin. In lanes
7–12, the cells were trypsinized and seeded into 6-well plates and
immediately incubated in the presence of IRES-inhibitor cpd_W or
vehicle control as indicated. Robust regeneration of
trypsin-catabolized IGF1R was observed within 24 h in
vehicle-treated cells, however, this was completely blocked in the
presence of cpd_W (10 µg/ml as shown; IC50, 2 µg/ml).
The asterisk (*) marks the position of trypsin-catabolized IGF1R.
In lanes 13–17, the cells were treated as described for lanes 7–12,
except that following trypsinization, cells were transferred to
low-adherence plates, forcing cells to adapt to a state of
anchorage-independence. The results confirmed the activity of cpd_W
against the endogenous IRES in genetically-unmodified tumor cells.
Similar results were obtained with IRES inhibitor lead cpd_P
(11). |
The inhibition of protein synthesis has been proven
to be a highly effective therapeutic strategy. Many of our
antibacterial antibiotics target the prokaryotic ribosome and
interfere selectively with its function (8,9).
Anticancer agents targeting the mTOR function and thereby
inhibiting conventional cap-dependent translation are showing
considerable promise in ongoing clinical trials (10). Given its role in promoting
tumor-cell survival under adverse conditions, there is reason to
anticipate that selective inhibition of IRES-mediated translation
could represent a highly effective anticancer strategy.
Our lab has been working on the development of a
series of inhibitors of IRES-mediated translation, which could be
used to probe the contribution of IRES-mediated translation to
cancer pathogenesis and to assess the potential use of IRES
inhibition as a therapeutic intervention. We recently described
(11) the first of three
IRES-inhibitor lead compounds (cpd_P), comparing and contrasting
the effects on the IGF1R and MYC IRESs. We found that sustained
IRES inhibition induced terminal differentiation in triple-negative
breast cancer and other highly undifferentiated tumor types
(12).
In the present study we examined the consequences of
IRES inhibition on models of ER-positive human breast cancer, using
the second of these lead compounds (cpd_W), which was found to be
particularly effective against these cells. We observed that IRES
inhibition forced these tumor cells to abandon the undifferentiated
phenotype, resulting in terminal differentiation, incapacitation,
or death of the malignant cells. Marked changes in ERα, CDK1,
connexin 43 and Myc were observed which correlated with these
detrimental outcomes. In addition, potential biomarkers of the
IRES-mediated translation were characterized which made it possible
to directly visualize these phenotypic transitions. These findings
have important implications for the biology of ER-positive breast
cancer and reveal what may be accomplished by modulating
IRES-mediated translation.
Materials and methods
Reagents and antibodies
IRES inhibitor lead compound W [cpd_W: Ethyl
2-{(2-(1,3-benzoxazol-2-ylthio)butanoyl)amino}-4-methyl-1,3-thiazole-5-carboxylate,
MW 405] was originally identified from a high throughput screen of
135,000 compounds using T47D cells genetically engineered with a
bicistronic construct containing the human IGF1R IRES (11). The compound was purchased from
Chembridge Corporation (San Diego, CA, USA), subsequently
resynthesized (97.9% purity) and the new stock precisely
recapitulated the biological activity of the original stock.
Cycloheximide and puromycin were obtained from Sigma-Aldrich (St.
Louis, MO, USA).
The primary antibodies used in these experiments are
listed in Table I. The secondary
antibodies for indirect immunofluorescence staining were AlexaFluor
488 or 594-conjugated goat anti-rabbit IgG, anti-mouse IgG, or
anti-mouse IgM (for RACK1) (highly cross-adsorbed; Life
Technologies; Thermo Fischer Scientific, Waltham, MA, USA).
4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) was obtained
from Sigma-Aldrich.
 | Table I.Primary antibodies used in the
present study. |
Table I.
Primary antibodies used in the
present study.
| Protein | Clone | Host | Source | Application
(dilution) |
|---|
| IGF1R | N20 | Rabbit | Santa Cruz
Biotechnology Inc. (Dallas, TX, USA) |
| IGF1R | C20 | Rabbit | Santa Cruz
Biotechnology Inc. | WB (1:400) |
| Myc | N262 | Rabbit | Santa Cruz
Biotechnology Inc. | WB (1:400) |
| RACK1 | 20 | Mouse IgM | BD Biosciences (Jan
Jose, CA, USA) | WB (1:2,500), IF
(1:100) |
| ERα | D8H8 | Rabbit mono | Cell Signaling
Technology, Inc. (Danvers, MA, USA) | WB (1:1,000), IF
(1:500) |
| GRP78 | 40 | Mouse | BD Biosciences | WB (1:500), IF
(1:100) |
| ZO-1 | Mid | Rabbit | Life Technologies;
Thermo Fisher Scientific (Waltham, MA, USA) | IF (1:100) |
| E-cadherin | HECD1 | Mouse | Invitrogen; Thermo
Fisher Scientific (Waltham, MA, USA) | WB (1:1,000), IF
(1:200) |
| CLIMP-63 | ab152154 | Rabbit | Abcam (Cambridge,
MA, USA) | IF (1:160) |
| CHOP | L63F7 | Mouse | Cell Signaling
Technology, Inc. | WB (1:1,000) |
| CDK1 | 17 | Mouse | Santa Cruz
Biotechnology Inc. | WB (1:400) |
| Connexin 43
(Cx43) | 3512 | Rabbit | Cell Signaling
Technology, Inc. | WB (1:1,000) |
| α-tubulin | B-5-1-2 | Mouse | Sigma-Aldrich (St.
Louis, MO, USA) | WB (1:4,000), IF
(1:500) |
Cells, cell culture and treatment
conditions
T47D cells were obtained from the American Type
Culture Collection (ATCC, Manassas, VA, USA) and propagated using
standard techniques in RPMI-1640 medium containing 10% fetal calf
serum (FCS) and 10 µg/ml insulin. ZR-75-1 cells were a generous
gift from Dr Patsy Oliver and propagated in RPMI-1640 with 20% FCS.
T47D and ZR-75-1 cells are human ER-positive breast carcinoma cell
lines. T47D cells were used in our initial characterization of the
IGF1R IRES (4) and in the
high-throughput screen for small molecule IRES inhibitors (11). We subsequently found that MYC IRES
was particularly sensitive to chemical inhibition in ZR-75-1 cells
(11).
Population (standard) density assays were performed
by seeding cells in full serum-containing medium, allowing 48 h for
recovery, before initiating treatment with cells at ~75% confluency
(~200,000 cells/cm2). Low (clonogenic)-density assays
were performed by seeding cells in full serum medium at ~700
cells/cm2 and allowing 48 h for recovery before
initiating treatment. cpd_W was solubilized in 100% dimethyl
sulphoxide (DMSO) to a concentration of 10 mg/ml and used
immediately or stored at −20°C. Stock solutions were diluted a
minimum of 1:1,000 in media until final DMSO concentration did not
exceed 0.1%, which was matched in vehicle-only control samples. The
compound was thoroughly dispersed in media before being added to
the cells. Low serum conditions (0.5% FCS, no supplemental insulin)
were used during treatment to simulate suboptimal
microenvironmental conditions to which tumor cells are exposed
in vivo and to increase dependence on IRES-mediated
translation. For the washout and recovery assays, treatment was
terminated by replacing with fresh media (without compound, but
maintaining low serum conditions).
Indirect immunofluorescence staining
and confocal imaging
The cells were seeded in 8-well chamber slides
(Nunc; Nalge Nunc International, Penfield, NY, USA) and allowed 48
h to recover and resume proliferation prior to treatment with cpd_W
or vehicle control (0.1% DMSO) as indicated in the corresponding
Figure legends. The cells were fixed with freshly prepared 2%
paraformaldehyde for 15 min at room temperature, followed by
permeabilization with 0.2% Triton X-100 for 10 min. After washing
in PBS with 75 mM glycine and blocking in 5% normal goat serum, the
primary antibody was added and incubated for 35 min to 1 h at room
temperature. Following two washes in PBS and a 10 min reblocking
step, secondary antibodies (1:200) were added and incubated for
25–45 min. Following two additional PBS washes, nuclei were stained
with DAPI (0.2 µg/ml) and mounted using ProLong Gold (Life
Technologies; Thermo Fischer Scientific).
Images were captured using a Nikon A1 confocal
instrument with 40X 1.3 NA objective (Nikon Corporation, Tokyo,
Japan). Fields were randomly selected for imaging on the basis of
the DAPI staining pattern alone. Paired images of control and
experimental wells were acquired sequentially and all settings
including laser power, PMT voltage and pinhole were held constant
between samples.
Cell viability assays
Viability was assessed based on ATP content
(CellTiter-Glo; Promega Corporation, Madison, WI, USA). Appropriate
negative controls (lysis buffer alone) were subtracted from the
readings.
Western blot analysis
Whole cell lysates were prepared from treated cells
as previously described (11) using
lysis buffer containing 4% SDS and 720 mM 2-mercaptoethanol,
pre-heated to 100°C. For cells at population density, equivalent
aliquots of protein from each sample were separated by SDS-PAGE,
transferred to 0.2 µm nitrocellulose membranes and probed with
antibodies using standard immunoblotting procedures.
For western blot analysis of protein recovered from
cells at clonogenic density, maximally-concentrated whole cell
lysates were prepared by rapid serial transfer of a minimum volume
(100 µl) of lysis buffer to three adjacent replicate wells (each 10
cm2, standard 6-well plates). Western blot analysis of
these samples was performed by loading equivalent aliquots (by
volume) into each lane, without normalizing for total protein
content.
In vitro translation
Standard in vitro translation reactions were
set up using micrococcal nuclease treated rabbit reticulocyte
lysate (Promega) at a final concentration of 50% (vol/vol). An
intermediate aqueous dilution (1:50) of IRES inhibitor stock
solution was prepared to allow for small volume addition to the
translation reaction without excessive (<0.2% final
concentration) DMSO. Reactions were initiated by adding in
vitro transcribed RNA (7 nM final concentration) prepared from
the bicistronic reporter construct pDualIGF1R(951–1040) containing
the core functional IGF1R IRES. Reactions were incubated at 30°C
for 100 min and firefly and Renilla luciferase activities
were assessed using the standard dual luciferase assay
protocol.
Results
Inhibition of IRES-mediated
translation in vitro and in cells
IRES inhibitor lead compound W (cpd_W, Fig. 1B) was initially identified on the
basis of its ability to selectively interfere with the expression
of firefly luciferase from a bicistronic (IRES reporter) vector in
breast tumor cells genetically engineered with this construct
(11). As an even more specific
test of mechanism of action, we titrated cpd_W in a cell-free in
vitro translation assay, using rabbit reticulocyte lysate as a
source of ribosomes, supplemented with amino acids and provided the
bicistronic RNA which had been synthesized and purified in
vitro. The results (Fig. 1C)
indicated that cpd_W selectively inhibited the synthesis of firefly
luciferase (product of the second cistron, mediated by the IRES) in
a concentration-dependent manner, with no significant impact on the
synthesis of Renilla luciferase (product of the first
cistron, translated by conventional ribosome scanning). These
results confirmed the mechanism of action in a simple reconstituted
system in which potentially confounding factors such as
transcription or signal transduction were not in play. The results
also distinguished cpd_W from the universal inhibitors of protein
synthesis cycloheximide or puromycin, which completely block
translation of both cistrons.
In addition to its activity against the IRES
reporter, it was critical that we determined whether cpd_W would
also inhibit the function of the endogenous IGF1R IRES in
untransfected cells. T47D (human ER-positive breast tumor) cells
express IGF1R at a high level. To assess the impact of IRES
inhibition on IGF1R protein level, the cells were incubated with
cpd_W and simultaneously subjected to acute serum deprivation (0.5%
FCS). Limiting access to soluble growth factors in this manner
simulated the adverse microenvironmental conditions tumor cells are
typically exposed to in vivo and caused cells to become more
dependent on IRES-mediated translation. Following 24 h exposure to
cpd_W, ~50% decrease in IGF1R protein is observed (Fig. 1D, lanes 4 and 6). This relatively
slow decline was consistent with the long half-life (>24 h) of
pre-existing IGF1R protein molecules.
We found that the ability of the IRES inhibitor to
block IGF1R synthesis could be more readily ascertained if the
equilibrium was perturbed. When the cells were trypsinized (using
the standard procedure for subculturing cells), all IGF1R molecules
on the surface of the cell were digested, resulting in a complete
shift (downward) on western blot analysis (Fig. 1D, lane 7). This forced the cells to
regenerate the entire population of IGF1R molecules and provided an
opportunity to clearly discriminate whether IGF1R synthesis would
be inhibited by cpd_W. Robust regeneration of full-length IGF1R to
baseline (pre-trypsin) level was observed within 24 h in
vehicle-treated cells (lanes 8,9,11), however, this was completely
blocked in the presence of cpd_W (lanes 10 and 12). Identical
results were obtained when cells were simultaneously deprived of
adhesion (forced anchorage-independence, lanes 13–17). These
results confirmed the activity of cpd_W against the endogenous
IGF1R IRES and matched the findings obtained with our first lead
compound cpd_P (11).
ER-positive breast tumor cells exist
in two major interconvertible phenotypic states
Subsequently, we developed imaging strategies which
may allow us to directly monitor IRES-mediated translation in
native (genetically-unmodified) cells. We anticipated that these
imaging analyses may allow us to distinguish in which cells
IRES-mediated translation is taking place and to correlate changes
in phenotype with changes in translational activity at the level of
the individual cell. Furthermore, we anticipated that such staining
patterns may help to assess the consequences of IRES
inhibition.
We first selected an antibody to IGF1R (N20) which
recognizes the N-terminal region of the protein in denatured form
(i.e. on western blotting) but fails to detect the mature IGF1R
protein at the cell surface. We reasoned that this antibody may
recognize nascently-translated IGF1R at the rough endoplasmic
reticulum where it is synthesized, prior to adoption of its native
three-dimensional conformation and in this manner, may serve as an
indicator of active IRES-mediated translation. At the same time, we
stained the cells with an antibody to RACK1, which is an integral
component of the 40S ribosomal subunit, to mark the sites where
active translation could take place. RACK1 resides near where
interactions with the 5′-untranslated regulatory region of the mRNA
take place (13) and is suspected
of being involved in regulating IRES-mediated translation (14).
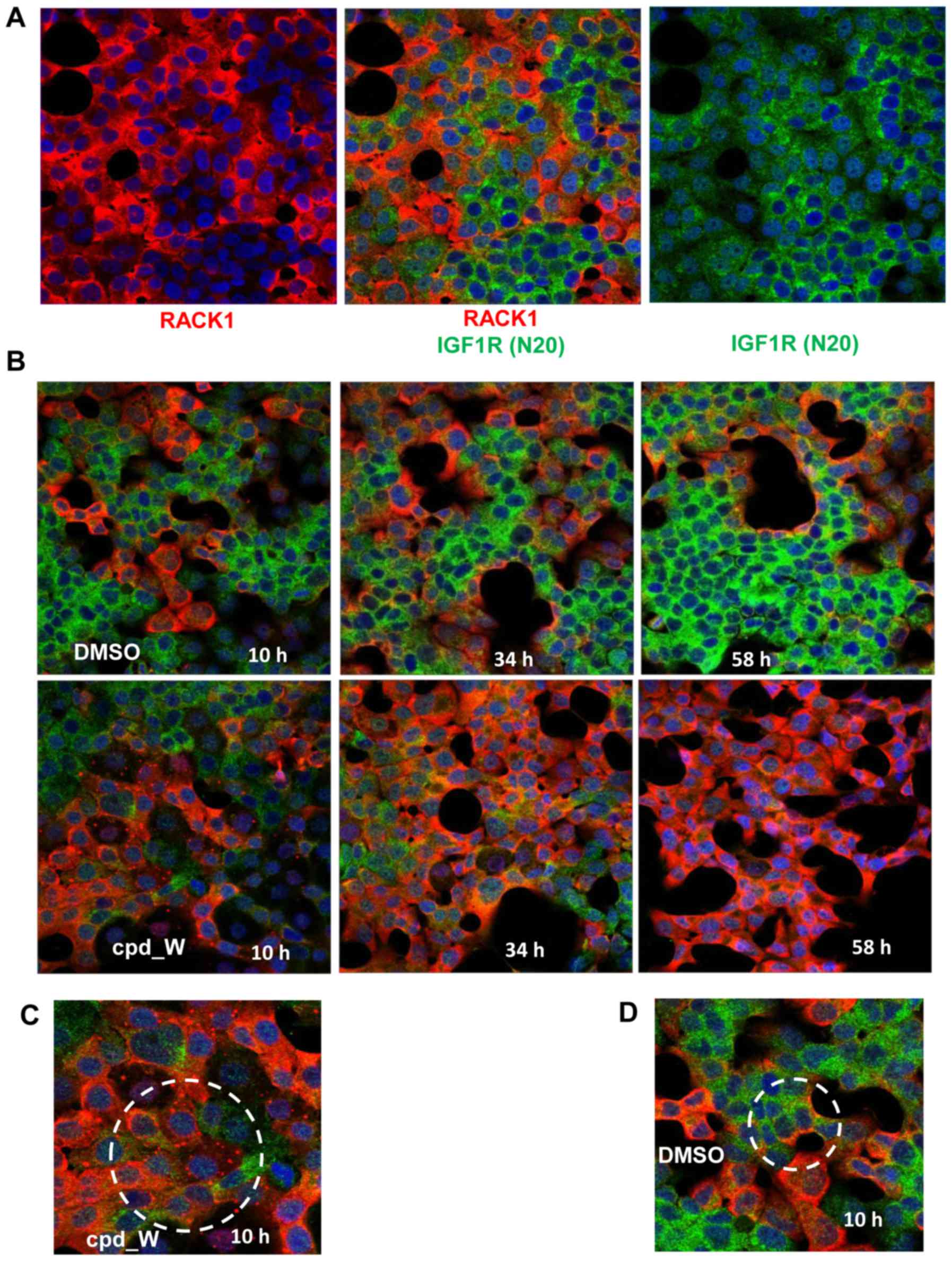
Applying this immunocytochemical staining strategy
to the T47D ER-positive breast tumor cell line, two phenotypically
distinct populations of cells were observed (Fig. 2A). One population of cells displayed
high intensity staining for nascently-translated IGF1R, but very
low intensity of RACK1 immunofluorescence. These cells tend to be
arranged in clusters located within the interior of a mass of
contiguous cells i.e. without exposed lateral surfaces. The other
population of cells exhibited precisely the inverse pattern: High
RACK1 intensity and very low staining for nascently-translated
IGF1R. These cells were consistently found bordering open spaces,
with concave lateral surfaces (planar polarity). The IGF1R N20 and
RACK1 staining patterns were almost perfectly mutually exclusive,
i.e. the majority of cells were either red or green, with
essentially no overlapping.
At baseline, both phenotypes were present. However,
if the cells were subjected to acute serum deprivation, over time a
progressively larger proportion of cells adopted the IGF1R
N20-positive/RACK1-negative (green) phenotype (Fig. 2B). In contrast, cells subjected to
serum deprivation but simultaneously treated with the IRES
inhibitor cpd_W exhibited just the opposite effect, with
essentially all cells shifting to the IGF1R
N20-negative/RACK1-positive (red) phenotype.
These findings suggested that the IGF1R N20-positive
cells represent a relatively undifferentiated population of cells
in which IRES-mediated translation is active, proliferating without
evidence of a higher-ordered structural organization. Transition to
the undifferentiated IRES-active phenotype could be expected to
facilitate cell survival under challenging microenvironmental
conditions. By contrast, the RACK1-positive cells appeared to
represent a more differentiated population of tumor cells, with
planar polarity and growth restraint and little or no IRES
activity. The apparent inverse correlation between RACK1
immunoreactivity and IRES-mediated translation was unexpected.
Western blot analysis revealed no significant change in RACK1
abundance under these conditions (Fig.
5). It rather appeared that the RACK1 epitope was masked when
the ribosomes were actively engaged in IRES-mediated translation.
The RACK1 immunoreactivity returned when IRES-mediated translation
was curtailed in association with the physiological transition to
the more differentiated phenotype, or when the IRES mechanism was
rendered non-functional by the IRES inhibitor.
Early during the incubation with the IRES inhibitor,
numerous cells were observed (circled in Fig. 2C) which contained discrete bright
RACK1-positive foci in place of the diffuse RACK1 cytoplasmic
staining pattern observed at later time-points or in control
fields. These foci were present only transiently and only in
cpd_W-treated cells, indicating that they arose as a direct
consequence of IRES inhibition and may represent sequestered
dysfunctional IRES-ribosome complexes. In addition, it is
noteworthy that, while colocalization of RACK1 and IGF1R N20 was
almost never observed, there were cells in the control fields
(circled in Fig. 2D) exhibiting a
biphenotypic pattern, in which the portion of the cytoplasm
bordering an acellular space was stained distinctly positive for
RACK1 (IRES-off), while the remainder of the cytoplasm (facing the
interior of a cell cluster) stained positive for IGF1R N20
(IRES-active).
Additional biomarkers of the
undifferentiated/IRES-active and differentiated/IRES-off
phenotypes
Subsequently, it was important to identify
additional markers of these two distinct phenotypes, in order to
confirm our findings and further investigate the relationship
between IRES-mediated translation and differentiation status in the
ER-positive breast tumor cells. We examined additional antibodies
to proteins relevant to translation and/or differentiation and
found several which allowed these two phenotypic states to be
readily distinguished.
Foremost among these was estrogen receptor a (ERα,
ESR1; Fig. 3A). Confocal images
revealed that cytoplasmic localization of ERα correlated with the
clustered/undifferentiated IRES-active phenotype, precisely
mirroring the IGF1R N20 staining pattern and reciprocal to RACK1.
It appeared that ERα was completely excluded from the nuclei of
these undifferentiated cells, where it was prevented from
transcriptionally programming cells to differentiate. IRES
inhibition was accompanied not only by an increase in RACK1
immunoreactivity, but also by a marked decrease in overall
intensity of ERα staining. ERα itself is known to be translated via
an IRES (15), thus these results
suggested that the ESR1 IRES may be sensitive to cpd_W.
 | Figure 3.Biomarkers of the
undifferentiated/IRES-active and differentiated/IRES-off
phenotypes. (A) T47D cells were treated with IRES inhibitor cpd_W
as described in Fig. 2. At the 48 h
time-point, the cells were fixed and stained for ERα (D8H8) and
RACK1. (B) T47D cells were treated with cpd_W for 15, 24 or 48 h.
Exposure to cpd_W was terminated by replacing with fresh media and
cells were fixed and stained for ERα and RACK1 at the 48 h
time-point. (C) ZR-75-1 cells were treated and stained for ERα and
RACK1 as abovedescribed in (A). (D-J) T47D (D, F, H and J) or
ZR-75-1 (E, G and I) cells were treated with cpd_W or vehicle
control for 48 h, then stained for IGF1R N20 and GRP78 (D and E),
RACK1 and ZO-1 (F and G), RACK1 and CLIMP-63 (H), ERα and
E-cadherin (I), or IGF1R N20 and α-tubulin (J). |
The progressive transition of cells from the
undifferentiated IRES-active (cytoplasmic ERα-positive) phenotype
to the differentiated IRES-off (RACK1-positive) state is displayed
in Fig. 3B, where the parallel gain
in structural organization, with the cells distinctively arranged
to surround open spaces, is also evident. With-24 h exposure to the
IRES inhibitor, the majority of cells became RACK1-positive and ERα
had shifted almost entirely to the nucleus. Following 48 h
continuous IRES inhibition, only a small quantity of ERα remained.
A similar pattern, including shifting of ERα to the nucleus upon
differentiation, was observed in another ER-positive breast tumor
cell line, ZR-75-1 (Fig. 3C).
GRP78 (HSPA5, BiP) is a prosurvival molecule
integrally involved in translation quality control. GRP78 nuclear
immunoreactivity exhibited a strong correlation with the
differentiated IRES-off state (Fig. 3D
and E). While the vehicle-treated field included
undifferentiated IGF1R N20-positive cells in which nuclear GRP78
was not observed, following treatment with cpd_W, the cells became
uniformly negative for IGF1R N20 and positive for nuclear GRP78.
De novo appearance of the tight junction protein ZO-1 within
the nuclei of the cpd_W-treated cells was also observed (Fig. 3F and G). The translocation of all
three of these molecules: ERα, GRP78 and ZO-1 to the nucleus is
consistent with the nuclear (transcriptional) dominance of the
differentiated state.
The images displayed in Fig. 3H confirmed that RACK1 colocalized
extensively with CLIMP-63 (CKAP4), an established marker of the
rough endoplasmic reticulum (16,17),
consistent with its tight association with ribosomes and confirming
that RACK1 staining (when its epitope is not masked) marked the
sites where translation could take place. Notably this is the only
pair of antibodies used in the present studies for which any
significant degree of colocalization was observed, further
highlighting the mutual exclusivity of the two distinct phenotypes
displayed by the ER-positive breast tumor cells.
Marked changes in the distribution of E-cadherin and
reorganization of the α-tubulin cytoskeleton were observed
accompanying the differentiation induced by the inhibition of
IRES-mediated translation (Fig. 3I and
J). Notably, these phenotypic changes persisted even after the
compound was removed from the media, indicating that a finite
period during which synthesis of critical IRES-driven proteins was
blocked may have a long-lasting impact on the phenotype of those
cells.
ER-positive breast tumor cells
tolerate forced differentiation however reproductive capacity is
severely compromised by IRES inhibition
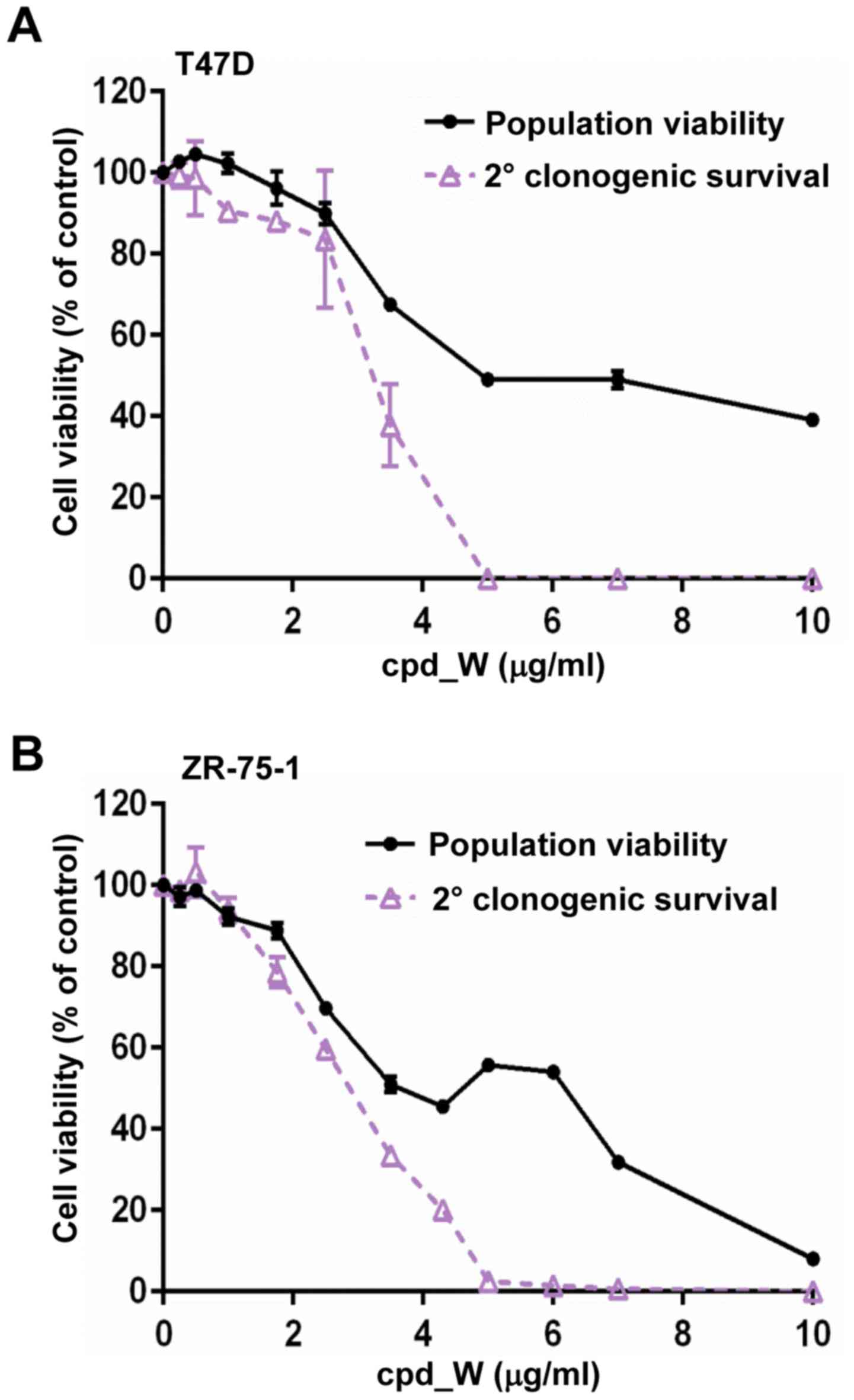
Although marked phenotypic changes were observed in
the ER-positive breast tumor cells subjected to IRES inhibition, it
did not appear that cell viability was markedly affected. To
document this, cell viability assays were performed on cells
following 96 h exposure to cpd_W at varying concentrations ranging
from <1 to 10 µg/ml. The results (black lines) obtained for both
T47D (Fig. 4A) and ZR-75-1 cell
lines (Fig. 4B) indicated that
these cells tolerated prolonged IRES inhibition with relatively
modest decreases in the number of viable cells.
Subsequently we examined whether the forced
transition to the differentiated phenotype brought about by the
inhibition of IRES-mediated translation may be irreversible, i.e.
whether the tumor cells may be hindered in their ability to
transition back to the undifferentiated state and resume
proliferation. To examine whether the reproductive capacity of the
breast tumor cells subjected to IRES inhibition may have been
altered, secondary clonogenic survival assays were performed.
Following 96-h treatment and 24-h recovery period in the absence of
compound, the cells were trypsinized and reseeded at low density.
The cells were provided a 6-day period to proliferate (in the
absence of compound) and then cell viability was assessed (Fig. 4A and B; purple lines). The cells
which had been treated with cpd_W at <2.5 µg/ml demonstrated a
repopulating ability that very closely matched the viability
readout assessed immediately after treatment (i.e. minimal
separation of the black and purple curves). However, a sharp
decline in the reproductive capacity was observed at 3.5 µg/ml
cpd_W and for the cells which had been treated with >5 µg/ml
cpd_W, the ability to repopulate the well was almost completely
lost (<2.5% of control). The same pattern was observed in both
T47D and ZR-75-1 cell lines. Thus, the secondary clonogenic
survival assay revealed the severe detrimental impact of IRES
inhibition on the ER-positive breast tumor cell population, which
was not readily apparent from the initial viability readout. In
essence, these cells were metabolically alive, yet reproductively
dead at the end of the treatment period, consistent with terminal
differentiation.
Alterations to critical
phenotype-determining proteins in ER-positive breast tumor cells
subjected to IRES inhibition
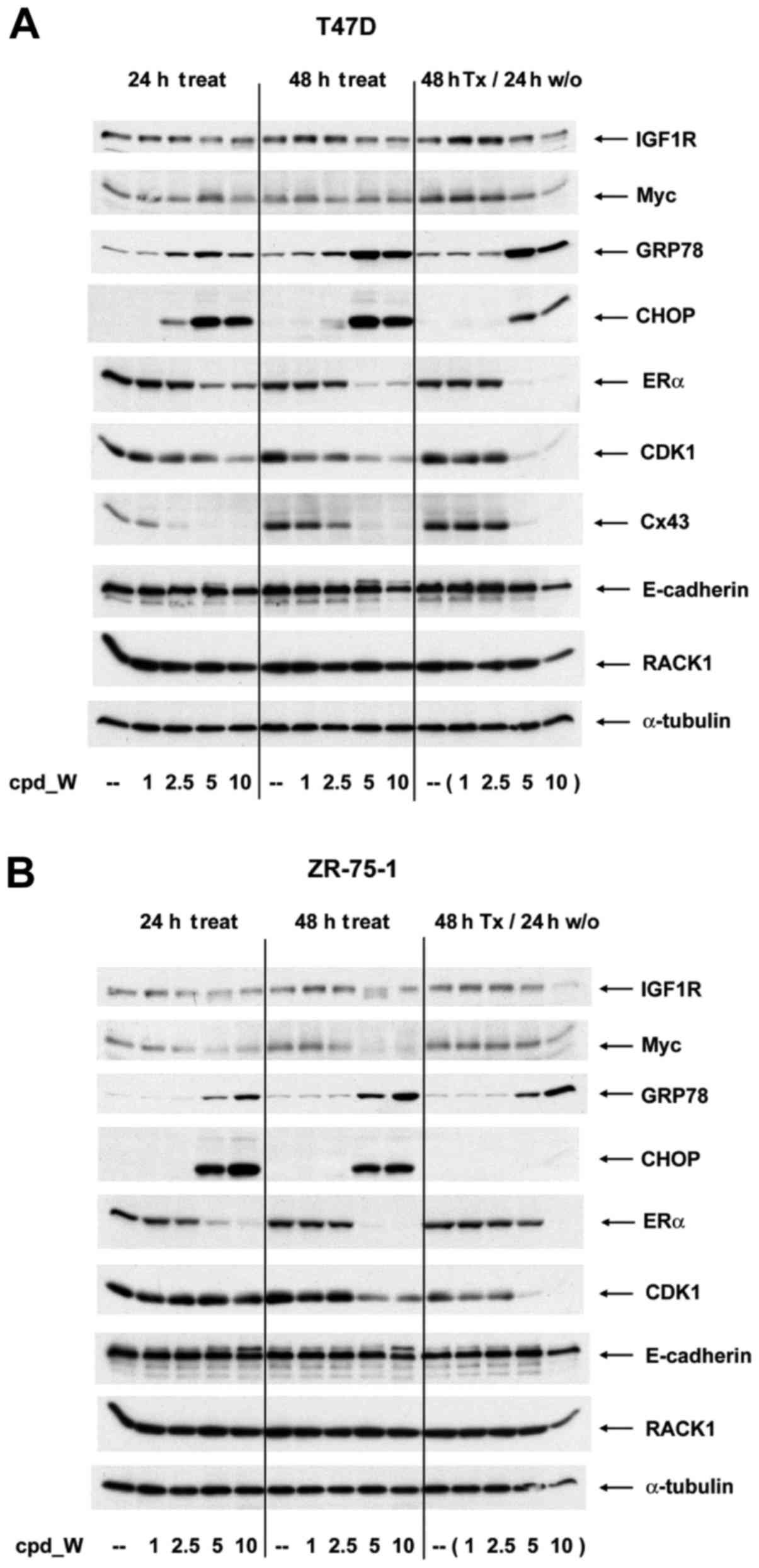
A series of western blot analyses were performed to
examine changes taking place in abundance of individual proteins as
a consequence of IRES inhibition (Fig.
5A and B). Although the long half-life of pre-existing IGF1R
molecules precluded a more rapid or more substantial impact, a
modest decrease in IGF1R is observed in cells treated with >5
µg/ml cpd_W (consistent with the results of Fig. 1). A marked decrease in Myc protein
brought about by IRES inhibition was observed in the ZR-75-1 cells,
which expressed Myc at a high level. In addition, ERα, CDK1 and
connexin 43, each of which is known to be translated via an IRES
(15,18,19),
were decreased substantially in a concentration-dependent manner
upon exposure to cpd_W.
The loss of ERα protein from the treated cells was
consistent with the imaging results. Notably, there are two
dynamics in play for ERα: i) a shift from the cytoplasm to the
nucleus in association with phenotypic transition to the
differentiated phenotype; and ii) a decrease in the synthesis of
ERα protein as a result of the inhibition of IRES-mediated
translation. Likewise, CDK1 levels declined rapidly, especially in
T47D cells and connexin 43 essentially disappeared from the treated
cells within 24 h. In contrast, a marked induction of both GRP78
and CHOP was observed in response to IRES inhibition, consistent
with the involvement of these molecules in the translation quality
control.
ERα, CDK1 and connexin 43 were actively being
translated at the time the IRES inhibitor was added and the
synthesis of these proteins did not rapidly rebound when the IRES
inhibitor was removed from the media (though there was evidence of
recovery of ERα during the washout period in ZR-75-1 cells
following treatment with 5 µg/ml cpd_W). This suggests that either
these mRNAs remained sequestered in dysfunctional IRES-ribosome
complexes, or that molecular changes induced during the period in
which IRES-mediated translation was inhibited altered the phenotype
so significantly that IRES-mediated translation of these proteins
was no longer a priority for these cells. Notably, GRP78 remained
elevated during washout, however CHOP dissipated quite rapidly
following the removal of the IRES inhibitor. For RACK1 and
E-cadherin, in spite of major changes in epitope accessibility and
structural organization respectively, no significant alterations
were observed in abundance of either of these proteins.
ER-positive breast tumor cells at low
density (with limited paracrine support and no intercellular
contact) exhibit enhanced susceptibility to IRES inhibition
Primary clonogenic survival assays revealed a
significant decrease in colony formation at 0.1–0.25 µg/ml cpd_W
and almost complete loss of colony formation at >0.5 µg/ml cpd_W
(Fig. 6A). This was a much lower
concentration of cpd_W than required to eliminate reproductive
capacity of the high-density tumor cell population, indicating that
sensitivity to IRES inhibition was enhanced for isolated or
low-density ER-positive breast tumor cells. To follow up on this
observation, a series of such experiments was performed using cell
viability readouts as a quantitative surrogate for colony
formation. The primary clonogenic survival dose-response curves for
both T47D and ZR-75-1 (Fig. 6B and
C; green lines) were superimposed on the population viability
and secondary clonogenic survival curves from Fig. 4. The marked increase in
susceptibility to IRES inhibition for the cells treated at low
density was apparent from the wide separation between these curves.
In fact, the viability readout tended to underestimate the negative
impact of IRES inhibition on clonogenic survival, as many of the
cells remaining in the treated wells and scoring as viable based on
ATP content were visualized microscopically as individual or pairs
of cells, apparently unable to generate colonies even following
prolonged incubation in absence of compound.
Thus, the ER-positive breast tumor cells responded
to IRES inhibition very differently when isolated or present at low
density compared to their response as part of a high-density
population. Both the nature of the response and quantitative
outcome were different. The cells at high density undergo terminal
differentiation when IRES-mediated translation was inhibited,
however at low density, where cells depend on IRES-mediated
translation and the undifferentiated phenotype to survive, IRES
inhibition led to acute cell death.
The time course assays presented in Fig. 7A and B revealed the progressive loss
of viability in these cells, reaching 80–90% cell loss by 96 h with
5 µg/ml cpd_W and >98% cell loss at 10 µg/ml cpd_W. Fig. 7C is a plot of the trajectories of
cell survival when the duration of exposure to cpd_W was titrated
and cell viability was assayed 96 h following removal of the
compound. Cells treated for >72 h uniformly exhibited a
continued downward trend in viability after the compound was
removed from the media. In fact, cell numbers declined an
additional 1–2 logs during the washout period, indicating that the
detrimental impact of IRES inhibition persisted well beyond the
treatment period. The results obtained for low-density ZR-75-1
cells (Fig. 7D) indicated that as
little as 5–15 h interruption in IRES-mediated translation had a
lasting impact on the proliferative capacity of the cells, but
72–96 h continuous exposure to cpd_W was required to ensure that
the loss of cell viability was extensive and cell recovery was
minimal. Phase contrast images illustrated the predominant
morphology exhibited by low-density cells succumbing to IRES
inhibition, characterized by gross osmotic swelling and widespread
detachment (Fig. 7E), very
different from the morphology observed with terminal
differentiation at high cell density (Fig. 3) (12).
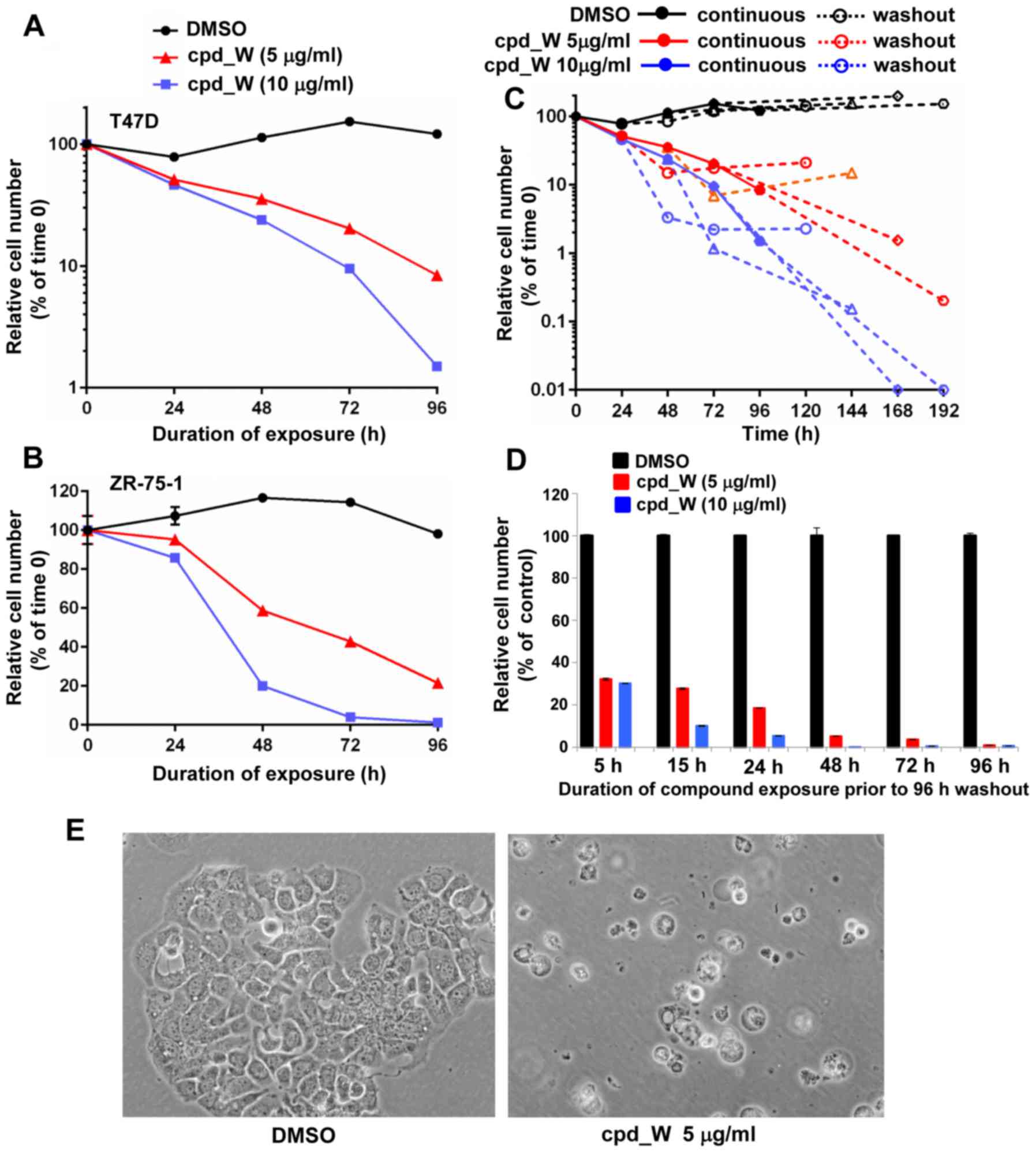 | Figure 7.Massive cell death as a consequence
of IRES inhibition in low-density ER-positive breast tumor cells.
T47D (A) or ZR-75-1 (B) cells were plated at clonogenic density,
allowed 48 h to recover, then treated with IRES inhibitor cpd_W at
5 or 10 µg/ml or vehicle control under low serum conditions for up
to 96 h and then viability was assessed at 24 h intervals. (C) T47D
cells were seeded at clonogenic density then treated with cpd_W at
5 µg/ml (red lines) or 10 µg/ml (blue lines) or vehicle (DMSO)
control (black lines) for 24, 48, 72 or 96 h. The cells were then
provided fresh media without compound and allowed 96 h to recover
and proliferate before viability was again assessed. Solid symbols
and lines are indicative of continuous treatment, while open
symbols and dashed lines are indicative of washout period. The
trajectories of the dashed lines allowed us to gauge the impact of
IRES inhibition on cell survival following removal of the compound.
(D) ZR-75-1 cells were seeded at clonogenic density then treated
with cpd_W at 5 or 10 µg/ml for 5, 15, 24, 48, 72 or 96 h, after
which all samples were provided fresh media and allowed 96 h to
recover in the absence of compound. The graph plotted relative
viability at the end of the 96 h washout period. (E) Phase contrast
images of T47D cells treated at low density with cpd_W at 5 µg/ml
for 96 h. |
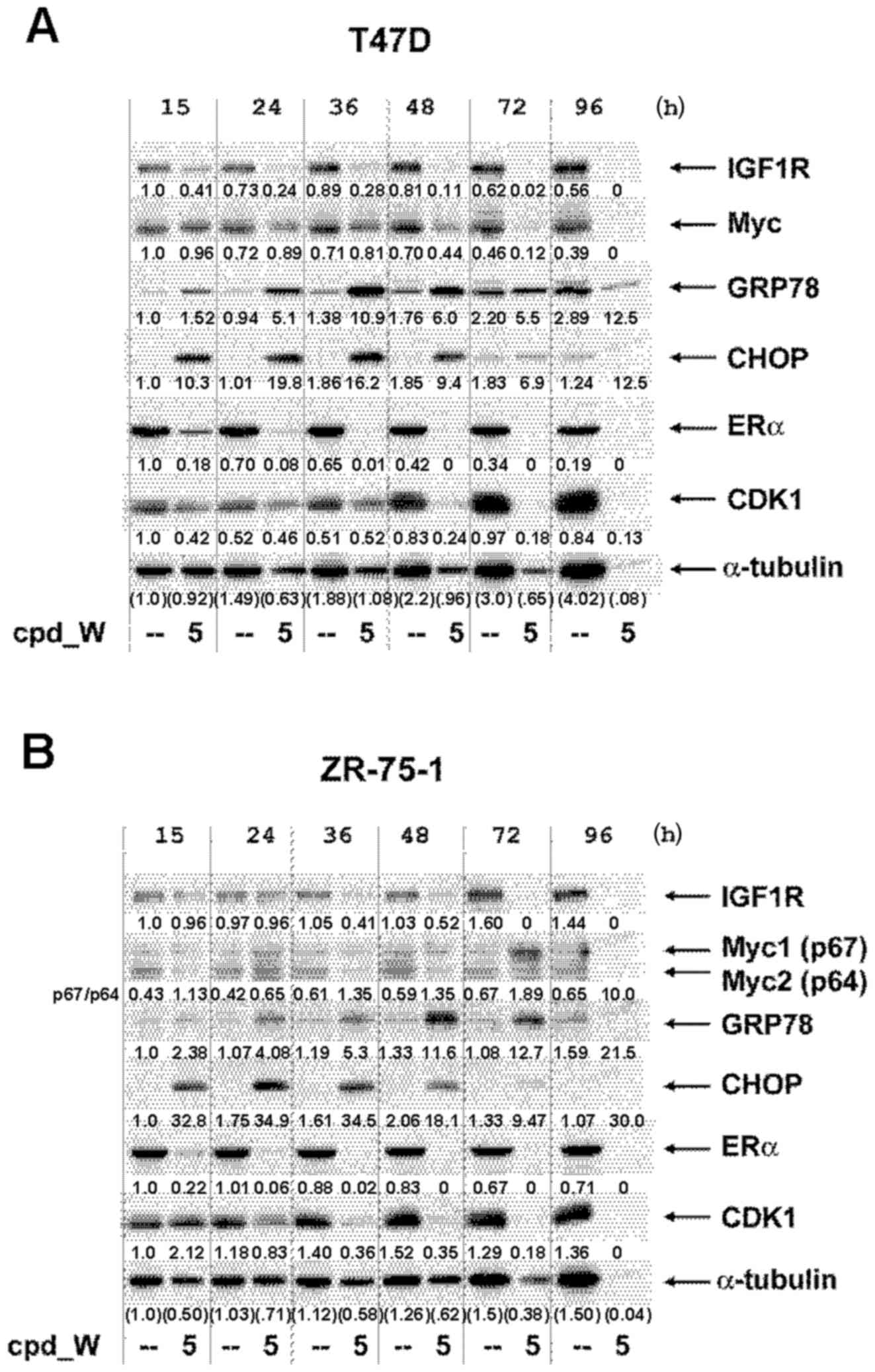
Western blot analyses were performed on low
(clonogenic) density cells by pooling lysates obtained from
multiple wells, as described in the Materials and methods section.
We deliberately elected not to normalize for total protein content
in these very limited samples, but instead loaded a constant
proportion (by volume) of the material recovered from the control
and treated wells (Fig. 8A and B).
The progressive decrease in the number of viable cells from
cpd_W-treated wells (vs. the expansion of the vehicle-treated
control cells) could be assessed by the relative intensities of the
α-tubulin bands and fitted well with the quantitative cell
viability data presented in the abovementioned graphs. However, the
intensities of bands representing IRES-driven proteins declined
even more rapidly as a consequence of the inhibition of
IRES-mediated translation and more rapidly than they did in the
standard western blot analyses performed on high-density tumor
cells. ERα declined to less than half the intensity of the control
cells by 15 h and was barely detectable by 24 h. Notable decreases
in IGF1R and CDK1 were observed in both cell lines, with only low
levels remaining at or beyond the 36 and 48 h time-points. Changes
in Myc protein were accentuated in ZR-75-1 cells, with a pronounced
induction of the alternative isoform of Myc (p67), particularly at
the 72 h time-point, as cell death was accelerating. The dominant
isoform of Myc (p64) is a major contributor to breast oncogenesis
(20). However, the p67 isoform of
Myc, which is known to be generated via use of an alternative
upstream initiation codon, has been attributed potent growth
inhibitory and pro-death properties (21,22)
and was also observed in the triple-negative breast tumor cells
subjected to IRES inhibition, where it correlated with terminal
differentiation and comprehensive death of those cells (11,12).
These decreases in IGF1R, CDK1 and ERα and the modulation of Myc
translation (including shift to the p67 isoform) may be key
molecular events that contributed to the stochastic death of
low-density tumor cells in which IRES-mediated translation has been
inhibited.
Incapacitation of low-density
ER-positive breast tumor cells treated with very low concentrations
of IRES inhibitor cpd_W
Finally, we further characterized the impact of very
low concentrations of cpd_W on the ER-positive breast tumor cells,
in an attempt to understand the basis for clonogenic survival being
so exquisitely sensitive to IRES inhibition. Time course assays
indicated that exposure of low-density ER-positive breast tumor
cells to a very low concentration (1 µg/ml) of cpd_W did not result
in rapid or massive cell death (Fig.
9A). In fact the cell number was relatively stable throughout
the 96 h treatment period, as cells surviving with limited
paracrine support and low soluble growth factors attempted to
proliferate and generate colonies. A gradual separation of the
curves between vehicle control and cpd_W-treated samples was
observed over the course of the experiment, with much of this
separation occurring after the compound had been removed from the
media. At the final endpoint, the vehicle controls have established
viable colonies and were accelerating proliferation, while the
cells which had been exposed to the IRES inhibitor at the beginning
of the experiment continued to exhibit a slow progressive decline
in cell number and few or no viable multi-cellular colonies.
Phase contrast images captured in the course of
these experiments revealed that, although the cells exposed to 1
µg/ml cpd_W retain the ability to divide, the cells produced were
dysmorphic and lacked the cohesiveness typical of ER-positive
breast tumor cells (Fig. 9B). The
cpd_W-treated cells exhibited very limited intercellular contact,
in stark contrast to the extensively interconnected cobblestone
morphology displayed by the control cells. Collectively, these
findings indicated that the potent inhibition of clonogenic
survival by very low concentrations of the IRES inhibitor resulted
not from acute cell death, but rather from incapacitation, i.e.
inability to generate viable, cohesive colonies. Without the
support of a cohesive colony, the majority of these cells
eventually died, though tumor cell death under these conditions was
insidious rather than acute. Notably, this concentration of cpd_W
had no discernible adverse effects on the established populations
of cells (Fig. 4).
Western blot analyses were performed on the
low-density cells exposed to 1 µg/ml cpd_W, in an attempt to
discern factors contributing to the failure of these cells to form
viable colonies (Fig. 9C). The
intensities of the α-tubulin bands matched the quantitation of
viable cells as displayed in the abovementioned time course graph.
However, three notable variations from this pattern were observed.
Firstly, induction of GRP78 and CHOP at a cpd_W concentration as
low as 1 µg/ml served as an indication that sensitivity to IRES
inhibition was heightened for cells at low density (5–10 µg/ml
cpd_W was required for the induction of these molecules in
high-density cells). Secondly, connexin 43, which is known to be
translated by an IRES and appeared in the western blot analyses of
the high-density tumor cell populations to be among the most
sensitive proteins to the IRES inhibitor (Fig. 5), was completely undetectable at all
time-points in the cpd_W-treated cells. Loss of the expression of
connexin 43 and resulting impairment in gap junctional
intercellular communication may have contributed significantly to
failure of the treated cells to establish viable cohesive colonies.
Thirdly, at the latest stage of the washout period, there was again
evidence of a shift in Myc translation favoring synthesis of the
p67 isoform, which has previously been associated with growth
inhibition, differentiation and cell death.
Discussion
ER-positive breast tumor cells
transition between two interconvertible phenotypic states
distinguished by the degree of differentiation and use of
IRES-mediated translation
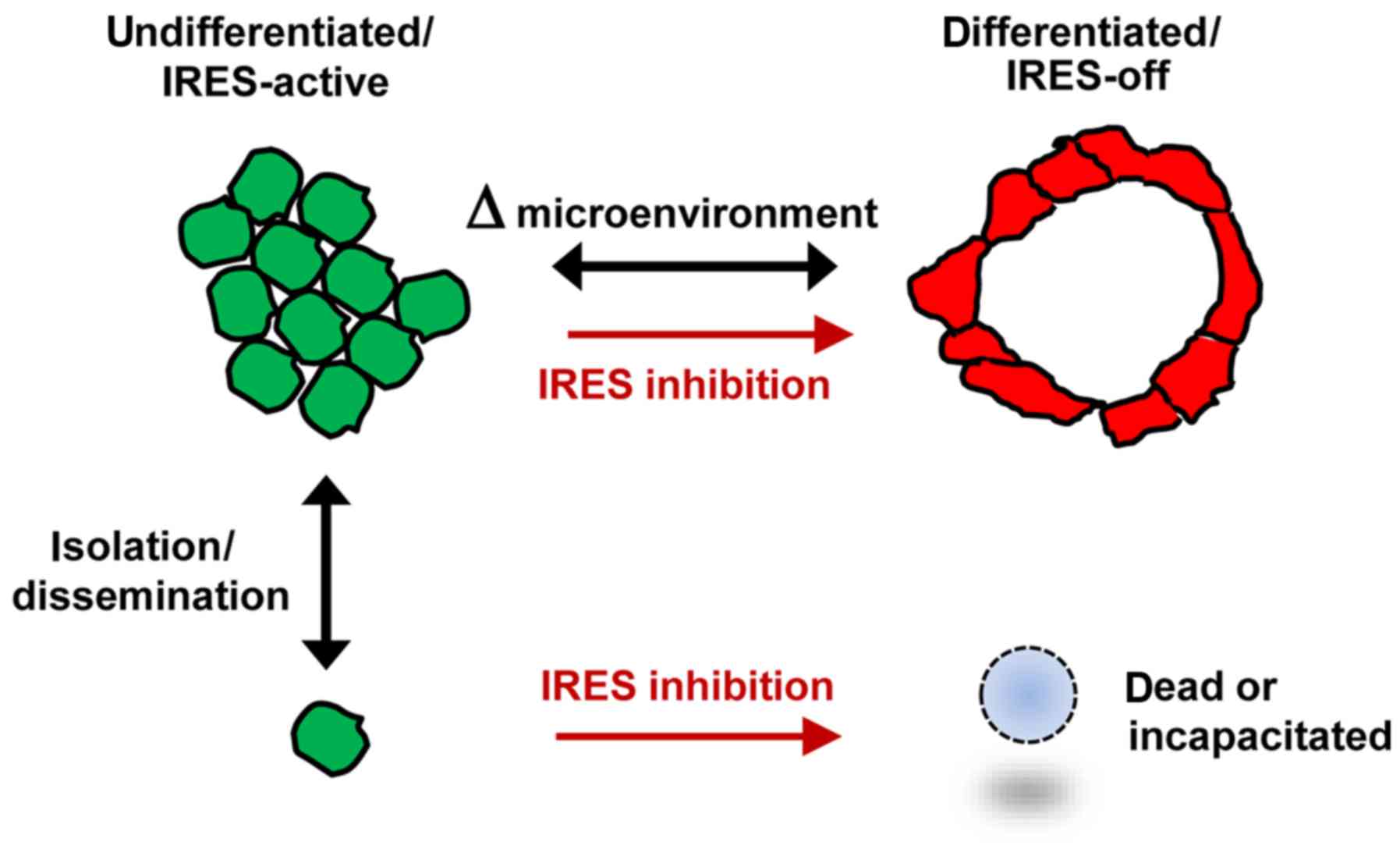
A working model illustrating two distinct phenotypic
states for ER-positive breast tumor cells, the transitions between
them and the relationship to IRES-mediated translation is displayed
in Fig. 10. Further investigation
of these parameters in additional ER-positive breast tumor models
and primary breast tumor specimens is warranted.
The results indicated that in ER-positive breast
tumor cells, differentiation status is dynamic, subject to a
reversible phenotypic transition between a moderately
differentiated state and a relatively undifferentiated state.
Imaging analyses allowed us to discern these variations in
phenotype at the level of individual cells or groups of cells. The
cells in the moderately differentiated state line acellular spaces,
adopted specific spatial orientations with respect to adjacent
cells and form highly contoured surfaces. Cells in the
undifferentiated state exhibited no structural organization other
than the propensity to cluster among themselves. The ER-positive
breast tumor cells readily transitioned between these two distinct
phenotypes, as a means of adapting to changes in microenvironment.
The cells may be capable of shifting to the more resilient
undifferentiated state when faced with challenging
microenvironmental conditions such as limiting growth factors (e.g.
serum deprivation). IRES-mediated translation is active and appears
to be essential, for the undifferentiated state. Under more
favorable conditions, a fraction of the tumor cells may adopt the
differentiated state in which IRES-mediated translation is
apparently not active. Chemical inhibition of IRES-mediated
translation forced the cells to transition unilaterally to the
differentiated IRES-off phenotype and this transition became
irreversible. Thus, translational regulation and IRES-mediated
translation in particular, may be an integral factor that
distinguishes these phenotypic states.
Multiple biomarkers were capable of distinguishing
these two distinct phenotypes. Each of the candidate biomarkers
aligned precisely with one of these phenotypic states. The
undifferentiated/IRES-active phenotype was IGF1R N20-positive and
RACK1-negative and ERα was restricted to the cytoplasm. The
differentiated/IRES-off phenotype was RACK1-positive and IGF1R
N20-negative and ERα, GRP78 and ZO-1 were all readily detected in
the nucleus. These molecules and staining patterns may ultimately
serve as diagnostic biomarkers applied to primary breast tumor
specimens, to help gauge differentiation status and the degree to
which an individual tumor or population of tumor cells relies on
IRES-mediated translation. These candidate biomarkers could also
potentially be used to investigate how IRES-mediated translation
and differentiation status change in response to the adverse
microenvironmental conditions experienced in vivo.
Of particular importance was the finding that
cytoplasmic ERα may serve as a biomarker for IRES-mediated
translation in ER-positive breast tumor cells. Cytoplasmic ERα
correlated with the undifferentiated phenotype in which the
IRES-mediated translation was active, mirroring precisely the
staining pattern observed with the antibody recognizing nascently
translated IGF1R and inversely correlated with RACK1 epitope
accessibility (as an indication of the IRES-off state). When cells
transitioned to the differentiated phenotype, IRES-mediated
translation was curtailed and ERα shifted entirely to the nucleus.
ERα has two functional nuclear export sequences along with one
nuclear localization signal, allowing it to shuttle between the
nuclear and cytoplasmic compartments (23). Export of ERα to the cytoplasm is
required for S-phase entry and cell proliferation (24) and blocking ERα function decreases
colony forming potential (25).
Accumulation of ERα in the cytoplasm also correlates with
resistance to hormonal therapy (26,27).
Characterization of non-nuclear (transcription-independent)
functions of ERα is an active area of investigation, however, to
our knowledge, a relationship between cytoplasmic ERα localization
and IRES-mediated translation has not been previously described.
The functional significance of this relationship is not yet clear,
however, it is of note that cytoplasmic ERα interacts with HSP90
(28), which appears to be involved
in regulating IRES-mediated translation (29). Additional research will be needed to
determine whether ERα may actively promote the undifferentiated
phenotype and/or IRES-mediated translation from its location in the
cytoplasm.
The stark contrast in RACK1 epitope accessibility
between the two phenotypic states indicated that RACK1 conformation
and/or intermolecular interactions were distinctly different in the
context of the clustered cells in which IRES-mediated translation
was active and the differentiated cells in which IRES-mediated
translation appeared to have been shut down. Cryo-EM images have
placed RACK1 in close proximity to the mRNA exit site on the
platform of the 40S subunit, where interactions with the 5′upstream
regulatory region of the mRNA take place (13). Although RACK1 is an integral
component of the 40S ribosomal subunit, it is not required for
general protein synthesis, but selectively promotes translation of
specific mRNAs (several of which are known to be translated via an
IRES, including MYC (30)). RACK1
is also required for the translation of certain viral IRESs (e.g.
HCV) and RACK1 facilitates IRES-mediated translation in in
vitro systems as well (14).
GRP78 and ZO-1 accompanied ERα in shifting from the
cytoplasm to the nucleus upon cellular differentiation. GRP78
interacts with matrin-3 (a component of nuclear matrix) (31), although its function in the nucleus
has not yet been thoroughly characterized. ZO-1 contains three
nuclear localization signals and localizes to the nucleus of cells
that lack full circumferential contact with other cells (32). ZO-1 interacts closely with the Y-box
transcription factor ZONAB, which binds to the promoters of genes
whose expression varies with cell density (33). ZO-1 has been observed in the nuclei
of normal terminally differentiated epithelial cells at the tips of
intestinal villi, post-mitotic cells which are preparing to undergo
programmed cell death and exfoliation (32).
Immunofluorescence staining and confocal imaging
demonstrated a marked increase in structural organization of
E-cadherin and the microtubule cytoskeleton, consistent with the
enhanced differentiation in the cells treated with the IRES
inhibitor. Yet the western blot results indicated no net increase
in the E-cadherin protein level. Thus, it appears that although
some of the structural proteins needed for the transition to the
terminally differentiated phenotype were already present in
sufficient quantity, the cells may have been prevented from fully
executing the differentiation program by one or more IRES-driven
factors which perpetuated the undifferentiated state.
Notably, these two distinct phenotypes were at times
observed even within an individual cell, where one region of
the cytoplasm (bordering an open space, i.e. characteristic of a
normal epithelial cell) revealed evidence of enhanced RACK1 epitope
accessibility (indicative of the more differentiated/IRES-off
state), while the cytoplasm on the other side of the cell
(bordering a cluster of undifferentiated cells) showed evidence of
active IRES-mediated translation (IGF1R N20 or cytoplasmic ERα
immunoreactivity and RACK1 epitope masking). These biphenotypic
cells were observed in vehicle-treated wells (i.e. independent of
IRES inhibition), therefore this subcellular phenotypic and
translational specialization apparently has a physiological basis
and is something these tumor cells readily employ. This pattern
also suggests that, at least to some degree, phenotype is
controlled at the cytoplasmic (i.e. translational) level.
Fundamental relationship between
IRES-mediated translation and the undifferentiated state
We previously reported (12) that highly undifferentiated
triple-negative breast tumor cells (as well as glioblastoma and
osteosarcoma cells) were forced into terminal differentiation when
continuously exposed to IRES inhibitor cpd_P for >72 h. The
triple-negative breast tumor cells did not tolerate this transition
to a fully differentiated phenotype and underwent a synchronized
population-wide cell death event which was non-apoptotic and shared
a number of features with cornification. In the ER-positive breast
tumor cells, as little as 15–24 h exposure to cpd_W led to a
terminal differentiation outcome which very closely resembled that
observed in the triple-negative cells, with similar increases in
polarity and structural organization. Unlike the triple-negative
breast tumor cells however, the ER-positive breast tumor cells
tolerated forced differentiation with only modest loss of
viability, although their ability to transition back to the
undifferentiated state and resume proliferation, which is of
integral importance to their malignant behavior, was substantially
compromised.
Thus, the impact of IRES inhibition on ER-positive
and triple-negative breast tumor cells was actually quite similar.
In both cases, the tumor cell population was forced to abandon the
undifferentiated phenotype. The differences in timing of response
and final outcome reflected the inherent differences in the
phenotypic starting point for these cells. ER-positive breast tumor
cells are intrinsically capable of transitioning reversibly to a
differentiated state and may not require as great a stimulus to
initiate terminal differentiation, while the triple-negative breast
tumor cells are phenotypically further removed from the terminally
differentiated state and therefore required prolonged deprivation
of IRES-mediated translation in order to revoke the
undifferentiated phenotype. Collectively, these results provided
further evidence that IRES-mediated translation is critical for the
maintenance of the undifferentiated state.
Clinical implications and potential
therapeutic applications of IRES inhibition in ER-positive breast
cancer
IRES inhibition exerted profound, detrimental
effects on ER-positive breast tumor cells, inducing terminal
differentiation in high-density tumor cell populations and complete
loss of clonogenic survival (either massive cell death or
incapacitation) in isolated or low-density tumor cells. In all
cases, the tumor cells lost the ability to perpetuate the malignant
clone. Thus, each of these could represent a clinically beneficial
outcome, illustrating the potential use of IRES inhibition as a
therapeutic strategy.
Low-density tumor cells were particularly vulnerable
to IRES inhibition because they rely on the undifferentiated
phenotype and IRES-mediated translation to survive when paracrine
support and intercellular contact were limited. The stringent
experimental protocols used in the present study were intended to
model the adverse microenvironmental conditions encountered by
disseminated (i.e. micrometastatic) tumor cells in vivo. The
fact that the ER-positive breast tumor cells were able to survive
and form colonies at low cell density concomitant with acute serum
deprivation was indicative of the extraordinary resiliency of these
cells. Although frequently tagged as ‘less aggressive’, we found
that the isolated ER-positive breast tumor cells are actually more
resilient than even triple-negative breast tumor cells, which do
not survive such conditions.
This resiliency is reflected in the natural history
of ER-positive breast cancer, where a substantial proportion of
patients fail to achieve long term remission with post-surgical
hormonal therapy and instead experience repeated recurrences and
progressive metastatic disease (34,35).
The potent inhibition of clonogenic survival by very low
concentrations of cpd_W suggests that IRES inhibition may be used
to effectively incapacitate or eliminate these otherwise highly
resilient ER-positive breast tumor cells, thereby filling a
critical gap in the therapeutic armamentarium.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Institutes of Health (grant no. R01CA108886), the American Society
for Clinical Oncology Young Investigator Award (to CV), Susan G
Komen Career Catalyst Award in Basic and Translational Research
(grant no. CCR15331062 to CV), the UAB Comprehensive Cancer Center
Drug Discovery and Development Program and the Alabama Drug
Discovery Alliance.
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
CV and SWB performed the experiments and wrote the
manuscript. ZM and HC made substantial contributions to conception,
design and intellectual content of the studies. KRZ, SLS, and WEG
made key contributions to analysis and interpretation of data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the UAB Institutional
Review Board (project no. X040506011).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ERα
|
estrogen receptor alpha
|
|
IGF1R
|
insulin-like growth factor 1
receptor
|
|
RACK1
|
receptor for activated C kinase 1
|
|
IRES
|
internal ribosome entry site
|
References
|
1
|
Prats AC and Prats H: Translational
control of gene expression: Role of IRESs and consequences for cell
transformation and angiogenesis. Prog Nucleic Acid Res Mol Biol.
72:367–413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baird SD, Turcotte M, Korneluk RG and
Holcik M: Searching for IRES. RNA. 12:1755–1785. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Spriggs KA, Stoneley M, Bushell M and
Willis AE: Re-programming of translation following cell stress
allows IRES-mediated translation to predominate. Biol Cell.
100:27–38. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meng Z, Jackson NL, Choi H, King PH,
Emanuel PD and Blume SW: Alterations in RNA-binding activities of
IRES-regulatory proteins as a mechanism for physiological
variability and pathological dysregulation of IGF-IR translational
control in human breast tumor cells. J Cell Physiol. 217:172–183.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagamachi A, Htun PW, Ma F, Miyazaki K,
Yamasaki N, Kanno M, Inaba T, Honda Z, Okuda T, Oda H, et al: A
5′untranslated region containing the IRES element in the Runx1 gene
is required for angiogenesis, hematopoiesis and leukemogenesis in a
knock-in mouse model. Dev Biol. 345:226–236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Blau L, Knirsh R, Ben-Dror I, Oren S,
Kuphal S, Hau P, Proescholdt M, Bosserhoff AK and Vardimon L:
Aberrant expression of c-Jun in glioblastoma by internal ribosome
entry site (IRES)-mediated translational activation. Proc Natl Acad
Sci USA. 109:E2875–E2884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dobson T, Chen J and Krushel LA:
Dysregulating IRES-dependent translation contributes to
over-expression of the Aurora A kinase onco-protein. Mol Cancer
Res. 11:887–900. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lynch SR and Puglisi JD: Structural
origins of aminoglycoside specificity for prokaryotic ribosomes. J
Mol Biol. 306:1037–1058. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hansen JL, Moore PB and Steitz TA:
Structures of five antibiotics bound to the peptidyl transferase
center of the large ribosomal subunit. J Mol Biol. 330:1061–1075.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baselga J, Campone M, Piccart M, Burris HA
III, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun
F, et al: Everolimus in postmenopausal hormone-receptor-positive
advanced breast cancer. N Engl J Med. 366:520–529. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vaklavas C, Meng Z, Choi H, Grizzle WE,
Zinn KR and Blume SW: Small molecule inhibitors of IRES-mediated
translation. Cancer Biol Ther. 16:1471–1485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vaklavas C, Grizzle WE, Choi H, Meng Z,
Zinn KR, Shrestha K and Blume SW: IRES inhibition induces terminal
differentiation and synchronized death in triple-negative breast
cancer and glioblastoma cells. Tumour Biol. 37:13247–13264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sengupta J, Nilsson J, Gursky R, Spahn CM,
Nissen P and Frank J: Identification of the versatile scaffold
protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol
Biol. 11:957–962. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Majzoub K, Hafirassou ML, Meignin C, Goto
A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F,
et al: RACK1 controls IRES-mediated translation of viruses. Cell.
159:1086–1095. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barraille P, Chinestra P, Bayard F and
Faye JC: Alternative initiation of translation accounts for a 67/45
kDa dimorphism of the human estrogen receptor ERalpha. Biochem
Biophys Res Commun. 257:84–88. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vedrenne C, Klopfenstein DR and Hauri HP:
Phosphorylation controls CLIMP-63-mediated anchoring of the
endoplasmic reticulum to microtubules. Mol Biol Cell. 16:1928–1937.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nikonov AV, Hauri HP, Lauring B and
Kreibich G: Climp-63-mediated binding of microtubules to the ER
affects the lateral mobility of translocon complexes. J Cell Sci.
120:2248–2258. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schiavi A, Hudder A and Werner R:
Connexin43 mRNA contains a functional internal ribosome entry site.
FEBS Lett. 464:118–122. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marash L, Liberman N, Henis-Korenblit S,
Sivan G, Reem E, Elroy-Stein O and Kimchi A: DAP5 promotes
cap-independent translation of Bcl-2 and CDK1 to facilitate cell
survival during mitosis. Mol Cell. 30:447–459. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Spandidos DA, Pintzas A, Kakkanas A,
Yiagnisis M, Mahera H, Patra E and Agnantis NJ: Elevated expression
of the myc gene in human benign and malignant breast lesions
compared to normal tissue. Anticancer Res. 7:1299–1304.
1987.PubMed/NCBI
|
|
21
|
Hann SR, Dixit M, Sears RC and Sealy L:
The alternatively initiated c-Myc proteins differentially regulate
transcription through a noncanonical DNA-binding site. Genes Dev.
8:2441–2452. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benassayag C, Montero L, Colombié N,
Gallant P, Cribbs D and Morello D: Human c-Myc isoforms
differentially regulate cell growth and apoptosis in Drosophila
melanogaster. Mol Cell Biol. 25:9897–9909. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tecalco-Cruz AC, Pérez-Alvarado IA,
Ramírez-Jarquín JO and Rocha-Zavaleta L: Nucleo-cytoplasmic
transport of estrogen receptor alpha in breast cancer cells. Cell
Signal. 34:121–132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lombardi M, Castoria G, Migliaccio A,
Barone MV, Di Stasio R, Ciociola A, Bottero D, Yamaguchi H, Appella
E and Auricchio F: Hormone-dependent nuclear export of estradiol
receptor and DNA synthesis in breast cancer cells. J Cell Biol.
182:327–340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Basak P, Chatterjee S, Weger S, Bruce MC,
Murphy LC and Raouf A: Estrogen regulates luminal progenitor cell
differentiation through H19 gene expression. Endocr Relat
Cancer. 22:505–517. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fan P, Wang J, Santen RJ and Yue W:
Long-term treatment with tamoxifen facilitates translocation of
estrogen receptor alpha out of the nucleus and enhances its
interaction with EGFR in MCF-7 breast cancer cells. Cancer Res.
67:1352–1360. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guest SK, Ribas R, Pancholi S,
Nikitorowicz-Buniak J, Simigdala N, Dowsett M, Johnston SR and
Martin LA: Src is a potential therapeutic target in
endocrine-resistant breast cancer exhibiting low estrogen
Receptor-mediated transactivation. PLoS One. 11:e01573972016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dhamad AE, Zhou Z, Zhou J and Du Y:
Systematic proteomic identification of the heat shock proteins
(Hsp) that interact with estrogen receptor alpha (ERα) and
biochemical characterization of the ERα-Hsp70 interaction. PLoS
One. 11:e01603122016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang HW, Kim TM, Song SS, Menon L, Jiang
X, Huang W, Black PM, Park PJ, Carroll RS and Johnson MD: A small
subunit processome protein promotes cancer by altering translation.
Oncogene. 34:4471–4481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ruan Y, Sun L, Hao Y, Wang L, Xu J, Zhang
W, Xie J, Guo L, Zhou L, Yun X, et al: Ribosomal RACK1 promotes
chemoresistance and growth in human hepatocellular carcinoma. J
Clin Invest. 122:2554–2566. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Osman AM and van Loveren H: Matrin 3
co-immunoprecipitates with the heat shock proteins
glucose-regulated protein 78 (GRP78), GRP75 and glutathione
S-transferase π isoform 2 (GSTπ2) in thymoma cells. Biochimie.
101:208–214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gottardi CJ, Arpin M, Fanning AS and
Louvard D: The junction-associated protein, zonula occludens-1,
localizes to the nucleus before the maturation and during the
remodeling of cell-cell contacts. Proc Natl Acad Sci USA.
93:10779–10784. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Balda MS, Garrett MD and Matter K: The
ZO-1-associated Y-box factor ZONAB regulates epithelial cell
proliferation and cell density. J Cell Biol. 160:423–432. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Clarke R, Tyson JJ and Dixon JM: Endocrine
resistance in breast cancer-An overview and update. Mol Cell
Endocrinol. 418:220–234. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Murphy CG and Dickler MN: Endocrine
resistance in hormone-responsive breast cancer: Mechanisms and
therapeutic strategies. Endocr Relat Cancer. 23:R337–R352. 2016.
View Article : Google Scholar : PubMed/NCBI
|