Introduction
Breast cancer is considered the most common
malignancy and the main leading cause of cancer-related deaths in
women (1,2). The pathogenesis and mechanisms of
breast cancer remain unclear. Therefore, further studies at the
molecular level are necessary. Various related molecular markers
[ER, PR and HER-2 (3,4)] are used in clinical practice to guide
the diagnosis, treatment and prognosis of breast cancer, however
they are only used to diagnose a small group type with poor
prognosis of breast cancer. Therefore, finding a tumour marker of
breast cancer that has high sensitivity and specificity, can be
used for early diagnosis, individualised treatment, monitoring the
curative effect and individual prognosis is highly important.
MicroRNAs (miRNAs) are highly conserved non-coding
small RNAs, 18–25 nucleotides in length, that widely exist in
plants and animals and can regulate cell differentiation,
proliferation, metabolism and apoptosis by inhibiting transcription
of protein coding genes or inducing their mRNA degradation
(5). miRNAs possess advantages such
as non-invasiveness, stability, high sensitivity and free from the
influence of blood cell components; hence, they have the
characteristics of ideal tumour markers (6). Therefore, further study of miRNAs is
expected to improve the diagnosis and treatment of breast
cancer.
The receptor for advanced glycosylation end products
(RAGE) is a type of membrane protein belonging to the
immunoglobulin superfamily. Our previous research revealed that
RAGE is an oncogene in breast cancer (7). Stoetzer et al considered RAGE
as a circulating immunogenic cell death biomarker (8). Therefore, targeting RAGE is considered
a novel strategy for clinical intervention in breast cancer. To
investigate the mechanism of RAGE and discover important miRNAs for
breast cancer, we screened miRNAs microarray after interfering with
the expression of RAGE in breast cancer cells.
In the present study, we found that miR-328-5p was
significantly upregulated after knockdown of RAGE. We investigated
the anti-oncogenic role of miR-328-5p and its correlation to
proliferation in breast cancer cells. We confirmed that there was
direct binding between miR-328-5p and RAGE. Furthermore, the
inhibitory functions of miR-328-5p in breast cancer cells were
positively correlated with RAGE.
Materials and methods
Cell line and culture
The human breast cancer cell line MDA-MB-231 was
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). The cells were cultured in RPMI-1640 medium
(HyClone, Hudson, NH, USA) supplemented with 10% fetal bovine serum
(FBS) at 37°C and 5% CO2.
Cell transfection
The knockdown of RAGE in MDA-MB-231 cells was
performed using siRNA. The sequence targeting RAGE was
5′-CACTGCAGTCGGAGCTAATGG-3′. A scrambled oligo siRNA (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) was used as a control at the
same concentration. The overexpression of RAGE was accomplished
using a pcDNA3.1 vector. The overexpression of miR-328-5p in
MDA-MB-231 cells was performed using miR-328-5p mimics, and
scramble mimics were used as the control at the same concentration.
MDA-MB-231 cells were initially plated in media containing 10% FBS.
After 24 h, the cells were washed once with phosphate-buffered
saline (PBS) and transfected with vector/mimics/siRNA or scrambled
siRNAs by Nucleofector (Lonza Group, Ltd., Basel, Switzerland)
according to the manufacturer's instructions.
miRNA microarray analysis
MDA-MB-231 cells transfected with siRAGE and
scrambled siRNAs were subjected to Agilent Human miRNA microarray
(8×60 K) analysis at the Oebitech Co. (Shanghai, China). Total RNA
was quantified using NanoDrop ND-2000 (Thermo Fisher Scientific,
Inc., Waltham, MA, USA), and the RNA integrity was assessed using
Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara,
CA, USA). The sample labelling, microarray hybridisation and
washing were performed according to the manufacturer's standard
protocols. Briefly, total RNA was dephosphorylated, denatured and
then labelled with cyanine-3-CTP. After purification, the labelled
RNAs were hybridised onto the microarray. After washing, the arrays
were scanned with the Agilent Scanner G2505C (Agilent Technologies
Inc.). Feature Extraction software (version 10.7.1.1; Agilent
Technologies, Inc.) was then used to analyse array images to obtain
the raw data. GeneSpring software (version 13.1; Agilent
Technologies) was employed to complete the basic analysis of the
raw data. The threshold set for upregulated and downregulated genes
was a fold change of ≥2.0 and a P-value ≤0.05. The target genes of
differentially expressed miRNAs were predicted with three databases
(TargetScan, microRNA.org and PITA). Gene Ontology
(GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) analyses
were performed to determine the roles of these target genes.
Hierarchical clustering was performed to reveal the distinguishable
miRNA expression patterns among samples.
RNA extraction and quantitative
real-time polymerase chain reaction (qPCR)
Total RNA was extracted from cell lines and tissues
using TRIzol (Dongshen Biotech, Co., Ltd. Guangzhou, China).
RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher
Scientific, Inc.) was used for reverse transcription. qPCR was
performed using an ABI Prism 7300 Sequence Detection System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with
SYBR-Green PCR mixture (Dongshen Biotech, Co., Ltd.). The miRNA and
U6 primers were purchased from GeneCopoeia (Guangzhou, China). The
RAGE primers were as follows: forward, GGTGCCTAATGAGAAGGGAGTA and
reverse, GAAGCTACAGGAGAAGGTGGG. The β-actin primers were as
follows: forward, AGGGGCCGGACTCGTCATACT and reverse,
GGCGGCACCACCATGTACCCT. The relative expression level was determined
using the 2−ΔΔCt analysis method, in which β-actin and
U6 were used as internal standards. All reactions were conducted in
triplicate, and all experiments were performed in three independent
replicates.
Tissue collection
The 13 paired patient tissues (breast cancer and
adjacent tissues) were collected during surgical resection at the
Third Xiangya Hospital (from May 2015 to September 2017). All
patient tissues were confirmed by histopathological evaluation. The
study was approved by the Ethical Committee of the Third Xiangya
Hospital of Central South University. No patients received
preoperative radiotherapy and/or chemotherapy. The collected
tissues were immediately stored at −80°C until use.
MTT assay
The cells were collected, and the concentration was
adjusted to 5×104 cells/well before culture. The cells
were cultured in 35-mm dishes at 5% CO2 and 37°C for 24,
48 and 72 h. For chemosensitivity detection, the cells were treated
with cisplatin (DDP; 0–200 µM) for 48 h. Then, 10 µl of MTT was
added to the cells and maintained at 5% CO2 and 37°C for
another 4 h before MTT was removed. The optical density value was
measured at 570 nm while the cells were suspended in 150 µl of
dimethyl sulfoxide (DMSO).
Colony formation assay
The clonogenicity of single cells was detected by
colony assay. Cells were collected by adding 0.25% trypsin and were
adjusted to a concentration of 400 cells/petri dish, which was
loaded with 2 ml of preheated culture media before culturing with
5% CO2 at 37°C for 2–3 weeks. Colony formation was
terminated when the colonies were visible to the naked eye.
Subsequently, the cells were washed twice with PBS, and then 4%
paraformaldehyde was added for 15 min to fix the cells before
staining with Giemsa for 10 min. Then the number of colonies were
counted, and the colony formation rate was calculated as follows:
Colony formation rate = (number of colonies/inoculation cell
number) × 100%.
Cell invasion assay
The invasion ability was determined using a
Transwell assay. MDA-MB-321 cells (5×104) were plated in
Transwell chambers. The chambers were placed in a 24-well plate and
incubated for 24 h at 37°C. The cells that invaded the membrane
were stained using a crystal violet staining solution kit (Beyotime
Institute of Biotechnology, Shanghai, China). The stained cells
were photographed using an inverted microscope (Motic Inc., Ltd.,
Causeway Bay, Hong Kong). After capturing the images, the stain
from the membrane was eluted in 500 µl of DMSO. The eluted staining
agent was then shifted into a 96-well plate and the absorbance was
assessed using a microplate reader (Thermo Fisher Scientific, Inc.)
at 570 nm.
Flow cytometric analysis of the cell
cycle and cell apoptosis
The cell cycle was assessed via flow cytometry.
Cells were trypsinised (Auragene Bioscience Co., Changsha, China)
and washed twice with PBS before fixation in 500 µl 75% precooled
ethanol at 4°C overnight. The cells were then washed twice with PBS
and incubated in a water bath at 37°C for 30 min with 100 µl RNase
A (Auragene Bioscience Co.). The cell suspensions were incubated
for 30 min in the dark using propidium iodide (PI) (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) or for
15 min using an Annexin V/PI detection kit (Molecular Probes, Inc.;
Thermo Fisher Scientific, Inc., Eugene, OR, USA), according to the
manufacturer's instructions. Finally, the cell cycle and apoptosis
were analysed by FACScanto™ (BD Biosciences, Franklin Lakes, NJ,
USA).
Western blotting
Cells were lysed in cold RIPA buffer, and protein
was separated with 10–12% SDS-PAGE, which was then transferred to
NC membranes (Thermo Fisher Scientific, Inc.). The membranes were
blocked with 5% non-fat dried milk (Beijing Yili Fine Chemical Co.,
Ltd., Beijing, China) in PBS Tween-20 solution (0.1% v/v) for 3 h
at 4°C. Then, the membranes were incubated with primary antibodies
rabbit polyclonal rage (1:1,000; cat. no. ab37647), rabbit
polyclonal anti-NF-κB p65 (1:50,000; cat. no. ab32536), rabbit
monoclonal anti-cyclin D1 (1:10,000; cat. no. ab134175) and mouse
monoclonal anti-β-actin (1:1,000; cat. no. ab8226; all were from
Abcam, Cambridge, MA, USA) overnight at 4°C, and then with the
appropriate secondary antibodies goat anti-rabbit (1:5,000; cat.
no. ab6721) and goat anti-mouse (1:10,000; cat. no. ab6789; both
were from Abcam) for 1 h at room temperature. The immune complexes
were detected using ECL Western Blotting kit (Thermo Fisher
Scientific, Inc.). The relative protein expression was analysed
using Image-Pro Plus software 6.0, using β-actin as the internal
reference.
Luciferase reporter assay
The possibility of miR-328-5p and RAGE target
binding was predicted using the online software TargetScan
(http://www.targetscan.org/vert_71/).
The RAGE mRNA predicted binding site region and mutated site region
were cloned into the psi-CHECK2 vector. MDA-MB-231 cells were
co-transfected with psi-CHECK2 vectors and miR-122 mimics or
scrambled controls. After 48 h, the supernatants were collected and
the luciferase activities were assessed using a Dual-Luciferase
Reporter Assay system (Promega, Fitchburg, WI, USA).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA) or
SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). All experiments
were repeated at least thrice, and the data are shown as the mean ±
standard deviation. An unpaired two-tailed Student's t-test and
one-way analysis of variance with Bonferroni post hoc test were
used to analyse the data, depending on conditions and group number.
The relationship between RAGE and miR-328-5p expression was
determined by Pearson correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Analysis of differentially expressed
miRNAs
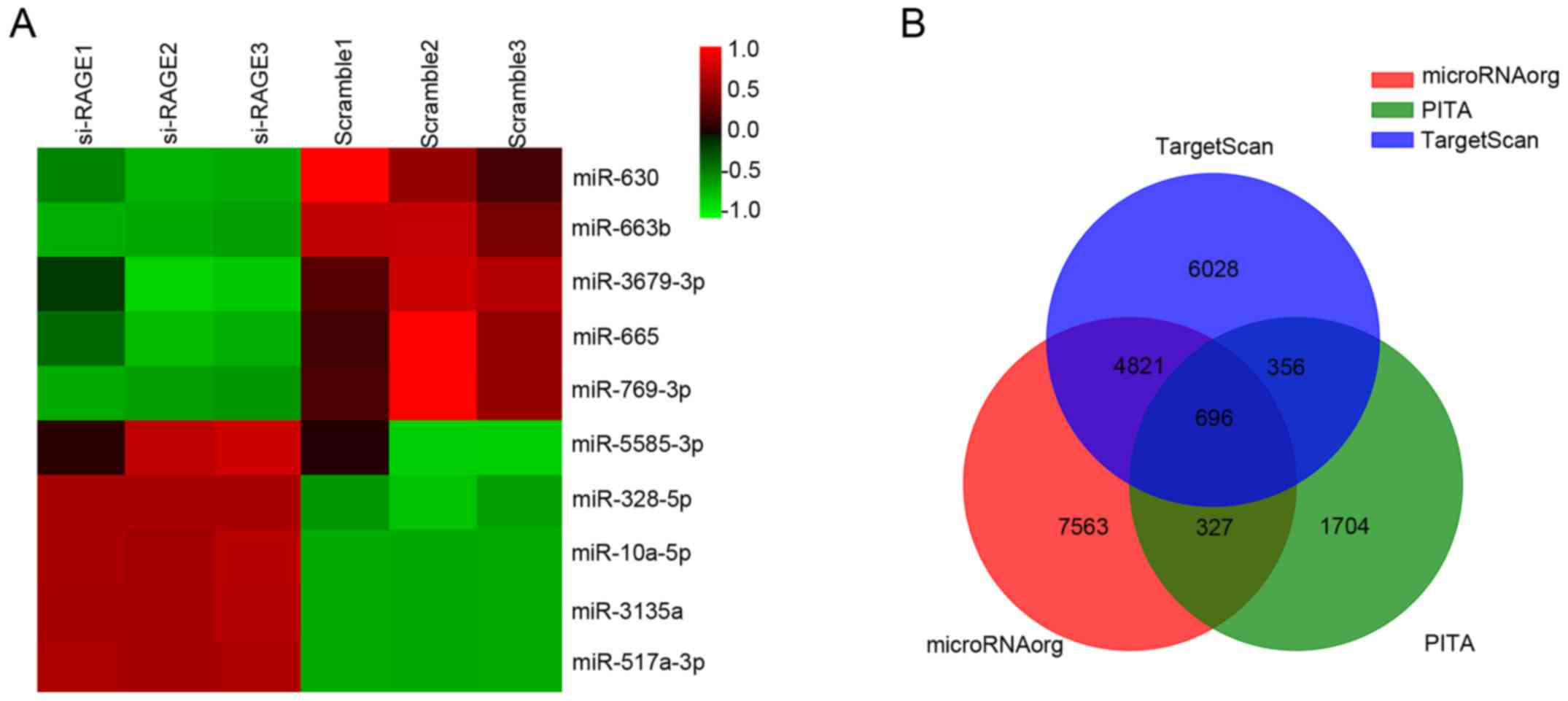
In total, 2,550 miRNAs were detected by Agilent
Human miRNA microarray. In addition, 10 miRNAs were found to be
differentially expressed after knockdown of RAGE, as shown in
Fig. 1A. The expression changes and
reported functions of the miRNAs are listed in Table I (9–18).
 | Table I.The fold changes and reported
functions of the 10 differently expressed miRNAs. |
Table I.
The fold changes and reported
functions of the 10 differently expressed miRNAs.
| Name | Fold change | Control vs. RAGE
siRNA | Reported in breast
cancer | Functions in
cancers | Refs. |
|---|
| miR-10a-5p | 27.93 | Up | Y | 1. Reduced in breast
cancer and may be a potential tumor suppressor in breast
cancer. | (9,10) |
|
|
|
|
| 2. Is deemed as a
valuable marker for prognostication of breast cancer patients. |
|
| miR-3135a | 141.20 | Up | N | N/A |
|
| miR-328-5p | 50.23 | Up | Y | Negatively
regulates the expression of breast cancer resistance protein
(BCRP/ABCG2) in human cancer cells. | (11) |
| miR-517a-3p | 258.76 | Up | N | 1. Induces cell
apoptosis in bladder cancer cell lines. | (12,13) |
|
|
|
|
| 2. Downregulation
of miR-517a promotes proliferation of hepatocellular carcinoma
cells. |
|
| miR-5585-3p | 8.82 | Up | N | N/A |
|
| miR-630 | 9.87 | Down | Y | Suppresses breast
cancer progression, inhibits cell migration and invasion. | (14) |
| miR-663b | 7.76 | Down | Y | Induces
chemotherapy resistance in human breast cancer cells. | (15) |
| miR-665 | 3.05 | Down | N | N/A |
|
| miR-769-3p | 3.47 | Down | Y | 1. Downregulates
NDRG1 and enhances apoptosis in MCF-7 cells during
reoxygenation. | (16,17) |
|
|
|
|
| 2. Promotes cell
proliferation in human melanoma by suppressing GSK3B
expression. |
|
| miR-3679-3p | 3.64 | Down | N | Promising biomarker
of different staging of lung squamous cell carcinoma. | (18) |
Analysis of the predicted target genes
of differentially expressed miRNAs
Target genes of differentially expressed miRNAs were
predicted using three databases (TargetScan, microRNA.org and PITA), as shown in Fig. 1B. GO and KEGG analyses were used to
determine the roles of these databases with crossed target genes.
According to the results shown in Fig.
2, we found that the following targets were principally
involved: i) biological processes (transcription from RNA
polymerase II promoter, response to mechanical stimulus and
regulation of insulin secretion); ii) cellular components (i.e.
nucleoplasm, cytoplasm and nuclear chromatin); and iii) molecular
functions (protein and chromatin binding, ionotropic glutamate
receptor binding). These genes are mainly involved in the following
pathways or conditions: Hippo signalling, alcoholism, Rap1 and
various cancer proliferation-related pathways.
Verification of the miRNA microarray
results
qPCR was used to assess the microarray results. The
results of all 10 differentially expressed miRNAs were the same
with the microarray results and had almost similar fold changes,
but only slightly lower (Fig.
3).
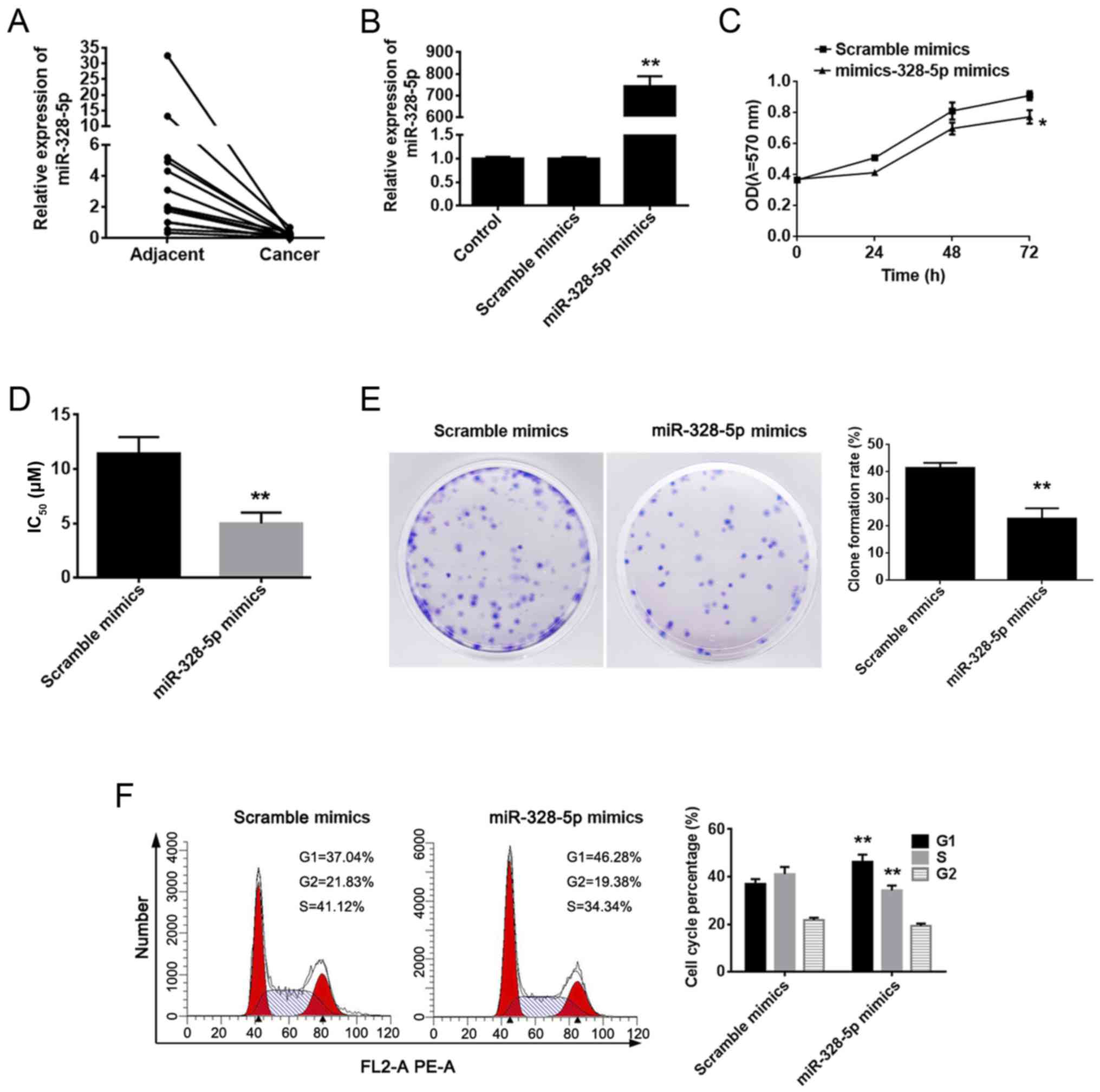
miR-328-5p is downregulated in breast
cancer tissues
Since miR-328-5p has been reported less in breast
cancer, it was chosen for further studies. To investigate the role
of miR-328-5p in breast cancer, we collected 13 samples of breast
cancer tissue and compared these with healthy adjacent tissues. The
qPCR results revealed that the expression of miR-328-5p was
significantly decreased in breast cancer tissue compared with
adjacent tissues (Fig. 4A). To
assess the function of miR-328-5p in breast cancer, we used
miR-328-5p mimics to construct a miR-328-5p overexpression cell
model in MDA-MB-231 cells, with scrambled mimics as a negative
control. The representative qPCR assay results, displayed in
Fig. 4B, indicated that our
miR-328-5p overexpression model was successfully constructed.
miR-328-5p mimics inhibit cell proliferation, drug
resistance and the cell cycle. To elucidate the functions of
miR-328-5p in breast cancer, MDA-MB-231 cells were transfected with
miR-328-5p and scramble mimics for 24, 48 and 72 h. MTT and colony
formation assays revealed that miR-328-5p mimics significantly
suppressed cell viability (Fig. 4C and
E). However, the Transwell assay revealed that miR-328-5p
mimics did not affect cell invasiveness (data not shown). Moreover,
chemotherapy resistance against DDP was suppressed in the
miR-328-5p mimic group, as previously described by Pan et al
(11) (Fig. 4D). To investigate the mechanisms of
miR-328-5p inhibition on cellular proliferation and drug
resistance, we analysed the effects of miR-328-5p on cell apoptosis
and the cell cycle. As shown in Fig.
4F, we found that the cell cycle was significantly arrested in
phase G1/S after overexpression of miR-328-5p. However, apoptosis
was not influenced (data not shown).
RAGE is a direct target gene of
miR-328-5p in MDA-MB-231 cells
The microarray was conducted after knockdown of
RAGE; hence, we analysed the relationship between RAGE and
miR-328-5p. We found that the relative expression level of RAGE was
markedly decreased when miR-328-5p was overexpressed in MDA-MB-231
cells (Fig. 5A and B). Then we
analysed the correlation between RAGE and miR-328-5p expression in
13 samples of breast cancer tissue. We found that there was a
negative correlation between RAGE and miR-328-5p expression in
breast cancer tissue (Fig. 5C).
Online (http://www.targetscan.org/)
bioinformatics predictions revealed that RAGE was a target of
miR-328-5p. The Wt-RAGE-3′UTR or Mut-RAGE-3′UTR luciferase reporter
vector was generated, respectively (Fig. 5D). We further performed a luciferase
reporter assay to confirm their relationship in MDA-MB-231 cells.
As shown in Fig. 5E, luciferase
activity was significantly downregulated only in cells
co-transfected with the miR-328-5p mimics and the Wt-RAGE-3′UTR
vector, which indicated that miR-328-5p could directly bind to the
3′ untranslated region (3′UTR) of RAGE in breast cancer cells.
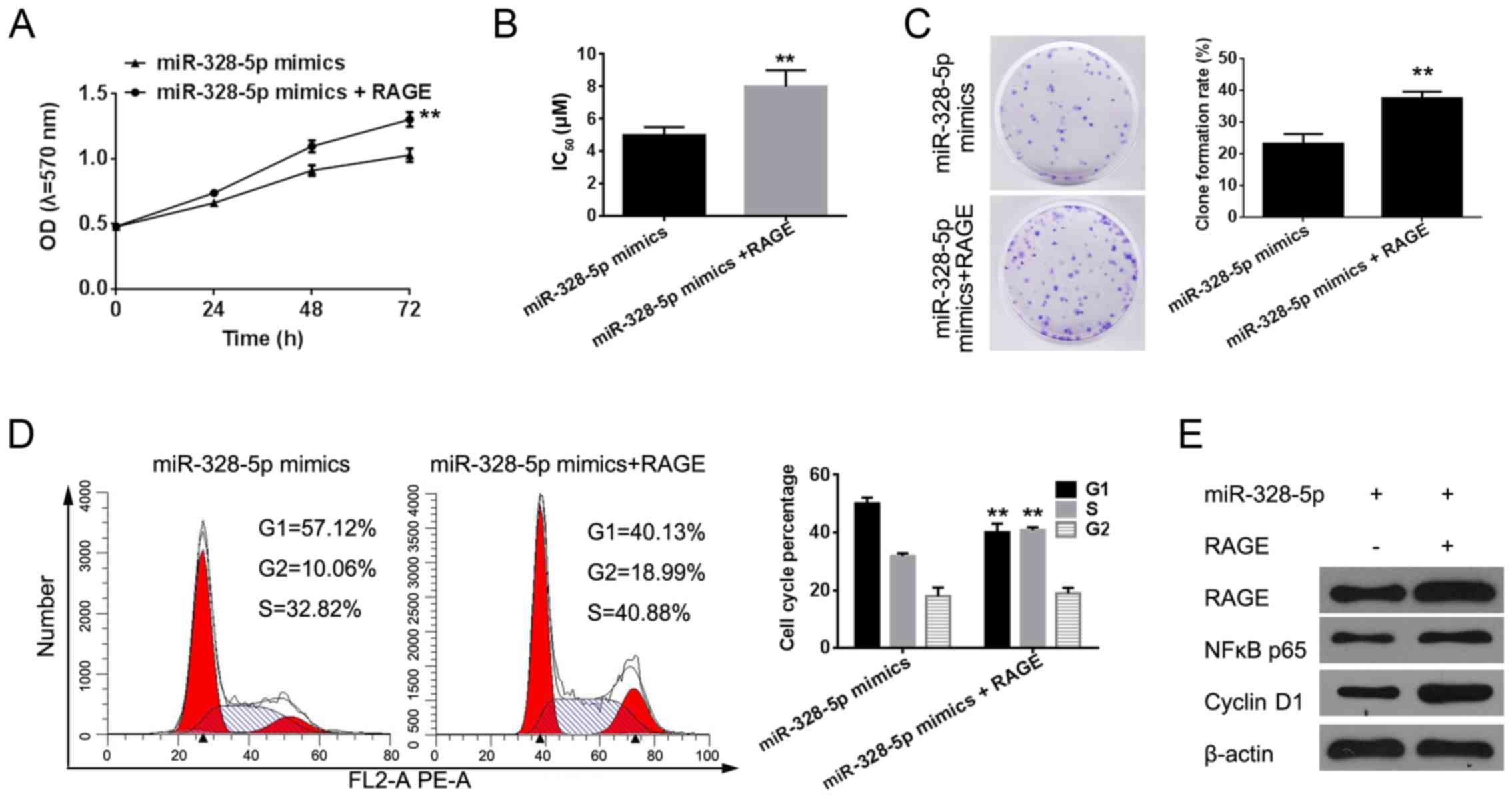
Effect of miR-328-5p in MDA-MB-231 is
achieved by targeting RAGE
After confirming that RAGE was the target of
miR-328-5p, we investigated whether the function of miR-328-5p in
MDA-MB-231 cells was associated with RAGE by co-transfecting the
overexpression vector of RAGE in cells overexpressing miR-328-5p to
further observe cellular function changes. We found that RAGE
overexpression could reverse the inhibition of cell growth,
chemotherapy resistance, clone formation ability and cell cycle
progression caused by the miR-328-5p mimics (Fig. 6A-D). We used western blotting to
assess the protein expression level of RAGE, nuclear factor
κ-light-chain-enhancer of activated B cells (NF-κB) and cyclin D1.
We found that RAGE enhanced the expression levels of these proteins
(Fig. 6E).
Discussion
The effects of miR-328-5p in breast cancer molecular
pathogenesis were thoroughly investigated in this study. Low
expression of miR-328-5p in breast cancer tissues and high
expression in RAGE-interfering cells were detected. Therefore, we
predicted that the expression of miR-328-5p plays an important role
in breast cancer development and may be regarded as a
tumour-suppressor factor. To confirm this hypothesis, a miR-328-5p
overexpression model was established in MDA-MB-231 cells. After
modelling, cell growth, cell proliferation, drug resistance and
cell cycle patterns were inhibited in the group of miR-328-5p
mimics. Moreover, we found that there was a target relationship
between miR-328-5p and RAGE. Therefore, we concluded that
miR-328-5p inhibits the development of breast cancer cells by
targeting RAGE.
RAGE is regarded as an oncogene and is overexpressed
in many types of cancers such as colon (19) and pancreatic cancer (20) and melanoma (21). In our previous research (7), we found that the RAGE expression level
was much higher in MDA-MB-231 cells than in other breast cancer
cells. Therefore, we chose MDA-MB-231 cells to assess the miRNA
microarray in order to explore the mechanism of RAGE as the main
research object in this study. MDA-MB-231 (triple-negative basal
subtype) exhibits markedly high proliferation (22–24)
and has more aggressive clinical behaviour, with a higher
recurrence rate, than other breast cancer subtypes (25–27).
Hence, it has been a great challenge to find another line of
treatment for this subtype.
In the miRNA microarray KEGG analysis results, we
found that interference by RAGE influences many cancer-related
pathways such as Ras, MAPK, PI3K/Akt and TGFβ signalling pathways.
The differentially expressed miRNAs are also closely related with
cancer, as shown in Table I.
Among the differentially expressed miRNAs,
miR-328-5p suppressed the survival of oesophageal cancer cells
(28), inhibited cervical cancer
cell proliferation and tumorigenesis (29) and was downregulated in colorectal
cancer (30). Although miR-328-5p
has been reported to be associated with cancer resistance, there
are few reports on breast cancer; hence, we chose miR-328-5p as our
target gene.
Notably, in the results of the cellular function
experiments, we found that the cellular functions produced by the
overexpression of miR-328-5p coincided with those produced by
interference with RAGE, as previously indicated by our study
(7). Then, the target relationship
between miR-328-5p and RAGE was confirmed by software prediction
and luciferase reporter assay. A previous study revealed that the
function of RAGE was associated with NF-κB and cyclin D1. NF-κB is
a protein complex that controls DNA transcription. Aberrant
regulation of NF-κB has been linked to cancer and NF-κB stimulates
proliferation in different cell types, including human breast
cancers (31). Cyclin D1 is a
protein involved in normal cell cycle regulation and is responsible
for the transition from the G1 (resting) phase to the S (DNA
synthesis) phase. In this study, the western blotting results in
Fig. 6E suggest that miR-328-5p
regulates breast cancer through these two pathways by targeting
RAGE.
For our future research, we will collect more
clinical samples and prognoses to analyse the value of miR-328-5p
as a diagnosis marker. Moreover, we would like to demonstrate
whether or not miR-328-5p plays the same role in other breast
cancer cell lines or in other cancers entirely. In addition, the
relationship between miR-328-5p and other signalling pathways
obtained by biological analysis needs to be addressed. Concerning
RAGE, there are many studies on the enhancement of invasive
function; however, in our study, the function of miR-328-5p was not
related with invasiveness in breast cancer cells. Furthermore, we
intend to continue researching the other miRNAs from the microarray
results and discover other pathways in which RAGE regulates
invasion.
We conclude that miR-328-5p overexpression reduced
the proliferation of breast cancer cells MDA-MB-231 by targeting
RAGE. Accordingly, overexpression of miR-328-5p or interference
with RAGE could be helpful in minimising breast cancer
proliferation. These data revealed that miR-328-5p represents a
novel strategy for the genetic therapeutic intervention of breast
cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Natural Science Foundation of Hunan Province, China (no.
2017JJ3423).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
TL and WW conceived and designed the study. TL, YY,
QH and XM performed the experiments. TL and WW wrote the paper. YY,
QH and XM reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be account for all aspects of
the research in ensuring that the accuracy or integrity of any part
of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethical Committee of the Third Xiangya Hospital of Central South
University (Changsha, China).
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNAs
|
microRNAs
|
|
RAGE
|
receptor for advanced glycosylation
end-product specific receptor
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Poellinger A: Near-infrared imaging of
breast cancer using optical contrast agents. J Biophotonics.
5:815–826. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Onitilo AA, Engel JM, Greenlee RT and
Mukesh BN: Breast cancer subtypes based on ER/PR and Her2
expression: Comparison of clinicopathologic features and survival.
Clin Med Res. 7:4–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lohrisch C and Piccart M: HER2/neu
as a predictive factor in breast cancer. Clin Breast Cancer.
2:129–135; discussion 136–137. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fabian MR, Sundermeier TR and Sonenberg N:
Understanding how miRNAs post-transcriptionally regulate gene
expression. Prog Mol Subcell Biol. 50:1–20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Radia AM, Yaser AM, Ma X, Zhang J, Yang C,
Dong Q, Rong P, Ye B, Liu S and Wang W: Specific siRNA targeting
receptor for advanced glycation end products (RAGE) decreases
proliferation in human breast cancer cell lines. Int J Mol Sci.
14:7959–7978. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stoetzer OJ, Fersching DM, Salat C,
Steinkohl O, Gabka CJ, Hamann U, Braun M, Feller AM, Heinemann V,
Siegele B, et al: Circulating immunogenic cell death biomarkers
HMGB1 and RAGE in breast cancer patients during neoadjuvant
chemotherapy. Tumour Biol. 34:81–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khan S, Wall D, Curran C, Newell J, Kerin
MJ and Dwyer RM: MicroRNA-10a is reduced in breast cancer and
regulated in part through retinoic acid. BMC Cancer. 15:3452015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang CH, Fan TC, Yu JC, Liao GS, Lin YC,
Shih AC, Li WH and Yu AL: The prognostic significance of RUNX2 and
miR-10a/10b and their inter-relationship in breast cancer. J Transl
Med. 12:2572014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan YZ, Morris ME and Yu AM: MicroRNA-328
negatively regulates the expression of breast cancer resistance
protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol.
75:1374–1379. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoshitomi T, Kawakami K, Enokida H,
Chiyomaru T, Kagara I, Tatarano S, Yoshino H, Arimura H, Nishiyama
K, Seki N, et al: Restoration of miR-517a expression induces cell
apoptosis in bladder cancer cell lines. Oncol Rep. 25:1661–1668.
2011.PubMed/NCBI
|
|
13
|
Liu RF, Xu X, Huang J, Fei QL, Chen F, Li
YD and Han ZG: Down-regulation of miR-517a and miR-517c promotes
proliferation of hepatocellular carcinoma cells via targeting Pyk2.
Cancer Lett. 329:164–173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou CX, Wang CL, Yu AL, Wang QY, Zhan MN,
Tang J, Gong XF, Yin QQ, He M, He JR, et al: MiR-630 suppresses
breast cancer progression by targeting metadherin. Oncotarget.
7:1288–1299. 2016.PubMed/NCBI
|
|
15
|
Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, Yao
H, Song E, Chen Y, Wang M, et al: The overexpression of
hypomethylated miR-663 induces chemotherapy resistance in human
breast cancer cells by targeting heparin sulfate proteoglycan 2
(HSPG2). J Biol Chem. 288:10973–10985. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo EC, Chang YC, Sher YP, Huang WY,
Chuang LL, Chiu YC, Tsai MH, Chuang EY and Lai LC: MicroRNA-769-3p
down-regulates NDRG1 and enhances apoptosis in MCF-7 cells
during reoxygenation. Sci Rep. 4:59082014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qiu HJ, Lu XH, Yang SS, Weng CY, Zhang EK
and Chen FC: MiR-769 promoted cell proliferation in human melanoma
by suppressing GSK3B expression. Biomed Pharmacother. 82:1172016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pu Q, Huang Y, Lu Y, Peng Y, Zhang J, Feng
G, Wang C, Liu L and Dai Y: Tissue-specific and plasma microRNA
profiles could be promising biomarkers of histological
classification and TNM stage in non-small cell lung cancer. Thorac
Cancer. 7:348–354. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fuentes MK, Nigavekar SS, Arumugam T,
Logsdon CD, Schmidt AM, Park JC and Huang EH: RAGE activation by
S100P in colon cancer stimulates growth, migration, and cell
signaling pathways. Dis Colon Rectum. 50:1230–1240. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takada M, Koizumi T, Toyama H, Suzuki Y
and Kuroda Y: Differential expression of RAGE in human pancreatic
carcinoma cells. Hepatogastroenterology. 48:1577–1578.
2001.PubMed/NCBI
|
|
21
|
Wagner NB, Weide B, Reith M, Tarnanidis K,
Kehrel C, Lichtenberger R, Pflugfelder A, Herpel E, Eubel J,
Ikenberg K, et al: Diminished levels of the soluble form of RAGE
are related to poor survival in malignant melanoma. Int J Cancer.
137:2607–2617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lerma E, Peiro G, Ramón T, Fernandez S,
Martinez D, Pons C, Muñoz F, Sabate JM, Alonso C, Ojeda B, et al:
Immunohistochemical heterogeneity of breast carcinomas negative for
estrogen receptors, progesterone receptors and Her2/neu (basal-like
breast carcinomas). Mod Pathol. 20:1200–1207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subhawong AP, Subhawong T, Nassar H,
Kouprina N, Begum S, Vang R, Westra WH and Argani P: Most
basal-like breast carcinomas demonstrate the same Rb-/p16+
immunophenotype as the HPV-related poorly differentiated squamous
cell carcinomas which they resemble morphologically. Am J Surg
Pathol. 33:163–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reis-Filho JS and Tutt ANJ: Triple
negative tumours: A critical review. Histopathology. 52:108–118.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nielsen TO, Hsu FD, Jensen K, Cheang M,
Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler
L, et al: Immunohistochemical and clinical characterization of the
basal-like subtype of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
El-Rehim Abd DM, Pinder SE, Paish CE, Bell
J, Blamey RW, Robertson JF, Nicholson RI and Ellis IO: Expression
of luminal and basal cytokeratins in human breast carcinoma. J
Pathol. 203:661–671. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan C, Oh DS, Wessels L, Weigelt B, Nuyten
DS, Nobel AB, van't Veer LJ and Perou CM: Concordance among
gene-expression-based predictors for breast cancer. N Engl J Med.
355:560–569. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han N, Zhao W, Zhang Z and Zheng P:
MiR-328 suppresses the survival of esophageal cancer cells by
targeting PLCE1. Biochem Biophys Res Commun. 470:175–180. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X and Xia Y: microRNA-328 inhibits
cervical cancer cell proliferation and tumorigenesis by targeting
TCF7L2. Biochem Biophys Res Commun. 475:169–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu XT, Xu Q, Tong JL, Zhu MM, Nie F, Chen
X, Xiao SD and Ran ZH: MicroRNA expression profiling identifies
miR-328 regulates cancer stem cell-like SP cells in colorectal
cancer. Br J Cancer. 106:1320–1330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jones RL, Rojo F, A'Hern R, Villena N,
Salter J, Corominas JM, Servitja S, Smith IE, Rovira A, Reis-Filho
JS, et al: Nuclear NF-κB/p65 expression and response to neoadjuvant
chemotherapy in breast cancer. J Clin Pathol. 64:130–135. 2011.
View Article : Google Scholar : PubMed/NCBI
|