Introduction
Esophageal carcinoma (EC) is a common
gastrointestinal tumor with leading cancer-related mortality
(1,2). Notably, as a main subtype of
esophageal carcinoma, the incidence of ESCC is particularly high in
China, with 1.5×105 cases of mortality annually reported
(3,4). The carcinogenesis of ESCC is
considered to be a multi-factor and multi-step process (5). Although medical and surgical
techniques have improved, the prognosis for ESCC is unsatisfactory,
and the 5-year survival rate of ESCC patients is in the range of
26.2–49.4% (6,7). Therefore, it is important to
understand the underlying molecular mechanisms and find novel
molecular markers associated with ESCC.
MicroRNAs (miRNAs) are non-coding RNAs, which
downregulate the gene expression of their targets by binding to
their 3′-untranslated regions (3′-UTRs) (8,9). It
has been revealed that miRNAs play important roles in multiple
cellular physiological processes, including cell proliferation,
apoptosis and differentiation (10,11).
Aberrantly expressed miRNAs are known to be closely associated with
tumor development and progression via the regulation of cell
growth, drug resistance and metastasis (12). miR-183, as a tumor-promoter, was
reported to enhance human ESCC cell proliferation and invasion
(13), while miR-486-5p was found
to exert antitumor effects by regulating the between cell
proliferation and apoptosis in cancers, such as NSCLC (14), breast cancer (15) and hepatocellular carcinoma (HCC)
(16). Collectively, miRNAs have
been recognized as diagnostic biomarkers for cancers.
miR-125b, a tumor-suppressor miRNA, suppressed cell
proliferation and differentiation by affecting Hedgehog signaling
in cerebellar neuronal progenitor and tumor cells (17). Overexpression of miR-125b also
suppressed the level of Bak1 and induced prostate cancer cell
proliferation (18). Additionally,
miR-125b exhibited its antitumor effects in colorectal (19) and breast cancer (20). Recent studies have revealed that
miR-125b expression was decreased in ESCC tissues (21). However, the correlation between
miR-125b and pathogenesis of ESCC still remains unknown. Herein, we
analyzed the underlying mechanisms of action of miR-125b, resulting
in tumor suppression in ESCC.
Materials and methods
ESCC tissue collection
Human esophageal tumor tissues and control normal
tissues were simultaneously isolated from 66 patients between
January 2015 and December 2016. All patients underwent
esophagectomy without chemotherapy or radiotherapy at the
Department of Gastroenterology at Jiangsu Cancer Hospital. All 66
patients provided written informed consent, and all experimental
protocols were approved by the Ethics Committee of Jiangsu Cancer
Hospital. Fresh tissues were stored at −80°C until future use.
Cell culture and transfection
In the present study, the human ESCC cell lines
(EC109 and EC9706) and human esophageal epithelial cells (HET-1A)
were obtained from Riken BioResource Center (Tsukuba, Japan). Cell
lines were cultured in Roswell Park Memorial Institute-1640 medium
(RPMI-1640; Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen;
Thermo Fisher Scientific, Inc.), 100 U/ml streptomycin and 100 U/ml
penicillin (Invitrogen; Thermo Fisher Scientific, Inc.).
miR-125b negative control (NC), mimics, inhibitors
and BCL-2-modifying factor (BMF) small interfering RNAs (siRNAs)
were purchased from GenePharma Company (Shanghai, China). The
aforementioned plasmids were transfected into ESCC cell lines
(EC109 and EC9706) and HET-1A using Lipofectamine 2000 (Thermo
Fisher Scientific, Inc.).
RNA isolation and quantitative
real-time PCR (qRT-PCR)
Total RNA was extracted from tissue or cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. RNA was
reversed-transcribed into cDNA using PrimeScript Reverse
Transcription kit (Takara Bio, Inc., Otsu, Japan). The qRT-PCR
reactions were performed using SYBR Premix Ex Taq (Takara Bio,
Inc.) on a 7500 ABI system (Applied Biosystems; Thermo Fisher
Scientific, Inc). U6-siRNA was used for normalization. Specific PCR
primers were synthesized at Invitrogen; Thermo Fisher Scientific,
Inc. The primer sequences used were as follows: miR-125b,
5′-TCCCTGAGACCTAACTTGTG-3′ (forward); U6,
5′-ACGCAAATTCGTGAAGCGTT-3′ (forward), a universal primer (reverse);
BMF, 5′-CCCATAAGCCAGGAAGACAA-3′ (forward), and
5′-CTGAAGCTTTCTGGCGATTCT-3′ (reverse).
Cell proliferation assay
The cell proliferation rate was evaluated using Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan), following the manufacturer's instructions.
Twenty-four hours after transfection, the cells were seeded into
96-well plates at a density of ~2×103 cells/well. The
absorbance of each sample was assessed using a microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) at three different
time-points. Each experiment was conducted in triplicate.
Cell apoptosis
A flow cytometer (BD FACSCalibur; BD Biosciences)
was used for quantifying the cell apoptosis rates. After
transfection for 48 h, the cells were collected by centrifugation
and were incubated in 500 µl binding buffer supplemented with 5 µl
FITC-Annexin V and 5 µl propidium iodide (PI). Fluorescence of the
stained cells was then analyzed using flow cytometry.
Cell cycle analysis
Cells were collected as previously described and
fixed in cold 70% ethanol overnight at 4°C. After staining with PI
and treatment with 100 µl RNase A, the cells were kept in the dark
at room temperature for 30 min. The signals were analyzed using
flow cytometry (BD FACSCalibur; BD Biosciences).
Tumor growth assay
All animal research was conducted using a protocol
approved by the Animal Care and Use Committee of Nanjing Medical
University (Nanjing, China). The animals were maintained at 25°C on
a 12-h light/dark cycle, and housed in a controlled environment and
received food and water ad libitum. The animals were
acclimated in rooms for 7 days before the initiation of the
experiment.
A total of 1×106 cells (EC109 cells with
stable expression of miR-125b, control cells or NC) were injected
the left forelimb in nude mice. The nude mice (25±5 g) purchased
from the Laboratory Animal Centre of Nanjing Medical University and
used in the experiment were both male and female, at a ratio of
1:1. Six mice were assigned per group. Tumor volumes were assessed
every 7 days. The mean tumor volume was calculated using the
formula: [0.5 × (length × width2)]. After 28 days, the
mice were sacrificed, and then tumors were dissected.
Mice were sacrificed using cervical dislocation
euthanasia, when the following humane endpoints were present: i)
tumor volumes reached ≥10% of the body weight of the mouse; ii)
tumor length ≥20 mm; iii) continuous body weight loss; iv) a rapid
growth of the tumor to ulceration, causing infection or necrosis;
and v) tumors of any size interfering with eating, drinking,
urinating, defecating, or walking. No mice exhibited any signs of
illness due tumor formation. No animals presented multiple tumors.
Animal health observations were performed twice daily at 10 and 3
pm by our husbandry staff.
Histological and immunohistochemical
analyses
Tumor tissues were dissected and stained with
hematoxylin and eosin (H&E). For immunohistochemistry, tumor
tissues were stained with Ki-67. The tissue apoptosis rate was
assessed using the TdT-mediated dUTP nick and labeling (TUNEL)
detection kit (Biotool, Houston, TX, USA), according to the
manufacturer's instructions. Images were obtained using an Olympus
microscope (Olympus, Tokyo, Japan).
Luciferase reporter assay
The mutant or wild-type 3′-UTR of BMF was amplified
and cloned into the vector psiCHECK-2 to construct luciferase
reporter plasmids. Cells (1×104/well) were
co-transfected with vectors and miR-125b mimics in each well. They
were incubated in RPMI-1640 medium supplemented with 10% FBS. The
luciferase activity was detected using the Dual-Luciferase Reporter
Assay System (Promega Corp., Madison, WI, USA) after incubation for
48 h. All the assays were performed in triplicate.
Western blotting
To assess the protein expression in tumor tissues
and cells, total proteins were extracted using RIPA lysis buffer.
The supernatant was collected by centrifugation at 13,282 × g for
15 min. Next, the extracted proteins were incubated with the
loading buffer at 100°C for 5 min. Protein concentration was
assessed using the Pierce BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.). Proteins were subjected to separation on 10%
SDS-PAGE gels and transferred onto polyvinylidene fluoride (PVDF)
membranes (Milipore, Billerica, MA, USA), which were blocked using
5% (w/v) non-fat dry milk. The membranes were probed using the
following antibodies: rabbit anti-Bax antibody (1:2,000; cat. no.
2744), rabbit anti-Bcl-2 antibody (1:2,000; cat. no. 4223; both
from Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit
anti-caspase-3 antibody (1:2,000; cat. no. ab13847; Abcam,
Cambridge, UK) and rabbit anti-p27 antibody (1:2,000; cat. no.
3688; Cell Signaling Technology). The membranes were visualized
using an ECL kit (Beyotime Institute of Biotechnology, Beijing,
China). The amount of protein was normalized with respect to GAPDH
and analyzed using the ImageJ software (NIH, Bethesda, MD, USA).
All experiments were performed independently in triplicate.
Statistical analysis
GraphPad Prism 6 software was used to carry out all
statistical analyses (GraphPad Software, Inc., La Jolla, CA, USA).
When only two groups were compared, a Student's t-test was
conducted. In addition, one-way ANOVA using Dunnett's multiple
comparison test was applied to compare differences between multiple
groups. A P-value of <0.05 was considered to indicate a
statistically significant difference. In addition, the relationship
between the expression of miR-125b and BMF was analyzed using
Pearson's correlation analysis. All quantitative data are expressed
as the mean ± SD.
Results
miR-125b is downregulated in ESCC
tissues and cells
The level of miR-125b in clinical ESCC and normal
tissues was assessed using real-time PCR. The results indicated
that miR-125b expression was downregulated in the ESCC tumor
tissues compared with paired normal tissues (Fig. 1A). Furthermore, compared to HET-1A,
a normal esophageal epithelial cell line, the expression of
miR-125b was lower in ESCC cell lines (EC109 and EC9706) (Fig. 1B). In addition, a decrease in the
level of miR-125b was markedly associated with lymphatic metastasis
in patients; Table I reveals the
detailed clinical characteristics of patients. These results
revealed that miR-125b may play a crucial role in ESCC
carcinogenesis.
 | Table I.The association between the
expression level of miR-125b with the clinical characteristics of
ESCC patients. |
Table I.
The association between the
expression level of miR-125b with the clinical characteristics of
ESCC patients.
| Clinical
characteristics | No. of
patients | Relative
expression | P-value |
|---|
| Age (years) |
|
| 0.681 |
|
≤60 | 35 | 0.364 |
|
|
>60 | 31 | 0.523 |
|
| Sex |
|
| 0.753 |
|
Male | 45 | 0.462 |
|
|
Female | 21 | 0.534 |
|
| Smoking |
|
| 0.712 |
|
Yes | 15 | 0.496 |
|
| No | 51 | 0.552 |
|
| Drinking |
|
| 0.806 |
|
Yes | 18 | 0.409 |
|
| No | 48 | 0.598 |
|
| pT stage |
|
| 0.668 |
|
T1+T2 | 30 | 0.460 |
|
|
T3+T4 | 36 | 0.517 |
|
| Lymphatic
metastasis |
|
| 0.035 |
|
Positive | 23 | 0.226 |
|
|
Negative | 43 | 0.654 |
|
| pTNM stage |
|
| 0.951 |
|
≤II | 26 | 0.554 |
|
|
>II | 40 | 0.512 |
|
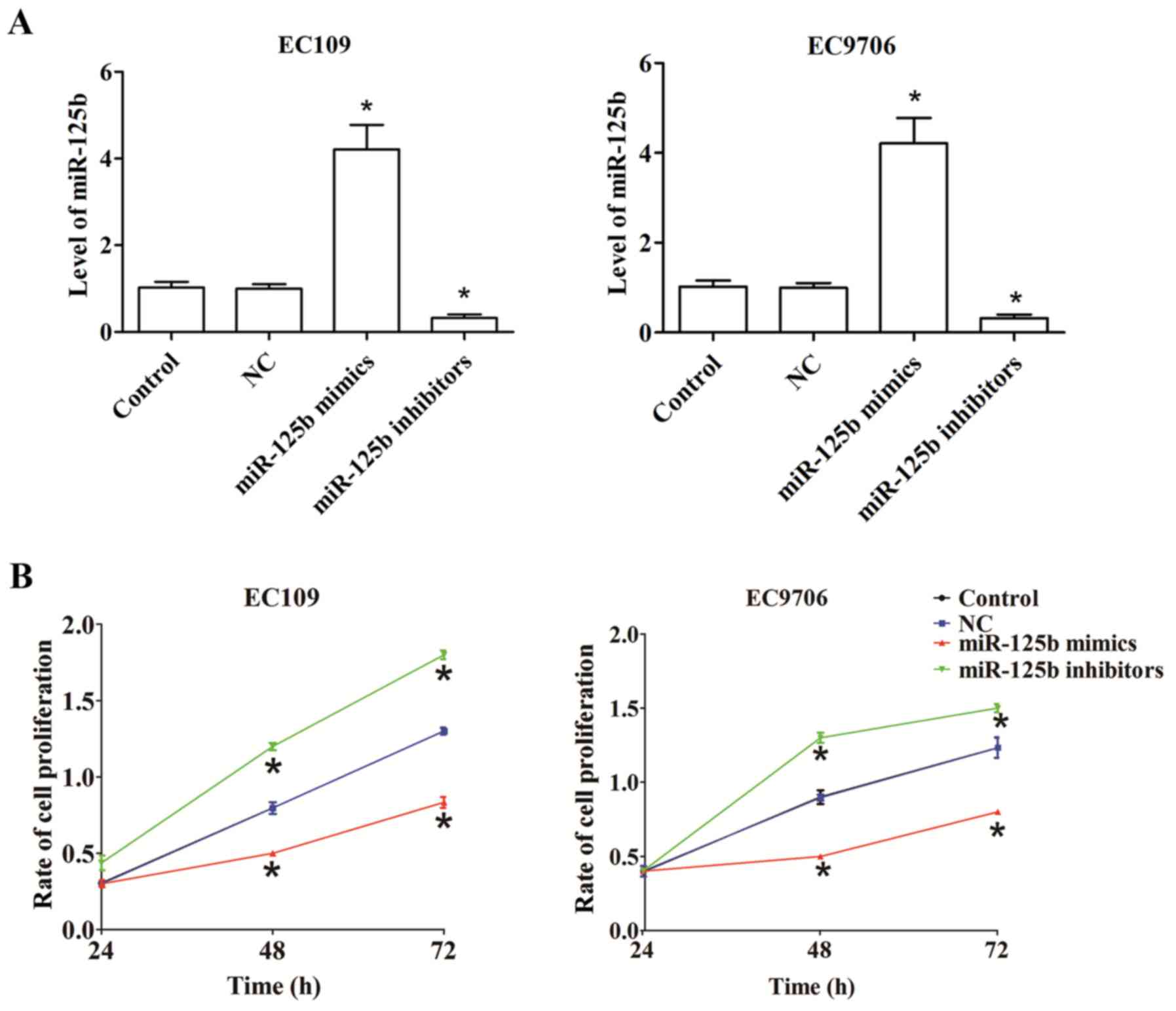
miR-125b suppresses proliferation in
ESCC cells
To explore the roles of miR-125b in ESCC, human
EC109 and EC9706 cells were transfected with miR-125b mimics or
inhibitors. Compared with the control, transfection with miR-125b
mimics significantly upregulated miR-125b expression, while
transfection with miR-125b inhibitors significantly downregulated
miR-125b expression (Fig. 2A).
A CCK-8 assay was performed to assess the
proliferation rate of EC109 and EC9706 cells transfected with
miR-125b mimics or miR-125b inhibitors for 24, 48 and 72 h.
Compared with the control, overexpressed miR-125b decreased cell
proliferation in EC109 and EC9706 cells, whereas the growth of
EC109 and EC9706 cells significantly increased post-transfection
with miR-125b inhibitors (Fig.
2B).
miR-125b induces cell cycle arrest in
the G1 phase in ESCC
Flow cytometric analysis was performed to detect the
effect of miR-125b on the cell cycle in EC109 and EC9706 cells. The
results revealed that upregulation of miR-125b significantly
increased the G1 phase of the cell cycle, while this phase was
significantly decreased when the EC109 and EC9706 cells were
transfected with miR-125b inhibitors compared with the control.
Collectively, it was observed that miR-125b could arrest the cell
cycle in the G1 phase in ESCC (Fig.
3).
miR-125b promotes apoptosis in ESCC
cells
To determine whether the decrease in cell
proliferation post-transfection with miR-125b mimic transfection
was the result of apoptosis in EC109 and EC9706 cells, flow
cytometric analysis was conducted to detect the apoptotic cells.
For EC109 cells, the the proportion of apoptotic cells (Q2 + Q3) in
the control group was 8.88±2.95%, while the proportion of apoptotic
cells (Q2 + Q3) was 51.50±9.47% in the miR-125b mimic group. This
revealed that the number of apoptotic cells markedly increased
after the EC109 cells were transfected with miR-125b mimics.
Similarly, the EC9706 cells after transfection with miR-125b mimics
exhibited increased cell apoptosis compared with the control, while
inhibition of miR-125b significantly decreased cell apoptosis
(Fig. 4A).
Furthermore, the expression of apoptosis-related
proteins, including Bax, Bcl-2, caspase-3 and p27, was analyzed. Of
these proteins, 3 of the pro-apoptotic proteins, namely Bax,
caspase-3 and p27, were observed to be upregulated in the EC109 and
EC9706 cells transfected with miR-125b mimics in comparison with
the cells of the control group. However, the anti-apoptotic protein
Bcl-2 was significantly downregulated in cells transfected with
miR-125b mimics. Additionally, the opposite results were obtained
when the cells were transfected with miR-125b inhibitors. Thus, the
levels of pro-apoptotic proteins were downregulated, while that of
an anti-apoptotic protein was upregulated (Fig. 4B).
miR-125b inhibits tumor growth in
vivo
We verified the role of miR-125b in ESCC using
miR-125b mimics and miR-125b inhibitors, in vitro. The
results revealed that overexpression of miR-125b inhibited cell
proliferation and induced cell apoptosis. Conversely,
downregulation of miR-125b promoted cell proliferation and
suppressed cell apoptosis. Thus, we next studied the inhibitory
role of miR-125b in ESCC in vivo.
To investigate the potential effect of miR-125b on
tumor growth in vivo, EC109 cells were transfected with
miR-125b mimics or NC and injected into nude mice. In this
experiment, one mouse reached a humane endpoint, and the percentage
of the total number of animals used in the study was 4.17%. After
the end of the experiment, the maximum tumor burden in mice was
9.84% (the tumor weight was 3.12 g and the animal body weight was
31.7 g), the longest diameter exhibited by a single subcutaneous
tumor was 1.90 cm, and none of the animals presented >1
tumor.
Compared with the control, miR-125b mimics markedly
inhibited tumor growth. Furthermore, compared to the control group,
the tumor weight of the miR-125b mimic group was remarkably lesser
than the NC group. Additionally, compared to the control group, a
lower expression of Ki-67 and a higher percentage of apoptotic
cells were observed in the miR-125b mimic group using
immunohistochemical staining (Fig.
5).
BMF is a direct target gene of
miR-125b
TargetScan was employed to search for the potential
target genes of miR-125b. It identified BMF as a candidate target
gene (Fig. 6A). The Dual-Luciferase
reporter assay was performed to confirm this hypothesis (Fig. 6B). Furthermore, we examined whether
the expression of BMF was affected by miR-125b in ESCC cells. In
our study, we observed that the level of BMF was significantly
reduced in the EC109 and EC9706 cells transfected with miR-125b
mimics (Fig. 6C). In the in
vivo tumor growth experiment, immunohistochemical analysis of
the tumor sections revealed decreased expression of BMF in the
miR-125b mimic group (Fig. 6D).
Silencing of BMF suppresses cell
proliferation and induces apoptosis in ESCC
To clarify whether BMF was involved in regulating
ESCC cell proliferation and apoptosis, we knocked down its
expression by transfecting the EC109 and EC9706 cells with si-BMF.
qRT-PCR and western blotting were performed to assess the
transfection efficiency. Compared to the control, the expression of
BMF was markedly downregulated in the EC109 and EC9706 cells
transfected with si-BMF (Fig. 7A and
B).
Cell proliferation was evaluated using
the CCK-8 assay
EC109 and EC9706 cells transfected with si-BMF
exhibited slower growth than the control cells (Fig. 7C). Moreover, compared to the
control, the si-BMF group exhibited an increase in the G1 phase of
the cell cycle in EC109. Similar results were obtained for the
EC9706 cells (Fig. 7D). BMF
silencing notably promoted cell apoptosis in EC109 and EC9706
cells. For EC109 cells, the proportion of apoptotic cells (Q2 + Q3)
was 8.09±1.96% in the control group, while the proportion of
apoptotic cells (Q2 + Q3) was 30.30±5.61% in the si-BMF group thus,
revealing a significant increase in apoptotic cells. Similar
results were obtained for the EC9706 cells (Fig. 8A). Western blot analysis indicated
that BMF silencing markedly increased the expression of Bax,
caspase-3 and p27, and decreased that of Bcl-2 in ESCC cells
(Fig. 8B). Collectively, these
results revealed that BMF participated in the miR-125b-mediated
regulation of ESCC cell proliferation, the cell cycle and
apoptosis.
The expression level of miR-125b is
negatively correlated with that of BMF in ESCC
The relationship between BMF and miR-125b was
further confirmed. We assessed the expression of BMF in tissues of
ESCC patients and ESCC cell lines. The results indicated that BMF
was increasingly upregulated in tumor tissues than in the adjacent
non-cancerous tissues (Fig. 9A and
C). We further observed that the levels of BMF in EC109 and
EC9706 were in accordance with the tissues (Fig. 9B and D). In addition, we also
explored the relationship between BMF and miR-125b. The result
revealed a negative correlation between miR-125b and BMF levels
(Fig. 9E).
Discussion
Accumulating evidence has revealed that miRNAs are
closely associated with the initiation and progression of ESCC by
activating or suppressing multiple malignant processes (22,23).
However, the mechanisms underlying ESCC pathogenesis have not been
fully elucidated. In the present study, miR-125b expression was
observed to be considerably low in ESCC tissues and cells.
Furthermore, low miR-125b expression in tumors was significantly
correlated with lymphatic metastasis in patients, thus revealing
that miR-125b may be a useful prognostic biomarker. It was also
observed that the level of miR-125b was closely associated with
ESCC tumorigenesis. We demonstrated that the overexpression of
miR-125b suppressed cell proliferation, and induced cell cycle
arrest, and promoted apoptosis in ESCC lines EC109 and EC9706,
which have been widely used in research of esophageal squamous cell
cancer (24,25). Additionally, overexpression of
miR-125b markedly inhibited tumor cell growth in vivo.
Furthermore, we observed that miR-125b directly targeted BMF in
ESCC cells. Notably, by decreasing the level of BMF similar
outcomes as with the overexpression of miR-125b were obtained.
Aberrant expression of miRNAs has been revealed in
several types of cancer (26,27).
Therefore, a better understanding of the mechanisms underlying
miRNA-mediated regulation networks is vital for developing
diagnostic and therapeutic strategies (28,29).
Herein, miR-125b expression was observed to be markedly decreased
in ESCC. Previous studies have reported the functions of miR-125b
in other types of cancers, including gallbladder and colorectal
cancer, and melanoma (30–32). miR-125b had the ability to regulate
cell proliferation, differentiation and invasion (33,34),
and potentially act as an tumor suppressor in some cancer types,
suppressing tumorigenesis and development (35,36).
We found that miR-125b was significantly reduced in thyroid cancer
and oral squamous cell carcinoma (37,38),
indicating that it may act as a tumor suppressor. miR-125b was
found to be markedly upregulated in the prostate, and it inhibited
cancer cell proliferation by suppressing the expression of BAK1.
However, the molecular mechanism of miR-125b in ESCC pathogenesis
remains largely unknown.
In the present study, BMF was validated as a direct
target of miR-125b. It was observed to be frequently overexpressed
in a variety of tumors (39). As an
oncogene, BMF participates in the pathogenesis of several tumors.
In human E-cadherin-negative breast cancer, BMF promoted tumor
growth and metastasis, and was activated by the transcription
factor FOXO3 (40). In colon
cancer, BMF promoted cell proliferation and inhibited apoptosis,
and was activated by the transcription factor Eomes (39). Consequently, BMF is a member of the
pro-apoptotic Bcl-2 family, which is mainly associated with cell
proliferation and apoptosis. In the present study, the results
revealed that silencing BMF could markedly inhibit cell growth and
promote cell apoptosis in EC109 and EC9706 cells.
In summary, the results indicated that knockdown of
miR-125b expression may prevent ESCC tumor initiation and
progression by controlling BMF levels. This may be used as a
potential therapeutic strategy to inhibit ESCC progression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YXF and XHB conceived and designed the study. PDQ,
ZZC, JW and QZ performed the experiments. YXF and QZ wrote the
manuscript. YHL and PWY reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the report work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethics Committee of Jiangsu Cancer Hospital (Nanjing, Jiangsu,
China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kollarova H, Machova L, Horakova D,
Janoutova G and Janout V: Epidemiology of esophageal cancer - an
overview article. Biomed Pap Med Fac Univ Palacky Olomouc Czech
Repub. 151:17–20. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bohanes P, Yang D, Chhibar RS, Labonte MJ,
Winder T, Ning Y, Gerger A, Benhaim L, Paez D, Wakatsuki T, et al:
Influence of sex on the survival of patients with esophageal
cancer. J Clin Oncol. 30:2265–2272. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao P, Dai M, Chen W and Li N: Cancer
trends in China. Jpn J Clin Oncol. 40:281–285. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J, Xie X, Zhou C, Peng S, Rao D and Fu
J: Which factors are associated with actual 5-year survival of
oesophageal squamous cell carcinoma? Eur J Cardiothorac Surg.
41:e7–e11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in Cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yi Y, Lu X, Chen J, Jiao C, Zhong J, Song
Z, Yu X and Lin B: Downregulated miR-486-5p acts as a tumor
suppressor in esophageal squamous cell carcinoma. Exp Ther Med.
12:3411–3416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferracin M, Veronese A and Negrini M:
Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev
Mol Diagn. 10:297–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren LH, Chen WX, Li S, He XY, Zhang ZM, Li
M, Cao RS, Hao B, Zhang HJ, Qiu HQ, et al: MicroRNA-183 promotes
proliferation and invasion in oesophageal squamous cell carcinoma
by targeting programmed cell death 4. Br J Cancer. 111:2003–2013.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33(1181–1189): 21042014.
|
|
15
|
Zhang G, Liu Z, Cui G, Wang X and Yang Z:
MicroRNA-486-5p targeting PIM-1 suppresses cell proliferation in
breast cancer cells. Tumour Biol. 35:11137–11145. 2104. View Article : Google Scholar
|
|
16
|
Huang XP, Hou J, Shen XY, Huang CY, Zhang
XH, Xie YA and Luo XL: MicroRNA-486-5p, which is downregulated in
hepatocellular carcinoma, suppresses tumor growth by targeting
PIK3R1. FEBS J. 282:579–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ferretti E, De Smaele E, Miele E, Laneve
P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E,
Screpanti I, et al: Concerted microRNA control of Hedgehog
signalling in cerebellar neuronal progenitor and tumour cells. EMBO
J. 27:2616–2627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ
and White RW: miR-125b promotes growth of prostate cancer xenograft
tumor through targeting pro-apoptotic genes. Prostate. 71:538–549.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fujino Y, Takeishi S, Nishida K, Okamoto
K, Muguruma N, Kimura T, Kitamura S, Miyamoto H, Fujimoto A,
Higashijima J, et al: Downregulation of miR-100/miR-125b is
associated with lymph node metastasis in early colorectal cancer
with submucosal invasion. Cancer Sci. 108:390–397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun Y, Liu X, Zhang Q, Mao X, Feng L, Su
P, Chen H, Guo Y and Jin F: Oncogenic potential of TSTA3 in breast
cancer and its regulation by the tumor suppressors miR-125a-5p and
miR-125b. Tumour Biol. 37:4963–4972. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu J, Wang Y and Wu X: MicroRNA in the
pathogenesis and prognosis of esophageal cancer. Curr Pharm Des.
19:1292–1300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakai NS, Samia-Aly E, Barbera M and
Fitzgerald RC: A review of the current understanding and clinical
utility of miRNAs in esophageal cancer. Semin Cancer Biol.
23:512–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mayne GC, Hussey DJ and Watson DI:
MicroRNAs and esophageal cancer - implications for pathogenesis and
therapy. Curr Pharm Des. 19:1211–1226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin YY, Chen QJ, Xu K, Ren HT, Bao X, Ma
YN, Wei Y and Ma HB: Involvement of microRNA-141-3p in
5-fluorouracil and oxaliplatin chemo-resistance in esophageal
cancer cells via regulation of PTEN. Mol Cell Biochem. 422:161–170.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han N, Zhao W, Zhang Z and Zheng P:
MiR-328 suppresses the survival of esophageal cancer cells by
targeting PLCE1. Biochem Biophys Res Commun. 470:175–180. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen L, Wang Q, Wang GD, Wang HS, Huang Y,
Liu XM and Cai XH: miR-16 inhibits cell proliferation by targeting
IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS
Lett. 587:1366–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q
and Ma B: MicroRNA-34a inhibits the proliferation and metastasis of
osteosarcoma cells both in vitro and in vivo. PLoS One.
7:e337782012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duan Z, Choy E, Harmon D, Liu X, Susa M,
Mankin H and Hornicek F: MicroRNA-199a-3p is downregulated in human
osteosarcoma and regulates cell proliferation and migration. Mol
Cancer Ther. 10:1337–1345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jianwei Z, Fan L, Xiancheng L, Enzhong B,
Shuai L and Can L: MicroRNA 181a improves proliferation and
invasion, suppresses apoptosis of osteosarcoma cell. Tumour Biol.
34:3331–3337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang D, Zhan M, Chen T, Chen W, Zhang Y,
Xu S, Yan J, Huang Q and Wang J: miR-125b-5p enhances chemotherapy
sensitivity to cisplatin by down-regulating Bcl2 in gallbladder
cancer. Sci Rep. 7:431092017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu X, Shi W, Zhang Y, Wang X, Sun S, Song
Z, Liu M, Zeng Q, Cui S and Qu X: CXCL12/CXCR4 axis induced
miR-125b promotes invasion and confers 5-fluorouracil resistance
through enhancing autophagy in colorectal cancer. Sci Rep.
7:422262017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pei G, Lan Y, Chen D, Ji L and Hua ZC: FAK
regulates E-cadherin expression via
p-SrcY416/p-ERK1/2/p-Stat3Y705 and
PPARγ/miR-125b/Stat3 signaling pathway in B16F10 melanoma cells.
Oncotarget. 8:13898–13908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bousquet M, Quelen C, Rosati R, Mansat-De
Mas V, La Starza R, Bastard C, Lippert E, Talmant P,
Lafage-Pochitaloff M, Leroux D, et al: Myeloid cell differentiation
arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid
leukemia with the t(2;11)(p21;q23) translocation. J Exp Med.
205:2499–2506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mizuno Y, Yagi K, Tokuzawa Y,
Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda
A, Amemiya T, et al: miR-125b inhibits osteoblastic differentiation
by down-regulation of cell proliferation. Biochem Biophys Res
Commun. 368:267–272. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Visone R, Pallante P, Vecchione A,
Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V,
Borbone E, et al: Specific microRNAs are downregulated in human
thyroid anaplastic carcinomas. Oncogene. 26:7590–7595. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Yan LX, Wu QN, Du ZM, Chen J,
Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, et al: miR-125b is
methylated and functions as a tumor suppressor by regulating the
ETS1 proto-oncogene in human invasive breast cancer. Cancer Res.
71:3552–3562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gefen N, Binder V, Zaliova M, Linka Y,
Morrow M, Novosel A, Edry L, Hertzberg L, Shomron N, Williams O, et
al: Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1
(TEL/AML1) leukemias and confers survival advantage to growth
inhibitory signals independent of p53. Leukemia. 24:89–96. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang R, Kang Y, Löhr CV, Fischer KA,
Bradford CS, Johnson G, Dashwood WM, Williams DE, Ho E and Dashwood
RH: Reciprocal regulation of BMF and BIRC5 (Survivin) linked to
Eomes overexpression in colorectal cancer. Cancer Lett.
381:341–348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hornsveld M, Tenhagen M, van de Ven RA,
Smits AM, van Triest MH, van Amersfoort M, Kloet DE, Dansen TB,
Burgering BM and Derksen PW: Restraining FOXO3-dependent
transcriptional BMF activation underpins tumour growth and
metastasis of E-cadherin-negative breast cancer. Cell Death Differ.
23:1483–1492. 2016. View Article : Google Scholar : PubMed/NCBI
|