Introduction
Pancreatic cancer (PC) is one of the most lethal
human malignancies, with an overall five-year survival rate of
<5% (1). The high mortality rate
associated with PC can be largely attributed to its highly
aggressive nature, wherein local invasion and remote metastasis may
occur during the early stages of carcinogenesis (2). Thus, the majority of patients
diagnosed with PC cannot undergo surgery and chemotherapy is thus
the main treatment option. At present, gemcitabine (GEM) is the
first-line drug used in the treatment of PC. However, its
therapeutic efficacy is far from satisfactory due to the inherent
chemoresistance of PC (3). A
previous study revealed that only 23.8% of GEM-treated patients
received therapeutic benefits in their early stages of treatment
(4). However, the majority of these
patients faced therapeutic failure due to the obtained
chemoresistance against GEM. Thus, a better understanding of the
molecular mechanisms underlying the development of GEM
chemoresistance is necessary to develop novel-targeted therapies to
‘flip the switch’ from drug resistance to susceptibility in PC.
Recently, a set of non-coding RNAs (ncRNAs),
including microRNAs (miRNAs or miRs) and long non-coding RNAs
(lncRNAs), have been found to be involved in PC pathogenesis.
Circular RNAs (circRNAs) are a special class of endogenously
expressed non-coding RNAs, which are featured with a covalently
closed loop structure without a 5′ to 3′ polarity and
polyadenylated tail (5). circRNAs
are highly conserved in mammals and are mainly expressed in a cell
type-specific or developmental stage-specific manner, indicating
their involvement in various physiological and pathological
processes (6–8). Currently, studies have confirmed that
circRNAs contain conserved miRNA binding sites and function as
miRNA sponges to modulate the expression of target genes. To date,
the dysregulation of circRNAs has been reported in a set of human
diseases, particularly in cancer development and progression
(9–16). A recent study demonstrated that
clusters of circRNAs were aberrantly expressed in PC compared with
normal samples (17); however, the
specific roles of circRNAs in PC pathogenesis remain unknown.
In the present study, to explore the roles of
circRNAs in the development of chemoresistance in PC, we first
developed a GEM-resistant PC cell line (SW1990/GZ) by exposing
SW1990 PC cells to gradient concentrations of GEM. We then
performed microarray analysis of SW1990/GZ cells, along with their
parental control cells. Our results demonstrated several
differentially expressed circRNAs that may be involved in the
transformation of the GEM resistance of PC and may provide
potential molecular biomarkers or therapeutic targets for PC in the
future.
Materials and methods
Establishment of GEM-resistant cell
line, SW1990/GZ
The PC cell line, SW1990, was purchased from the
Institute of Biochemistry and Cell Biology of the Chinese Academy
of Sciences (Shanghai, China). The GEM-resistant cell line,
SW1990/GZ, was established by repeated subcultures in the presence
of stepwise increases in GEM concentrations (Tocris Bioscience,
Ellisville, MO, USA) during the growth of SW1990 cells. First,
SW1990 cells were cultured in RPMI-1640 (HyClone Laboratories/GE
Healthcare, Chicago, IL, USA) containing 10% fetal bovine serum
(Gibco/Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
various concentrations of GEM. Subsequently, cell death was
observed, and the median lethal dose of SW1990 cells was set to
0.07 µg/ml. Subsequently, the SW1990 cells were cultured in medium
containing GEM at a concentration of 0.1 µg/ml. Following
incubation at 37°C for 48 h, the culture and dead cells were
replaced with fresh drug-free medium. The remaining cells could
then grow and probably enter the logarithmic phase of cell growth.
They were passaged twice and then re-cultured in medium containing
GEM at a concentration of 0.1 µg/ml. The medium was then replaced
with culture medium containing GEM at a concentration of 0.4 µg/ml,
and the cells were cultured with the aforementioned cycle progress,
according to four-fold increase in the drug concentration. Finally,
the cells were cultured in a medium with a drug concentration of
400 µg/ml. Therefore, the filter viable cells produced a stable
resistance to high concentrations of GEM. After 10 months, we had
successfully acquired a stable GEM-resistant cell line designated
as SW1990/GZ.
Drug sensitivity assay
Cells (3×103 cells/well) were seeded in
96-well plates. After 12 h, the cells were exposed to increasing
concentrations of GEM (0, 25, 50, 100, 150, 200, 250, 300, 350 and
400 µg/ml) and incubated at 37°C for 72 h to determine the
IC50 value using Cell Counting Kit-8 assay (CCK-8;
Dojindo Molecular Technologies, Inc., Kumamoto, Japan) to evaluate
the sensitivity to GEM.
RNA extraction, purification and array
hybridization
Total RNA was extracted from three samples of
SW1990/GZ and SW1990 cells using TRIzol reagent (Invitrogen,
Carlsbad, CA, USA) and treated with Rnase R (Epicentre, Madison,
WI, USA) to remove linear RNA. Subsequently, enriched circRNA
samples were amplified and transcribed into fluorescent cRNA by a
random priming method (Arraystar Super RNA Labeling kit; Arraystar,
Inc., Rockville, MD, USA). The labeled cRNAs were purified by the
RNeasy mini kit (Qiagen GmbH, Hilden, Germany). The concentration
and specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were
determined by NanoDrop ND-1000 (NanoDrop Technologies, Inc.,
Wilmington, DE, USA). Labeled cRNA (1 µl each) was fragmented by
adding 5 µl of 10X blocking agent and 1 µl of 25X fragmentation
buffer, and the mixture was then heated at 60°C for 30 min.
Subsequently, the labeled cRNA was diluted with 25 µl of 2X
hybridization buffer. Finally, the labeled cRNA was hybridized
using Human 8×15 K circRNA Array (Arraystar). The hybridized arrays
were washed, fixed and scanned using the Agilent DNA Microarray
Scanner G2505C (Agilent Technologies, Inc., Santa Clara, CA,
USA).
Microarray data analysis
Agilent Feature Extraction software (version
11.0.1.1; Agilent Technologies) was used to analyze scanned images
for raw data extraction. Quintile normalization and subsequent data
processing were performed using the R software package R version
3.1.2 (Agilent Technologies), and low intensity filtering was
performed. In the comparison of the cricRNA profiles of two groups
using Student's t-test, the differences with a fold change (FC) of
≥2 and P<0.05 were considered statistically significant.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cell samples using
TRIzol reagent (Invitrogen/Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Briefly, cDNA was synthesized from 1 µg of total RNA with
SuperScript III Reverse Transcriptase (Invitrogen Japan) samples.
Quantitative PCR (qPCR) was performed using a SYBR PrimeScript
RT-PCR kit (Takara, Kyoto, Japan) on a Rotor-Gene 6000 real-time
genetic analyzer (Corbett Life Science, Mortlake, Australia).
Primer sequences for candidate genes are listed in Table I. The PCR program was initiated for
10 min at 95°C before 40 thermal cycles, each at 10 sec at 95°C and
1 min at 60°C to collect fluorescent signals. The relative
quantification of circRNA expression levels was determined by
taking the average of the GAPDH-normalized 2−ΔΔCq values
(18).
 | Table I.The list of primers (F, forward and R,
reverse) used in this study. |
Table I.
The list of primers (F, forward and R,
reverse) used in this study.
| circRNA | Primer sequence |
|---|
| circ_101543 | F:
5′-AAAAAGCACAGGCAGTTACTCA-3′ |
|
| R:
5′-CATTCCAGTAGGCGCTAAGA-3′ |
| circ_000926 | F:
5′-TTGTGCTTTCTGGAGGGTCT-3′ |
|
| R:
5′-GCACAAATAAACCCCACATTTT-3′ |
| circ_003251 | F:
5′-TATTATTCCCCCAGCTGCTC-3′ |
|
| R:
5′-CTGCTGCAACAGAAACCTGA-3′ |
| circ_004077 | F:
5′-AAGATCCCGGATGACATGAG-3′ |
|
| R:
5′-GAGTCTTGGGAGGGTTGTCA-3′ |
| circ_101672 | F:
5′-GGTTCTGCACCATCTTCAGG-3′ |
|
| R:
5′-TGGTGGTGGTCTTGTAGTCG-3′ |
| circ_102747 | F:
5′-GTATCCTGGCCTGCCATC-3′ |
|
| R:
5′-TTGCCTCATCACCAACCA-3′ |
| GAPDH | F: 5′-
CATGAGAAGTATGACAACAGCCT-3′ |
|
| R:
5′-AGTCCTTCCACGATACCAAAGT-3′ |
GO and pathway analyses
The potential functions of the parental genes of
differential circRNAs were analyzed using the Database for
Annotation, Visualization and Integrated Discovery (DAVID;
https://david.ncifcrf.gov/). The
parental gene function was then predicted by GO functional
annotation in terms of biological process (BP), cellular component
(CC) and molecular function (MF). The results of the GO analysis
are presented in a scatter plot, and the related pathways of the
parental genes of differential circRNAs were analyzed by Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/).
miRNA prediction
The circRNA-miRNA interaction was predicted by using
Arraystar home-made miRNA target prediction software (Arraystar),
which is based on the TargetScan (http://www.targetscan.org/vert_72/) and miRanda
(http://www.microrna.org/) prediction algorithm.
The differentially expressed circRNAs in all the comparisons were
annotated in detail with the circRNA/miRNA interaction
information.
CircRNA-miRNA-target gene network
To further elucidate the associations between
circRNAs and miRNAs, potential circRNA-miRNA-target gene
interaction analysis was conducted by Cytoscape (version 3.6.1;
http://www.cytoscape.org/). The size of each node
represents the number of putative miRNA functionally connected to
each circRNA.
Statistical analysis
Statistical analysis was conducted using SPSS 13.0
software (SPSS, Inc., Chicago, IL, USA). The Student's t test was
used for comparisons between two groups. P<0.05 was considered
to indicate a statistically significant difference.
Results
Drug resistance index of
SW1990/GZ
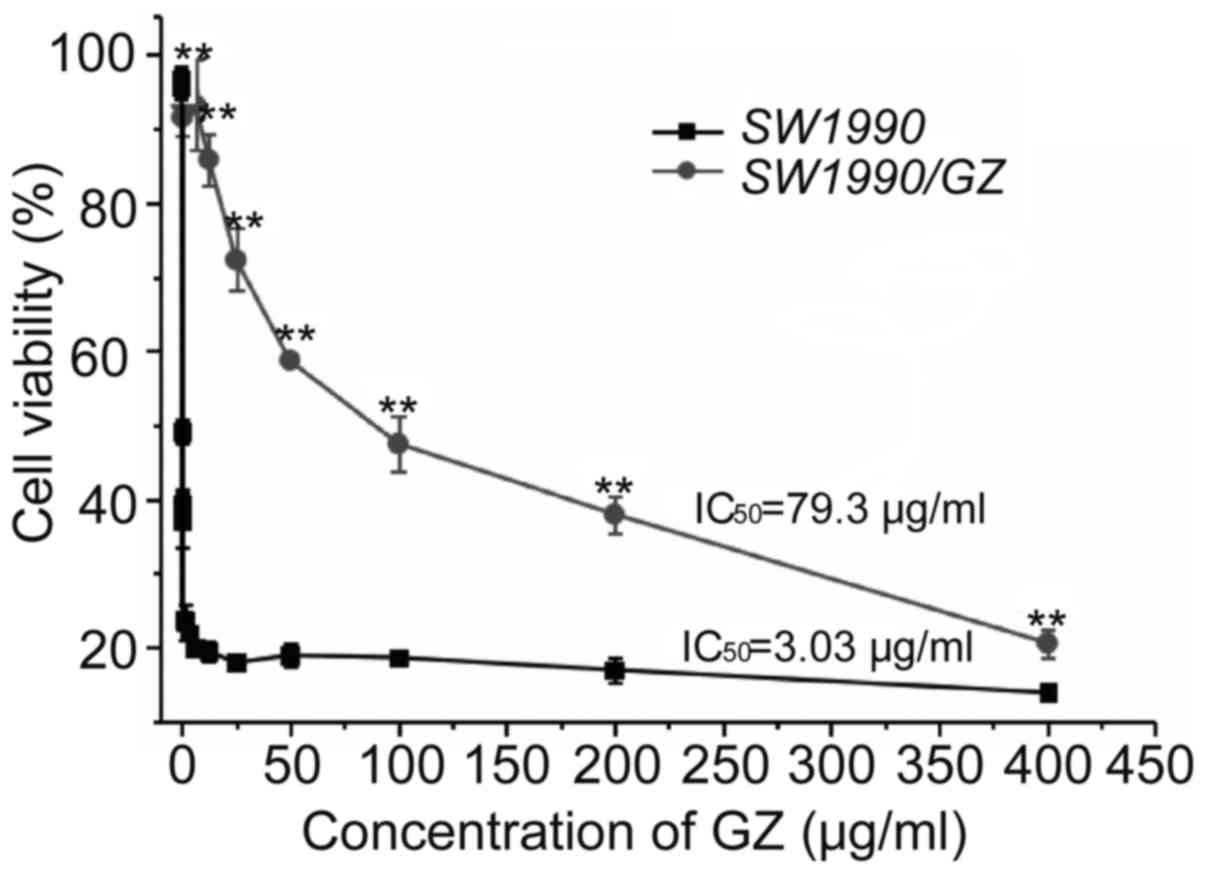
The GEM resistance of the SW1990/GZ cell line was
identified by determining its IC50 value against the
parental SWl990 cell line. The IC50 value of GEM for the
SW1990/GZ cells was 79.3±5.31 µg/ml, which was 26.2-fold higher
than that of the parental cell line, SWl990 (3.03±0.27 µg/ml),
indicating that the drug resistance index of SWl990/GZ cells
relative to the parental SW1990 cells was 26.2 (Fig. 1). The GEM-resistant cell line,
SW1990/GZ, was thus successfully established.
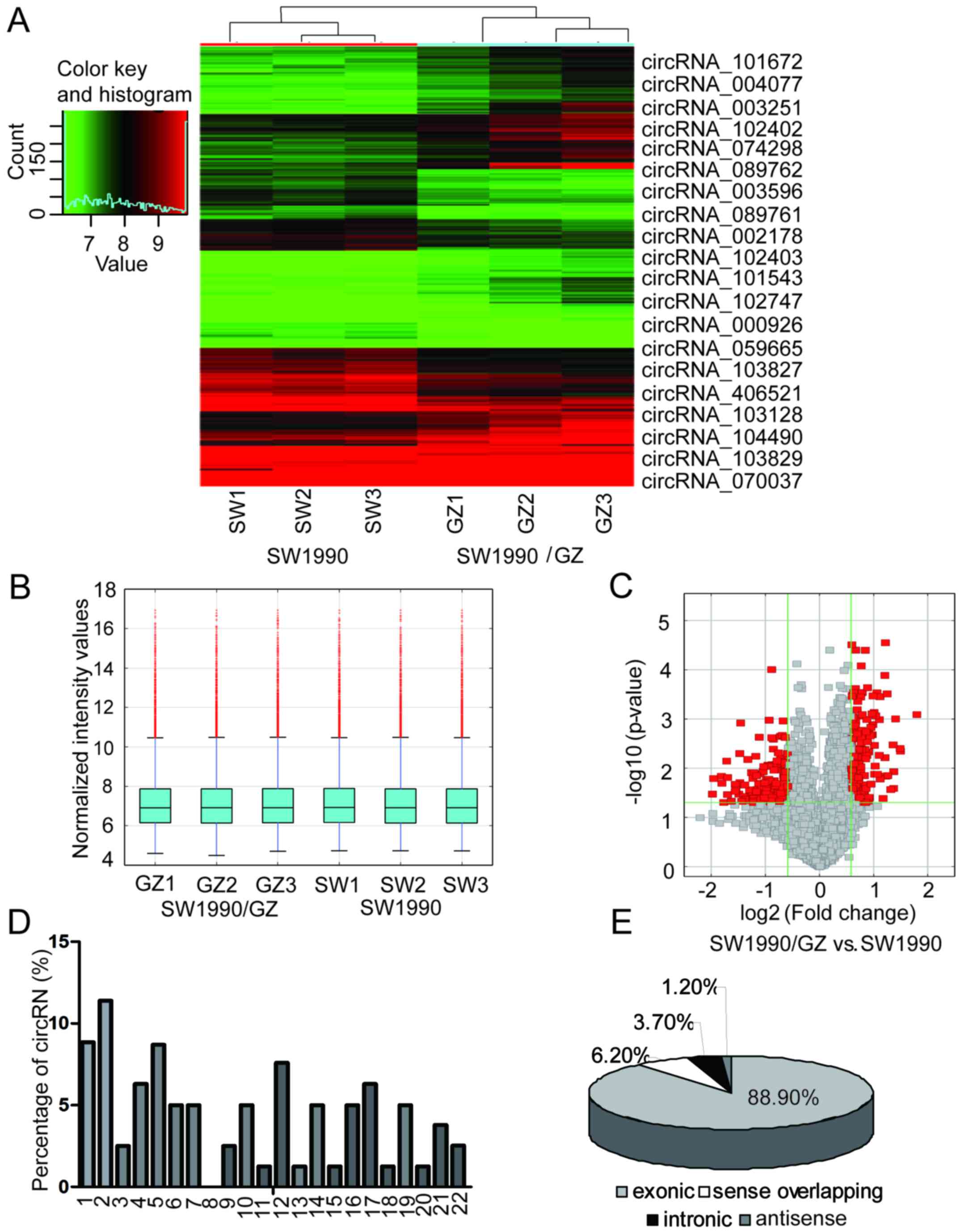
Overview of circRNA profiles
The expression of human circRNAs was screened in the
SW1990 and SW1990/GZ cell samples using a microarray platform.
Hierarchical clustering and box plot visualization revealed a
distinguishable circRNA expression pattern between SW1990 and
SW1990/GZ cells (Fig. 2A and B). In
total, 81 circRNAs were differentially expressed (fold change ≥2.0
and P<0.05) between the SW1990/GZ and SW1990 cells. Among these,
26 circRNAs were upregulated and 55 circRNAs were downregulated
>2-fold in SW1990/GZ cells. The 10 circRNAs with the most
signficant increased and decreased in expression in the SW1990/GZ
cells compared to the SW1990 cells are displayed in Fig. 2A and Table II. Significantly, the expression
levels of circRNA_101672, circRNA_004077 and circRNA_003251 were
upregulated in SW1990/GZ by 3.47-, 2.82- and 2.81-fold,
respectively. Furthermore, circRNA_101543, circRNA_102747 and
circRNA_000926 were downregulated by 3.94-, 3.88- and 3.51-fold,
respectively. We also analyzed the chromosome distribution of these
deregulated circRNAs. The differentially expressed circRNAs with
statistically significance between SW1990/GZ and SW1990 cells were
identified by volcano plot filtering (Fig. 2C). As displayed in Fig. 2D, each chromosome had circRNA
locations, while chromosomes 1, 2, 4, 5, 12 and 17 had considerably
more circRNA locations than the other chromosomes, indicating a
stronger association with GEM resistance in PC. The top two
upregulated circRNAs, circRNA_101672 and circRNA_004077, were both
located on chromosome 16, which may be the most important circRNA
in GEM resistance in PC. Among the deregulated circRNAs, there were
72 exonic, 1 antisense, 3 intronic and 5 sense overlapping
(Fig. 2E).
 | Table II.Top dysregulated circRNAs in
GEM-resistant PC cells. |
Table II.
Top dysregulated circRNAs in
GEM-resistant PC cells.
| circRNA | Gene symbol | Type | Chrom | P-value | FC (abs) |
|---|
| Upregulated
circRNAs |
| circ_101672 | RAB40C | Exonic | chr16 | 0.000815496 | 3.4745157 |
| circ_004077 | VAT1L | Exonic | chr16 | 0.003990298 | 2.8248131 |
| circ_003251 | WNK1 | Exonic | chr12 | 0.004520208 | 2.8156161 |
| circ_102402 | DAZAP1 | Exonic | chr19 | 0.000995122 | 2.646115 |
| circ_074298 | HARS | Exonic | chr5 | 0.011756552 | 2.5846195 |
| circ_089762 | JA760602 | Exonic | chrM | 0.025705535 | 2.5812949 |
| circ_003596 | COL5A1 | Exonic | chr9 | 0.003354102 | 2.5655559 |
| circ_089761 | JA760602 | Exonic | chrM | 0.027054021 | 2.4890659 |
| circ_002178 | RPPH1 | Sense
overlapping | chr14 | 0.014113033 | 2.3939466 |
| circ_102403 | DAZAP1 | Exonic | chr19 | 0.000309541 | 2.3735941 |
| Downregulated
circRNAs |
| circRNA_101543 | VPS13C | Exonic | chr15 | 0.033434975 | 3.9419461 |
| circRNA_102747 | ACTR2 | Exonic | chr2 | 0.016163204 | 3.8810596 |
| circRNA_000926 | ACTR2 | Sense
overlapping | chr2 | 0.014583979 | 3.5123931 |
| circRNA_059665 | ABHD12 | Exonic | chr20 | 0.020206272 | 3.4890827 |
| circRNA_103827 | HMGCS1 | Exonic | chr5 | 0.049403727 | 3.4092977 |
| circRNA_406521 | UGT8 | Sense
overlapping | chr4 | 0.007149841 | 3.283491 |
| circRNA_103128 | DYRK1A | Exonic | chr21 | 0.034078531 | 3.2488121 |
| circRNA_104490 | MKLN1 | Exonic | chr7 | 0.015401059 | 3.2463403 |
| circRNA_103829 | HMGCS1 | Exonic | chr5 | 0.046511499 | 3.0960301 |
| circRNA_070037 | NUP54 | Exonic | chr4 | 0.021180458 | 2.938704 |
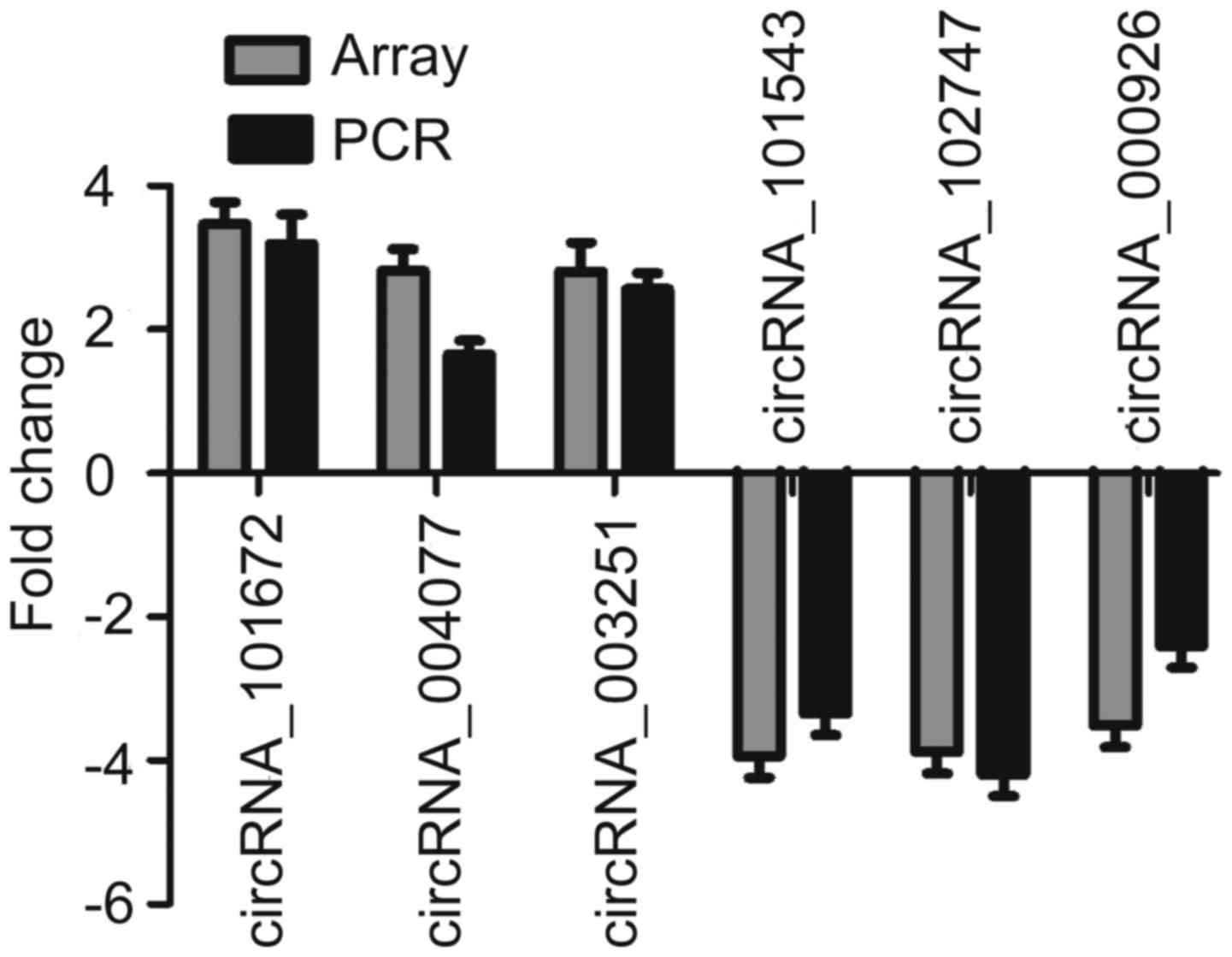
Validation of the microarray data by
RT-qPCR
To validate the microarray data, three upregulated
and three downregulated circRNAs were selected as representatives
for further validation by RT-qPCR. According to the data shown in
Fig. 3, four of the six tested
circRNAs yielded results quite similar to those of the microarray.
These well-validated circRNAs included two upregulated circRNAs
(circRNA_101672 and circRNA_003251) and two downregulated circRNAs
(circRNA_101543 and circRNA_102747). Although the other two
circRNAs were not well repeated, the direction of change was
similar to that noted in the microarray data. This result indicated
that most of the circRNAs identified by microarray were
reliable.
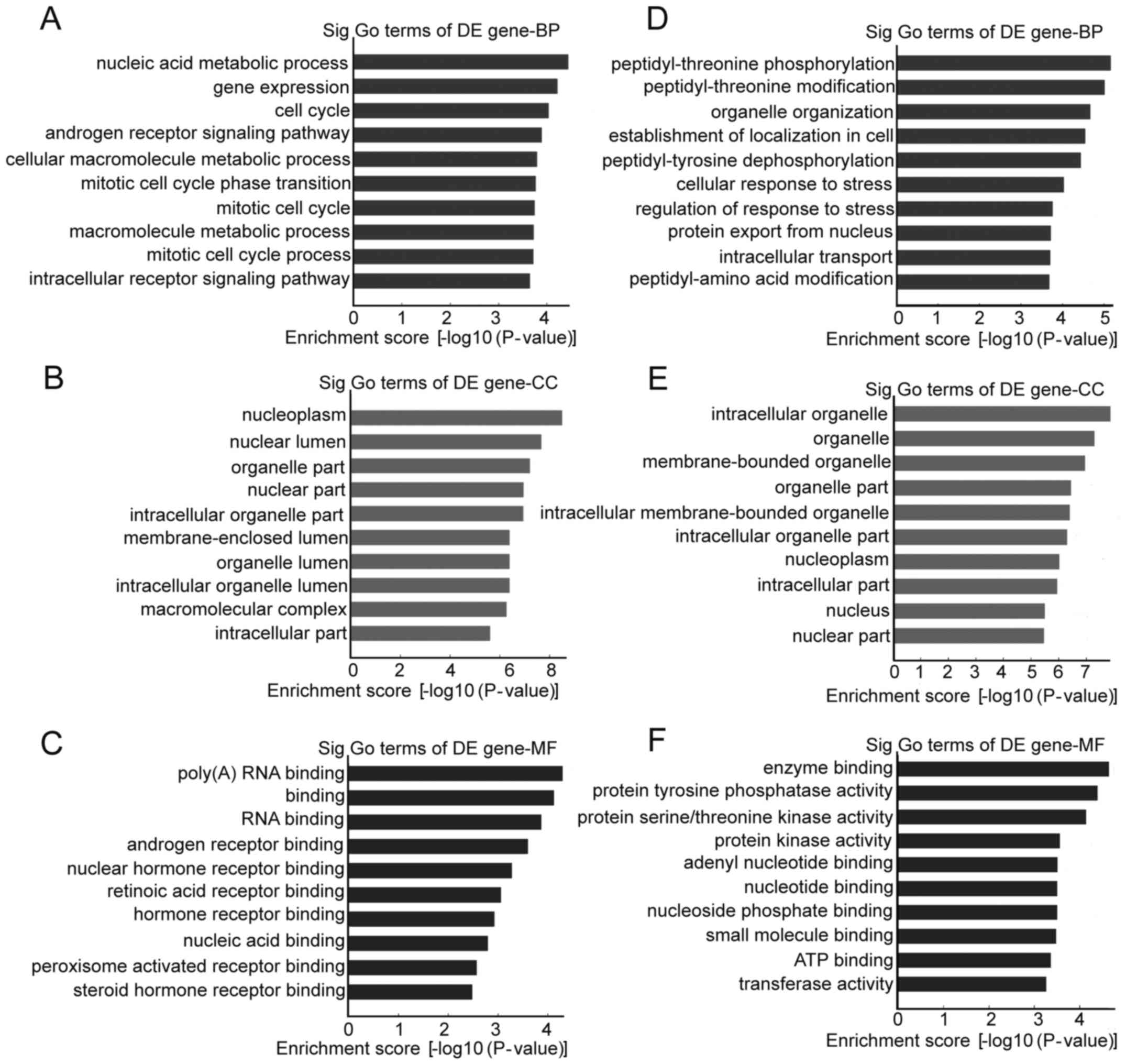
GO and pathway analysis of the
parental genes of circRNAs
Differentially regulated circRNAs and their parental
genes were further analyzed by GO analysis to speculate circRNA
potential functions on three different aspects: BP, CC and MF. The
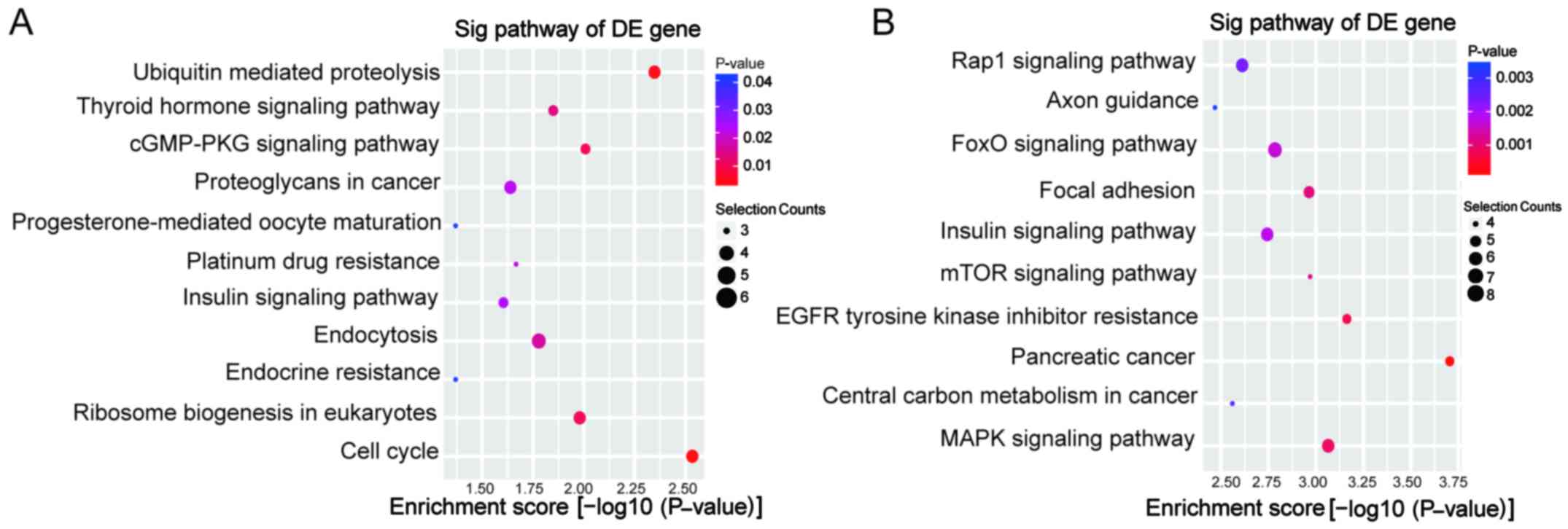
top 10 enrichment GO entries for upregulated and downregulated
circRNAs are displayed in Fig. 4.
We found that the upregulated circRNAs were mainly enriched for GO
terms related to nucleic acid metabolic process involved in BP,
poly(A) RNA binding linked with MF, and nucleoplasm in CC. For the
downregulated circRNAs, the most significantly enriched GO terms in
BP, MF, and CC were peptidyl-threonine phosphorylation, enzyme
binding and intracellular organelle, respectively. Furthermore,
KEGG pathway analysis was performed. The upregulated circRNAs were
involved in 11 pathways, while the downregulated circRNAs were
involved in 37 pathways. The predominant pathways are displayed in
Fig. 5. The top three enriched
pathways for upregulated circRNAs were cell cycle,
ubiquitin-mediated proteolysis and the cGMP-PKG signaling pathway.
The top three enriched pathways for downregulated circRNAs were
pancreatic cancer, EGFR tyrosine kinase inhibitor resistance and
the mitogen-activated protein kinase (MAPK) signaling pathway.
Among these enriched pathways, the MAPK and mammalian target of
rapamycin (mTOR) signaling pathways have been previously reported
to be involved in the chemoresistance of PC (19,20).
As shown in Fig. 5C, the potential
target genes of downregulated circRNAs incuding ATF2, BRAF, DUSP16,
MAPK8, NFATC3, RAF1, TAB2, TAOK1 are directly associated with the
MAPK signaling pathway.
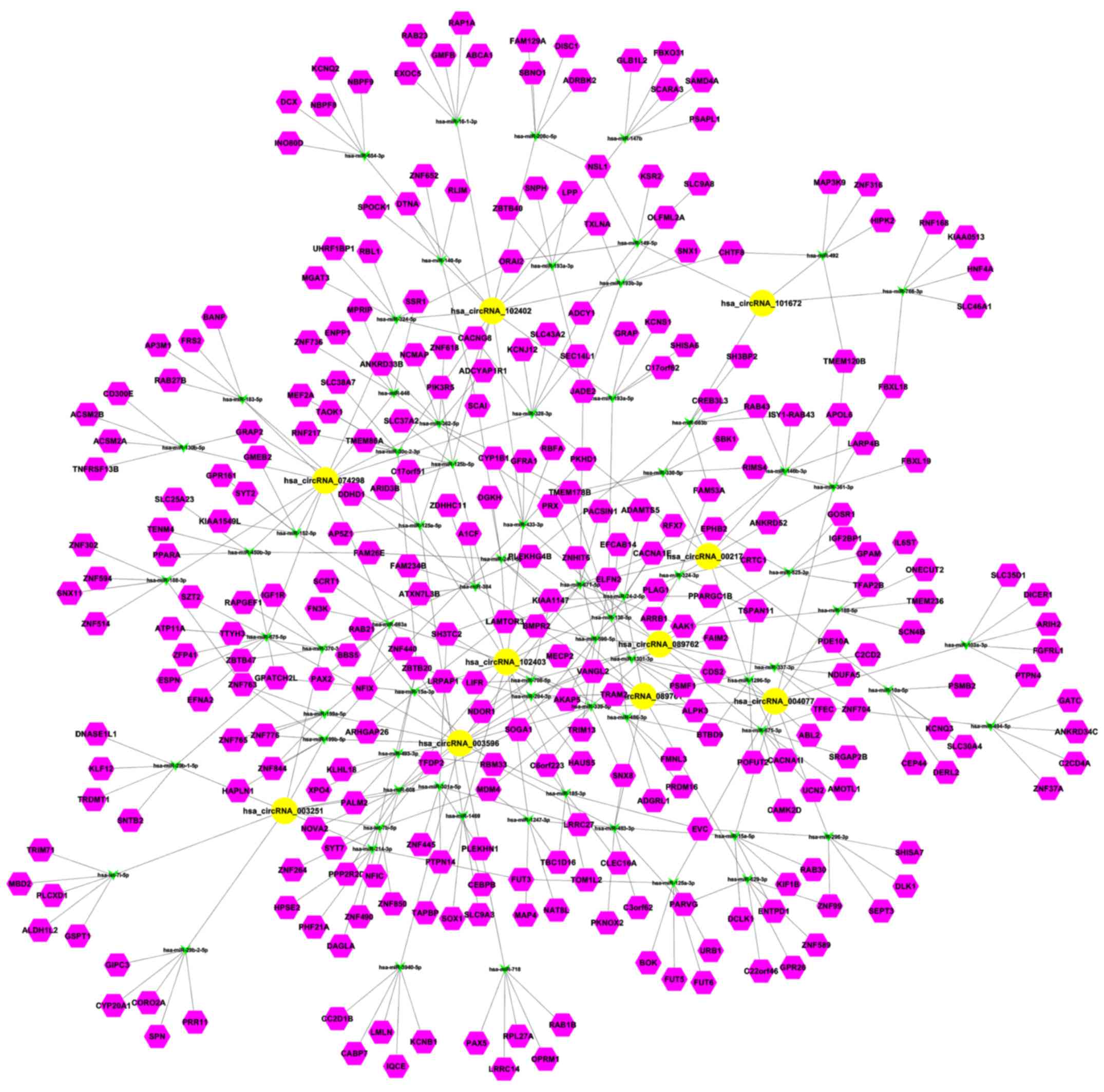
Prediction for circRNA-miRNA
interaction and circRNA-miRNA-target gene network
Given that circRNAs can function as sponges or
inhibitors of their interacting miRNAs to regulate gene expression,
circRNA-miRNA interaction was predicted with Arraystar homemade
miRNA target prediction software based on TargetScan and miRanda. A
total of 378 mature miRNAs were predicted to have docking sites in
the differentially expressed circRNAs. Therefore, they can interact
with the circRNAs. The circRNA-miRNA-target gene interacting
network of the top 10 upregulated circRNAs was established
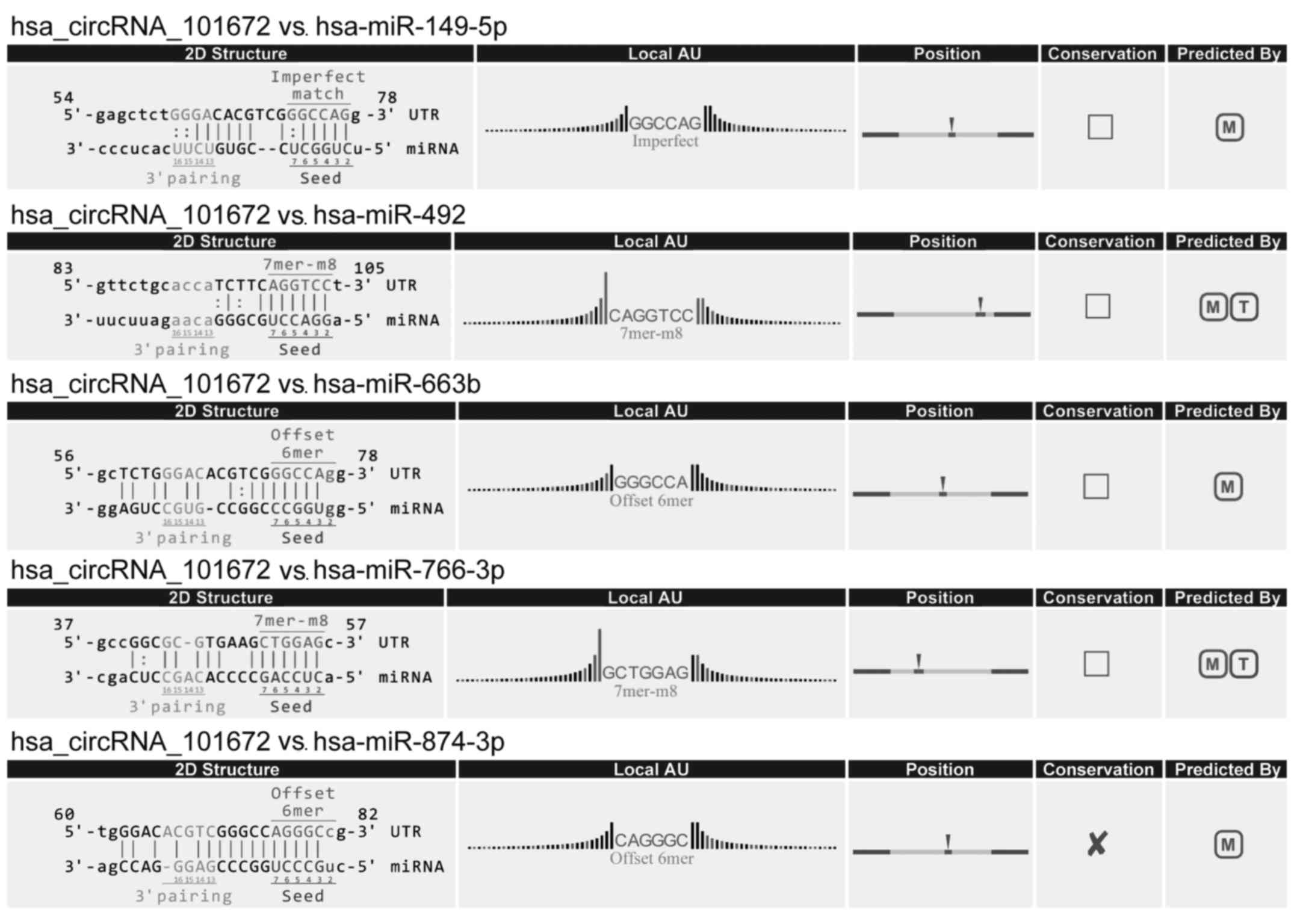
(Fig. 6). circRNA_101672 was
annotated in detail using the circRNA-miRNA interaction information
(Fig. 7).
Discussion
GEM is the most efficient mono-drug therapy for
treating PC. Thus, it is recommended as first-line therapeutic
option for advanced PC patients. However, acquired GEM resistance
contributes to treatment failure in a substantial number of PC
patients. Thus, deciphering the mechanisms underlying GEM
resistance is essential to overcome the problem. circRNAs were
recently identified as novel functional endogenous ncRNAs, which
can function as miRNA sponges, thereby interfering with the
post-transcriptional actions of miRNAs as suppressors of the target
genes (21). Accumulating evidence
has indicated that circRNAs are involved in a number of human
diseases, particularly in carcinomas, including hepatoma,
neuroglioma, bladder carcinoma and breast cancer (22–25).
Hence, circRNAs may serve as novel diagnostic and therapeutic
strategies in human diseases. However, the changes in the
expression of circRNAs and the related functional significance in
PC chemoresistance has been rarely reported.
In the present study, we first generated a
GEM-resistant PC cell line, SW1990/GZ, by stepwise selection and
then analyzed the circRNA expression profiles between the SW1990/GZ
and parental SW1990 cells with high-throughput circRNA microarrays
to investigate the mechanisms of acquired GEM resistance. We
observed that 26 circRNAs were upregulated, and 55 circRNAs were
downregulated by >2-fold in the SWl990/GZ cells compared with
the SW1990 cells. circRNA_101672, circRNA_004077 and circRNA_003251
were upregulated with top magnitudes. Conversely, circRNA_101543,
circRNA_102747 and circRNA_000926 were downregulated with top
magnitudes. The expression patterns of the above-mentioned circRNAs
were then validated by RT-qPCR, which revealed a high consistency
between the RT-qPCR results and microarray data. Although we only
used the SW1990 cell line as a cellular model in the present study,
we hypothesize that some of the deregulated circRNAs in SW1990/GZ
cells compared to SW1990 cells may be important and common
contributors in PC-acquired GEM resistance. Our next step will be
to confirm the results in other PC cell lines in our future
study.
circRNAs are primarily generated from exons or
introns of their parental genes and involved in the parental gene
expression regulation (26–28). Thus, we investigated the biological
functions and potential mechanisms of circRNAs in GEM resistance
based on the GO and KEGG pathway analyses. GO enrichment analysis
revealed that deregulated circRNAs were involved in the regulation
of some crucial biological processes, such as cellular response to
stress and cell cycle, which were important during the development
of chemoresistance. Markedly, among these enriched pathways, the
MAPK and mTOR signaling pathways have been shown to contribute to
GEM chemoresistance (29,30). Thus, some circRNAs may be involved
in the GEM resistance of PC by regulating the above-mentioned
signaling pathways.
Increasing evidence has demonstrated that circRNAs
can ‘sponge up’ miRNAs to promote the expression of miRNA target
protein-coding genes (31,32). Given the important roles of miRNAs
in the pathogenesis of PC, we hypothesized that some circRNAs may
contribute to the GEM resistance of PC by interacting with miRNAs.
Therefore, in this study, we performed in silico analyses to
predict miRNAs targeted by these dysregulated circRNAs. For
example, the upregulated circRNA with the largest fold change,
circRNA_101672, potentially binds miR-492. A previous study
indicated that miR-492 was involved in colon cancer chemoresistance
via regulating the expression of CD147 (33). In addition, one of the top
downregulated circRNAs, circ_102747, potentially binds miR-21.
Hwang et al (34) and Dong
et al (35) provided
experimental evidence for the role of miR-21 in PC GEM resistance
through modulation of apoptosis by directly regulating Bcl-2 and
PTEN expression. In order to confirm whether these circRNAs are
involved in PC chemoresistance, future studies, which will include
the overexpression and knockdown of circRNA, their interaction with
their potential targeted miRNAs and their involvement in GEM
resistance in clinical PC samples are required.
In conclusion, the present study revealed that
circRNAs were dysregulated in the GEM-resistant PC cell line
compared with its parental cell line. For these dysregulated
circRNAs, we conducted GO enrichment and pathway analyses for their
parental genes, which indicated that circRNAs may play important
roles in the development of GEM resistance. By predicting the
circRNA-miRNA interaction, we found several dysregulated circRNA,
i.e., circ_101672 and circ_102747, which can potentially bind some
miRNAs involved in cancer chemoresistance. These could be essential
molecular mechanisms underlying the function of circRNAs in the
chemoresistance of PC. Since our results were only based on an
in vitro cell line model, clinical sample and in vivo
validation is warranted in the future. On the whole, our findings
revealed the potential roles of circRNAs in PC chemoresistance and
potential therapeutic targets for circRNAs in PC treatment.
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science
Foundation of Anhui province (grant no. 1508085SQH224).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CX conceived and designed the study. CX and FD
performed the research. CX wrote the manuscript. CX, YY and FD
analyzed the data. All authors have read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yeo TP, Hruban RH, Leach SD, Wilentz RE,
Sohn TA, Kern SE, Iacobuzio-Donahue CA, Maitra A, Goggins M, Canto
MI, et al: Pancreatic cancer. Curr Probl Cancer. 26:176–275. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rosenberg L: Pancreatic cancer: A review
of emerging therapies. Drugs. 59:1071–1089. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mittal A, Chitkara D, Behrman SW and
Mahato RI: Efficacy of gemcitabine conjugated and miR-NA-205
complexed micelles for treatment of advanced pancreatic cancer.
Biomaterials. 35:7077–7087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16:582017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bonizzato A, Gaffo E, Kronnie Te G and
Bortoluzzi S: CircRNAs in hematopoiesis and hematological
malignancies. Blood Cancer J. 6:e4832016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Yang J, Zhou P, Le Y, Zhou C, Wang
S, Xu D, Lin H and Gong Z: Circular RNAs in cancer: Novel insights
into origins, properties, functions and implications. Am J Cancer
Res. 5:472–480. 2015.PubMed/NCBI
|
|
9
|
Taborda MI, Ramírez S and Bernal G:
Circular RNAs in colorectal cancer: Possible roles in regulation of
cancer cells. World J Gastrointest Oncol. 9:62–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016.PubMed/NCBI
|
|
12
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200.
2016.PubMed/NCBI
|
|
13
|
Ahmed I, Karedath T, Andrews SS, Al-Azwani
IK, Mohamoud YA, Querleu D, Rafii A and Malek JA: Altered
expression pattern of circular RNAs in primary and metastatic sites
of epithelial ovarian carcinoma. Oncotarget. 7:36366–36381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
15
|
Fan X, Weng X, Zhao Y, Chen W, Gan T and
Xu D: Circular RNAs in cardiovascular disease: An overview. Biomed
Res Int. 2017:51357812017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Van Rossum D, Verheijen BM and Pasterkamp
RJ: Circular RNAs: Novel regulators of neuronal development. Front
Mol Neurosci. 9:742016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li HM, Hao XK, Wang HM, Liu ZC, He Y, Pu
M, Zhang HT, Yu HC, Duan JL and Qu SB: Circular RNA expression
profile of pancreatic ductal adenocarcinoma revealed by microarray.
Cell Physiol Biochem. 40:1334–1344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin X, Pan YQ, Wang LG, Ma T, Zhang LZ,
Tang AH, Billadeau DD, Wu HS and Huang HJ:
Fructose-1,6-bisphosphatase Inhibits ERK activation and bypasses
gemcitabine resistance in pancreatic cancer by blocking IQGAP1-MAPK
interaction. Cancer Res. 77:4328–4341. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kagawa S, Takano S, Yoshitomi H, Kimura F,
Satoh M, Shimizu H, Yoshidome H, Ohtsuka M, Kato A, Furukawa K, et
al: Akt/mTOR signaling pathway is crucial for gemcitabine
resistance induced by Annexin II in pancreatic cancer cell. J Surg
Res. 178:758–767. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang WS,
Gu C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin
signaling pathway. Oncotarget. 7:63449–63455. 2016.PubMed/NCBI
|
|
24
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang W, Du WW, Li X, Yee AJ and Yang BB:
Foxo3 activity promoted by non-coding effects of circular RNA and
Foxo3 pseudogene in the inhibition of tumor growth and
angiogenesis. Oncogene. 35:3919–3931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
27
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sui X, Kong N, Ye L, Han W, Zhou J, Zhang
Q, He C and Pan H: p38 and JNK MAPK pathways control the balance of
apoptosis and autophagy in response to chemotherapeutic agents.
Cancer Lett. 344:174–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kagawa S, Takano S, Yoshitomi H, Kimura F,
Satoh M, Shimizu H, Yoshidome H, Ohtsuka M, Kato A, Furukawa K, et
al: Akt/mTOR signaling pathway is crucial for gemcitabine
resistance induced by Annexin II in pancreatic cancer cells. J Surg
Res. 178:758–767. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peng L, Zhu H, Wang J, Sui H, Zhang H, Jin
C, Li L, Xu T and Miao R: MiR-492 is functionally involved in
Oxaliplatin resistance in colon cancer cells LS174T via its
regulating the expression of CD147. Mol Cell Biochem. 405:73–79.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hwang JH, Voortman J, Giovannetti E,
Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH,
et al: Identification of microRNA-21 as a biomarker for
chemoresistance and clinical outcome following adjuvant therapy in
resectable pancreatic cancer. PLoS One. 5:e106302010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dong J, Zhao Y, Zhou L, Zhang TP and Chen
G: Bcl-2 upregulation induced by miR-21 via a direct interaction is
associated with apoptosis and chemoresistance in MIA PaCa-2
pancreatic cancer cells. Arch Med Res. 42:8–14. 2011. View Article : Google Scholar : PubMed/NCBI
|