Introduction
Wilms tumor is the most common pediatric renal tumor
with a prevalence of ~1 in 10,000 children (1). Although combination therapy has
improved the prognosis for most patients, ~10% of patients with
Wilms tumors experience poor survival due to metastasis and
recurrence (2–5). Hence, it is essential to elucidate the
molecular mechanism underlying the tumorigenesis and metastasis of
Wilms tumors, which could provide predictive and therapeutic
targets for this childhood disease.
MicroRNAs (miRNAs) are a class of single-stranded,
highly conserved small non-coding RNAs that regulate gene
expression at the post-transcriptional level by binding to the
3′-unstranslated region (UTR) of target mRNAs, resulting in mRNA
silencing or degradation (6–8). In
tumors, various studies have confirmed that miRNAs function as
oncogenes or tumor suppressors which regulate tumor initiation,
progression and prognosis (9–11). The
expression of miRNAs has been shown to be stable and an excellent
marker for the early diagnosis of tumors (12).
miR-92a-3p is a member of the miR-17-92 family,
which plays a critical role in modulating cell viability, apoptosis
and metastasis of tumor cells (13,14).
In glioma, miR-92a-3p was found to exert various effects on tumor
stem-like cells by targeting the Notch-1/Akt pathway (15). In colorectal adenocarcinoma, the
expression level of miR-92a-3p was able to predict the prognosis of
patients (16). However, the
expression level, clinicopathological and biological functions of
miR-92a-3p in Wilms tumor and its underlying molecular mechanisms
remain unclear. In the present study, we aimed to investigate the
predicted miRNAs and the related molecular mechanisms in Wilms
tumor.
Materials and methods
Microarray data
The gene expression profiles of GSE50505 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50505),
GSE57370 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57370)
and GSE17342 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17342)
were downloaded from the GEO database. GSE50505, which was based on
GPL17667 platform (Luminex Homo sapiens bead-based microRNA
profiling platform), was submitted by Liu et al. The dataset
contained 28 samples, including 20 Wilms tumor samples and 3 normal
kidney samples. GSE17342 was based on GPL8367 platform
(LC_MRA-1001_miRHuman_10.0_070802), including 2 Wilms tumor samples
and 2 normal kidney samples. GSE57370 was based on GPL16770
platform (Agilent-031181
Unrestricted_Human_miRNA_V16.0_Microarray), including 62 Wilms
tumor samples and 4 normal kidney samples.
Patients and tissue samples
Wilms tumor tissues and the corresponding adjacent
non-tumor tissues were obtained from 68 patients who had Wilms
tumors and had undergone surgery at the Women and Chidren's
Hospital of Guangzhou Medical University (Guangzhou, China) between
July 2012 and July 2017. The age and sex distribution of WT
patients are shown in Table I.
Adjacent non-tumor tissues were obtained 3 cm away from the tumor,
and the lack of tumor cell infiltration was verified by
pathological examination. All tissue samples were frozen in liquid
nitrogen and stored at −80°C. All patients had not received
chemotherapy or radiotherapy before the surgery. Informed consent
was obtained from each patient, and the study protocol and consent
procedures were approved by the Ethics Committee of Guangzhou
Medical University (Guangzhou, China).
 | Table I.Relationship between miR-92a-3p and
the clinicopathological features of the Wilms tumor cases. |
Table I.
Relationship between miR-92a-3p and
the clinicopathological features of the Wilms tumor cases.
|
| miR-92a-3p
expression |
|
|---|
|
|
|
|
|---|
| Characteristics | High (%) | Low (%) | P-value |
|---|
| Sex |
|
| 0.061 |
| Male | 38.2 | 16.2 |
|
|
Female | 30.9 | 14.7 |
|
| Age (years) |
|
| 0.832 |
|
<4 | 41.2 | 11.8 |
|
| ≥4 | 27.9 | 19.1 |
|
| Tumor size (cm) |
|
| 0.710 |
|
<10 | 51.5 | 16.2 |
|
| ≥10 | 17.6 | 14.7 |
|
| Histologic type |
|
| 0.353 |
|
Triphasic | 11.8 | 5.9 |
|
|
Blastemal | 13.2 | 4.4 |
|
|
Stromal | 17.6 | 8.8 |
|
|
Epithelial | 16.2 | 10.3 |
|
|
Others | 5.9 | 4.4 |
|
| Lung metastasis |
|
| 0.042 |
| No | 23.5 | 35.5 |
|
| Yes | 8.8 | 32.4 |
|
| Survival status |
|
| 0.016 |
|
Alive | 38.2 | 26.5 |
|
|
Deceased | 7.4 | 33.8 |
|
Primary cell line and culture
The fresh tumor tissues were sliced into 0.1
cm3 pieces and washed with phosphate-buffered saline
(PBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
tissues were then incubated overnight with 2 U/ml dispase (Gibco;
Thermo Fisher Scientific, Inc.) at 4°C on a stirrer at 100 rpm,
followed by digestion with 160 µg/ml of collagenase A
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C for 3 h.
Thecollected and digested cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (FBS; Invitrogen;
Thermo Fisher Scientific, Inc.) until the cells had grown in a
confluent monolayer at 37°C in a humidified chamber supplemented
with 5% CO2. A maintenance culture was carried out in a
25-ml flask with DMEM supplemented with 10% FBS and 100 units/ml of
streptomycin and 100 µg/ml penicillin (both from Gibco; Thermo
Fisher Scientific, Inc.). The culture medium was replaced every 2
days and the cells were propagated every 3 days. For
cryopreservation, the cells were frozen in DMEM containing 10%
dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA) and 90%
FBS and stored in liquid nitrogen.
Transient transfection
The miR-92a-3p mimic, miR-92a-3p inhibitor and
corresponding negative control (miR-NC) were purchased from
Shanghai GenePharma Co., Ltd. (Shanghai, China). The siRNA against
NOTCH1 (UGGCGGGAAGUGUGAAGCG) and its negative control were provided
by Takara Bio Inc. (Otsu, Japan). These molecular products were
transfected into cells using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) for the various experiments according to
the manufacturer's instructions. Briefly, the final concentration
of these products was 50 nM and the cells were harvested for
subsequent experiments at 24 h after transfection. Each experiment
was repeated three times each in triplicates.
RNA extraction and quantitative
real-time PCR
Total RNA was isolated from the Wilms tumor tissues,
matched adjacent normal tissues and WT cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Briefly, all samples were treated with TRIzol followed by
chloroform. The mixture was centrifuged at 14,000 rpm for 10 min at
4°C and 700 µl 75% ethanol was added to the aqueous layer. Finally,
the purified RNA was diluted with 30 µl of RNase-free water. cDNA
synthesis was performed with 2 µg total RNA using the PrimeScript™
RT reagent kit with gDNA Eraser (Takara Bio, Inc., Otsu, Japan) for
the next qPCR with Mir-X miRNA First-Strand Synthesis kit (Takara
Bio, Inc.) for microRNA according to the manufacturers'
instructions. The primers sequences of NOTCH1 were F,
TGCCAGACCAACATCAAC and R, CTCATAGTCCTCGGATTGC (Takara
Biotechnology, Co., Ltd., Dalian, China). The sequences of
miR-92a-3p were F: GGGGCAGTTATTGCACTTGTC and R:General reverse
primer for microRNA is purchased from RiboBio Co. Ltd. (Guangzhou,
China). The sequences of GAPDH were F: GCACCGTCAAGGCTGAGAAC and R:
TGGTGAAGACGCCAGTGGA (Takara Biotechnology). A qPCR was performed
using the SYBR Premix Ex Taq II kit (Takara Bio) and the Applied
Biosystems 7500 Fluorescent Quantitative PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The mixtures were
incubated at 95°C for 30 sec, followed by 40 amplification cycles
of 95°C for 5 sec and 60°C for 34 sec. The comparative cycle
threshold method was used to quantify the relative expression
levels of mRNA and microRNA. Expression levels of the housekeeping
gene GAPDH and U6 were used to normalize the expression levels of
the genes-of-interest, respectively. The relative mRNA levels were
calculated based on the Ct values and normalized using the relative
housekeeping gene expression.
Cell proliferation assay
In vitro cell proliferation was measured
using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT) method following the manufacturer's instructions
(Nanjing KeyGen Biotech, Nanjing, China). Briefly, the transfected
cells were seeded into 96-well plates (2×103 cells/well)
and cultured for 5 days. The MTT solution (formazan in DMSO) was
added to each well at the indicated time-points (1, 2, 3, 4 and 5
days) and incubated at 37°C for 4 h. The optical density value (OD)
of each well was measured at 450 nm using a microplate
spectrophotometer (BioTek Instruments, Inc., Winooski, VT,
USA).
Cell colony formation assay
The transfected cells were seeded into 6-well plates
at a density of 100 cells/well. After culture for 10 days, the
colonies were washed with PBS, fixed with 4% paraformaldehyde and
stained with 1% crystal violet. The colonies were imaged and
counted in five randomly selected fields under a light microscope
(Olympus Corp., Tokyo, Japan).
Wound healing assay
Briefly, cells (1×105) were seeded in
6-well plates and incubated overnight. A wound was created with a
10-µm pipette tip and images were obtained under a light microscope
(Olympus Corp.). The wound gaps were measured per time-point.
Transwell assay
The assays were carried out in Transwell chambers
(8-µm pore size) (Corning, Inc., Corning, NY, USA). Matrigel™
Matrix (BD Biosciences, San Jose, CA, USA) was diluted 1:7 using
serum-free basal medium and 50 µl Matrigel Matrix dilution was
added to the upper chamber of the Transwell inserts. Moreover, 100
µl transfected cell (2×105/ml) suspensions were seeded
in the upper chambers precoated with Matrigel Matrix dilution in
24-well plates and cultured in serum-free basal medium. A total of
500 µl medium with 10% FBS was also added to the lower chambers.
After 24 h, cells in the upper chambers were removed using cotton
swabs. The inserts was washed three times with PBS, and cells that
invaded to the bottom surface of the insert were fixed with 4%
paraformaldehyde and stained using 1% crystal violet. The invading
cells were countedunder a Leica DMI4000B microscope (Leica
Microsystems, Heidelberg, Germany) from randomly selected five
fields and photomicrographs were captured.
Luciferase reporter assay
The wild-type (WT) or mutant-type (MUT) seed region
at the 3′UTR of NOTCH1 was synthesized and cloned into the
downstream region of a firefly luciferase cassette in the
pGL3-promoter vector (Promega Corporation, Madison, WI, USA)
according to the manufacturer's instructions. The cells were
cotransfected with vectors carrying the WT 3′UTR or MUT 3′UTR
NOTCH1 and miR-92a-3p mimic or miR-NC by using Lipofectamine 2000
reagent according to the manufacturer's instructions. After a 48-h
transfection, the cells were harvested to detect luciferase
activity by using the Dual-Luciferase assay (Promega
Corporation).
Western blotting assay
Total proteins were extracted from cells or tissues
with RIPA buffer (10 mM Tris-HCl, pH 7.4, 1% Triton X-100, 0.1%
SDS, 1% NP-40 and 1 mM MgCl) containing protease inhibitors. The
total protein concentration was determined using a BCA Protein
Assay kit (Nanjiing KeyGen Biotech). A total of 30 mg of protein
was separated on a 10% SDS-polyacrylamide gel and then transferred
onto polyvinylidene fluoride (PDVF) membranes (Sigma-Aldrich; Merck
KGaA). The membranes were blocked with 5% milk and then incubated
with primary antibodies against NOTCH1 (rabbit IgG, 1:1,000; cat.
no. 3608S) and GAPDH (rabbit IgG, 1:1,000; cat. no. 5174) overnight
at 4°C. On the second day, the blots were washed with PBST and
incubated with secondary antibodies (anti-rabbit IgG, 1:2,000; cat.
no. 7074) for 2 h at room temperature. The antibodies were
purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA).
The protein band was visualized by chemiluminescence imaging system
(ChemiDoc Touch; Bio-Rad Laboratories, Hercules, CA, USA). GAPDH
was used as an internal control.
Statistical analysis
For comparisons, two-tailed Student's t-test,
Wilcoxon rank-test, Fisher's exact test, one-way analysis of
variance (ANOVA) test and the Kruskal-Wallis test were performed.
Overall survival (OS) was calculated and multivariate Cox's
proportional harzards model was performed to determine the
independent factors. Survival curves were performed by
Kaplan-Meier's method and calculated by log-rank test. For
correlation, Spearman's and Pearson's correlation were used.
Statistical analyses were conducted using SPSS 13.0 software (SPSS,
Inc., Chicago, IL, USA) with a two-sided significance level of
P<0.05.
Results
miR-92a-3p is downregulated in Wilms
tumors
To reveal the expression of miRNAs in Wilms tumor,
we downloaded the microarray chips concerning the miRNA gene
expression profiles of GSE50505, GSE57370 and GSE17342. We chose
the genes with P<0.05 and fold control (FC) 1.5 as criteria.
After analysis with InteractiVenn, we obtained a common gene
miR-92a-3p (Fig. 1A). To reveal the
role of miR-92a-3p in Wilms tumor, RT-qPCR was performed to examine
the expression levels of miR-92a-3p in tumor samples and adjacent
non-tumor tissues of 68 Wilms tumor patients. As revealed in
Fig. 1B, miR-92a-3p was frequently
downregulated in Wilms tumor, compared with that in the adjacent
tissues (P<0.001). To further investigate the clinical
significance of miR-92a-3p expression in Wilms tumor, we tested the
expression level of miR-92a-3p in patients with or without
metastasis and found that miR-92a-3p expression was lower in
patients with metastasis than patients without metastasis
(P<0.01) (Fig. 1C). We divided
the 68 patients into two groups according to the median value of
miR-92a-3p expression in the Wilms tumors: high miR-92a-3p
expression group and low miR-92a-3p expression group (Table I). In addition, Kaplan-Meier's
analysis (Fig. 1D) indicated that
Wilms tumor patients with low miR-92a-3p expression exhibited
poorer overall survival (P<0.05).
miR-92a-3p inhibits Wilms tumor cell
proliferation and colony formation
To examine the effect of miR-92a-3p on Wilms tumor
growth, we obtained the primary cells from a Wilms tumor (Fig. 2A). The Wilms tumor cells were then
transfected with the miR-92a-3p mimic, miR-NC and inhibitor.
miR-92a-3p expression was comfirmed by qPCR (Fig. 2B). The MTT assay (Fig. 2C) indicated that WT cells with
higher miR-92a-3p expression exhibited reduced proliferation
compared to the cells transfected with miR-NC. In contrast, the
Wilms tumor cells with low miR-92a-3p expression exhibited
increased proliferation (P<0.05). The colony formation assay
(Fig. 2D and E) demonstrated that
Wilms tumor cells transfected with the miR-92a-3p mimic developed
significantly lower rates of colony formation when compared with
the cells transfected with miR-NC. Additionaly, Wilms tumor cells
transfected with the miR-92a-3p inhibitor had a higher rate of
colony formation (P<0.05). These results suggest that miR-92a-3p
inhibitedthe proliferation and colony formation of Wilms tumor
cells.
miR-92a-3p inhibits Wilms tumor cell
migration and invasion
To measure the effect of miR-92a-3p on the migratory
and invasive capacities of the Wilms tumor cells, we used
Matrigel-coated Transwell experiments and wound healing assays. The
results revealed a significant decrease in the wound-healing
distance in the miR-92a-3p mimic-transfected WT cells after 24 h.
Meanwhile, the wound-healing distance of the miR-92a-3p
inhibitor-transfected Wilms tumor cells was more extensive when
compared with the miR-NC cells (Fig. 3A
and B). We observed that the miR-92a-3p mimic significantly
decreased the invasiveness of the Wilms tumor cells through
Matrigel. The miR-92a-3p inhibitor increased the invasion potential
of the Wilms tumor cells (Fig. 3C and
D). These results demonstrated that miR-92a-3p inhibits the
potential of Wilms tumor cells in terms of migration and
invasion.
NOTCH1 is a direct target of
miR-92a-3p in Wilms tumor
To explore the molecular mechanism by which
miR-92a-3p functions in Wilms tumor, we used bioinformatic
prediction software (TargetScan) to determine the potential target.
We identified that miR-92a-3p was able to bind the 3′-UTR of NOTCH1
(Fig. 4A). To further confirm this
binding, we performed a luciferase assay and demonstrated that
miR-92a-3p dramatically inhibited the luferase activity of the
wild-type (WT) 3′-UTR but not of the mutant-type (Mut) 3′-UTR and
blank vector of NOTCH1 (Fig. 4B).
Moreover, miR-92a-3p mimic significantly inhibited mRNA and protein
expression of NOTCH1 and the inhibitor promoted the NOTCH1
expression (Fig. 4C and D).
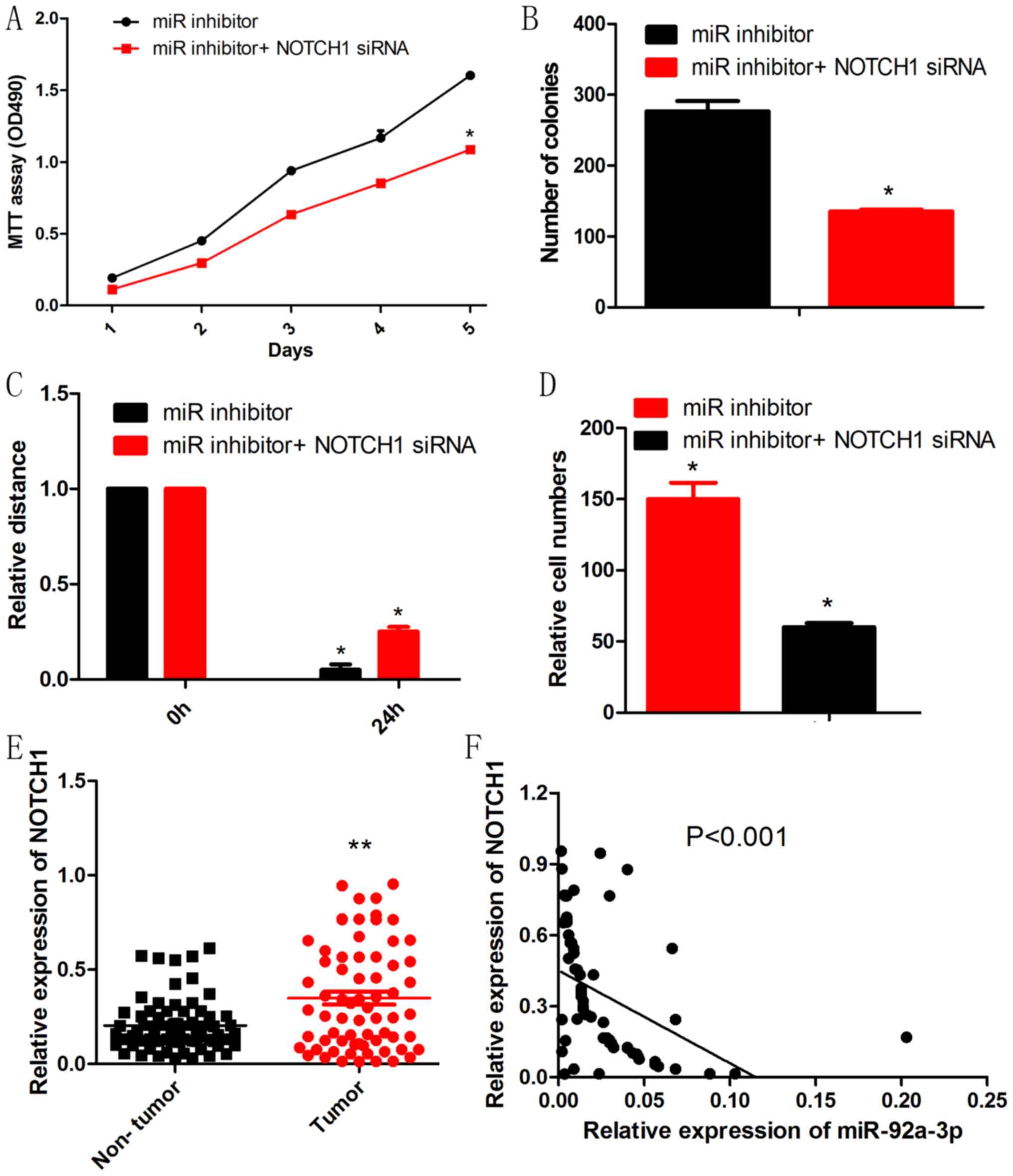
NOTCH1 knockdown rescued the effect of
miR-92a-3p inhibitor on Wilms tumor cells
To further determine whether NOTCH1 is a functional
target of miR-92a-3p in Wilms tumor, we performed a rescue
experiment. NOTCH1 siRNA reduced the promotive effects of the
miR-92a-3p inhibitor on proliferation, migration and invasion of
Wilms tumor cells (Fig. 5A-D). To
explore the relationship between miR-92a-3p and NOCTH1 in Wilms
tumortissues, RT-qPCR was performed to test the expression of
NOTCH1. As shown in Fig. 5E, Wilms
tumor tissues had significantly higher levels of NOTCH1 mRNA than
those of adjacent non-tumor tissues. Moreover, NOTCH1 mRNA had an
inverse correlation with miR-92a-3p expression in Wilms tumor
tissues (Fig. 5F).
Discussion
A series of microarray chips have been used to
detect the miRNA expression of Wilms tumors and these studies
reported the abnormal expression levels of various miRNAs in Wilms
tumor, such as the upregulated genes, miR-378 and miR-18b and the
downregulated genes, miR-193a-5p and miR-199a-5p (5,17).
However, studiesconcerning the cliniopathological and biological
mechanisms concerning miRNAs in Wilms tumors are sparse. In the
present study, we extracted the data from microarray profiles of
GSE50505, GSE57370 and GSE17342, including thousands of miRNA genes
in the human genome simultaneously, which has been widely used to
predict the potential therapeutic targets for tumors. Notably, we
identified a common downregulated gene miR-92a-3p in Wilms tumor.
The present study further confirmed the low expression of
miR-92a-3p in Wilms tumor and we also found that overexpression of
miR-92a-3p inhibited the proliferation, migration and invasion of
Wilms tumor cells. In addition, miR-92a-3p knockdown showed
contrary results. These results indicate that miR-92a-3p may serve
as a tumor suppressor.
NOTCH1 is a member of the Notch family, the
evolutionarily conserved family of transmembrane receptors, which
regulate cell fate, stem cell self-renewal and differentiation
during development (18,19). Recently, Notch1 was reported to take
part in diverse tumor processes including cell proliferation,
apoptosis, and cancer metastasis and angiogenesis in various types
of cancer (20,21). In addition, in kidney development,
Notch receptors were reported to regulate mesangial cell
specification, proliferation or survival (22). These results suggest that NOTCH
family receptors may play pivotal roles in Wilms tumor. In the
present study, we found that the expression of NOTCH1 was
downregulated by miR-92a-3p mimic, and NOTCH1 was upregulated by
the miR-92a-3p inhibitor. Moreover, the functions of miR-92a-3p
inhibitor on Wilms tumor were reversed by NOTCH1 knockdown. The
expression of NOTCH1 and miR-92a-3p had an obvious negative
correlation in Wilms tumor. The results above suggest that NOTCH1
is a direct target of miR-92a-3p and miR-92a-3p inhibits the
proliferation, migration and invasion of Wilms tumor by targeting
NOTCH1.
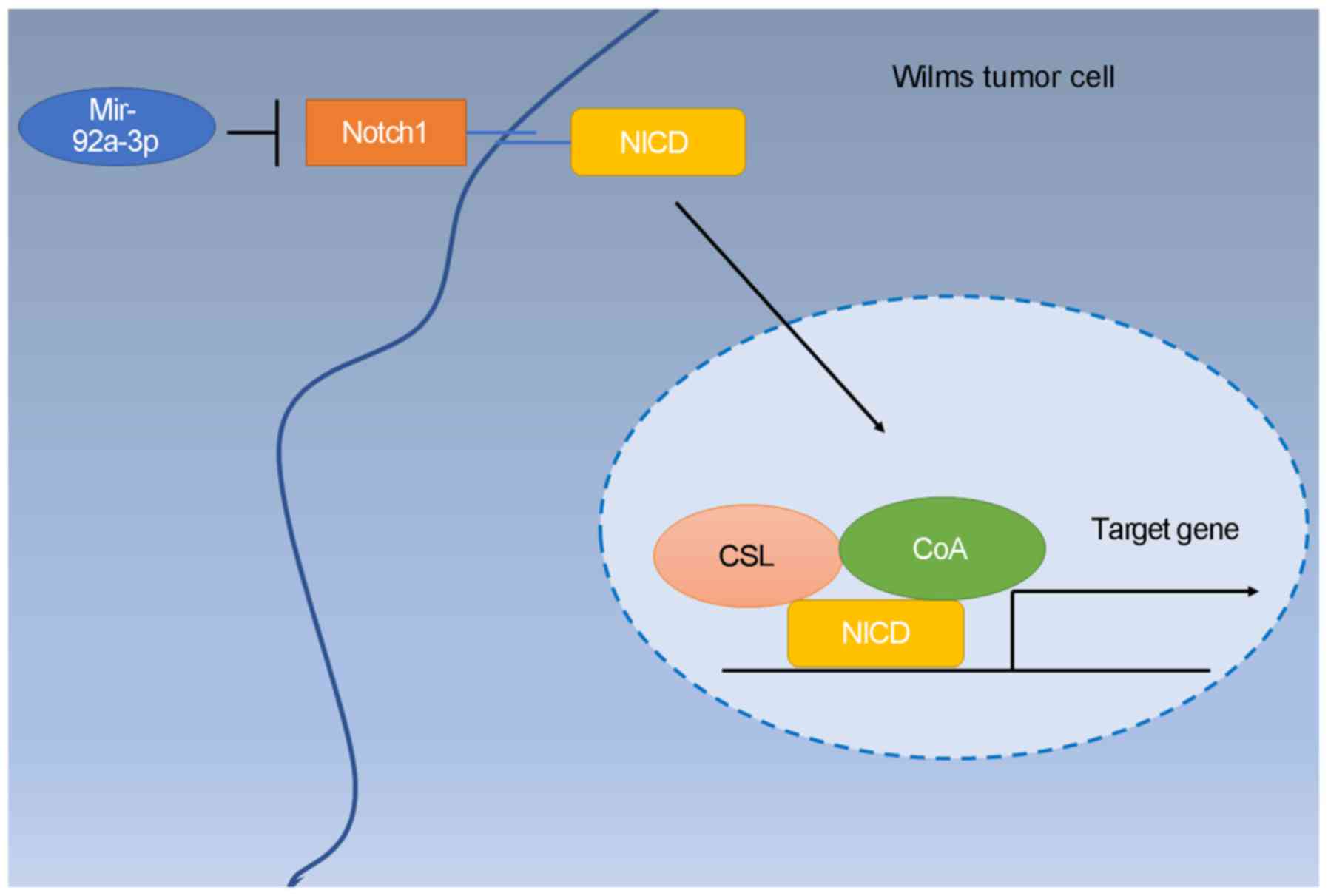
In conclusion, the present study demonstrated that
miR-92a-3p was downregulated inWilms tumor tissues and
significantly correlated with the lung metastasis of patients.
Furthermore, miR-92a-3p mimics suppressed Wilms tumor cell
proliferation, migration and invasion. Additionally, miR-92a-3p
knockdown promoted the progression. Moreover, NOTCH1 is a direct
target of miR-92a-3p and miR-92a-3p inhibits tumor progression by
targeting NOTCH1. Knockdown of NOTCH1 expression reversed the
promotive effect of the miR-92a-3p inhibitor on Wilms tumor
progression. In conclusion, miR-92a-3p blocks the progression of
Wilms tumor by targeting NOTCH1 (Fig.
6).
Acknowledgements
The authors thank Mr. zhongmin Li for his technical
support.
Funding
The present study was supported by a grant from the
Guangdong Provincial Department of Science and Technology
Foundation, P.R. China (no. 2016A020215009).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GL and WJ designed the study; SZ wrote the
manuscript, collected clinical information and performed
statistical analyses; ZZ and WF assisted with PCR, western blotting
and in vitro experiments; LZ assisted with the
Dual-Luciferase reporter assays. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Informed consent was obtained from each patient, and
the study protocol and consent procedures were approved by the
Ethics Committee of Guangzhou Medical University (Guangzhou,
China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Charlton J, Pavasovic V and
Pritchard-Jones K: Biomarkers to detect Wilms tumors in pediatric
patients: Where are we now? Future Oncol. 11:2221–2234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cone EB, Dalton SS, Van Noord M, Tracy ET,
Rice HE and Routh JC: Biomarkers for wilms tumor: A systematic
review. J Urol. 196:1530–1535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qi C, Hu Y, Yang F, An H, Zhang J, Jin H
and Guo F: Preliminary observations regarding the expression of
collagen triple helix repeat-containing 1 is an independent
prognostic factor for Wilms' tumor. J Pediatr Surg. 51:1501–1506.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brok J, Pritchard-Jones K, Geller JI and
Spreafico F: Review of phase I and II trials for Wilms' tumour e
Can we optimise the search for novel agents? Eur J Cancer.
79:205–213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu X, Li Z, Chan MT and Wu WK: The roles
of microRNAs in Wilms' tumors. Tumor Biol. 37:1445–1450. 2016.
View Article : Google Scholar
|
|
6
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bushati N and Cohen SM: MicroRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Valinezhad Orang A, Safaralizadeh R and
Kazemzadeh-Bavili M: Mechanisms of miRNA-mediated gene regulation
from common down-regulation to mRNA-specifc upregulation. Int J
Genomics. 2014:9706072014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tie J and Fan D: Big roles of microRNAs in
tumorigenesis and tumor development. Histol Histopathol.
26:1353–1361. 2011.PubMed/NCBI
|
|
10
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baltimore D, Boldin MP, O'Connell RM, Rao
DS and Taganov KD: MicroRNAs: New regulators of immune cell
development and function. Nat Immunol. 9:839–845. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang C, Wang C, Chen X, Yang C, Li K,
Wang J, Dai J, Hu Z, Zhou X, Chen L, et al: Expression profile of
microRNAs in serum: A fingerprint for esophageal squamous cell
carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma H, Pan JS, Jin LX, Wu J, Ren YD, Chen
P, Xiao C and Han J: MicroRNA-17-92 inhibits colorectal cancer
progression by targeting angiogenesis. Cancer Lett. 376:293–302.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou P, Ma L, Zhou J, Jiang M, Rao E, Zhao
Y and Guo F: miR-17-92 plays an oncogenic role and conveys
chemoresistance to cisplatin in human prostate cancer cells. Int J
Oncol. 48:1737–1748. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song H, Zhang Y, Liu N, Zhao S, Kong Y and
Yuan L: miR-92a-3p exerts various effects in glioma and glioma
stem-Like cells specifically targeting CDH1/v-catenin and
Notch-1/Akt signaling pathways. Int J Mol Sci. 17:pii: E1799. 2016.
View Article : Google Scholar
|
|
16
|
Zheng G, Du L, Yang X, Zhang X, Wang L,
Yang Y, Li J and Wang C: Serum microRNA panel as biomarkers for
early diagnosis of colorectal adenocarcinoma. Br J Cancer.
111:1985–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Watson JA, Bryan K, Williams R, Popov S,
Vujanic G, Coulomb A, Boccon-Gibod L, Graf N, Pritchard-Jones K and
O'Sullivan M: MiRNA profiles as a predictor of chemoresponsiveness
in Wilms' tumor blastema. PLoS One. 8:534172013. View Article : Google Scholar
|
|
18
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fortini ME: Notch signaling: The core
pathway and its posttranslational regulation. Dev Cell. 16:633–647.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolos V, Grego-Bessa J and de la Pompa JL:
Notch signaling in development and cancer. Endocr Rev. 28:339–363.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu P, Qiu M, Zhang Z, Kang C, Jiang R, Jia
Z, Wang G, Jiang H and Pu P: The oncogenic roles of Notch1 in
astrocytic gliomas in vitro and in vivo. J Neurooncol. 97:41–51.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boyle SC, Liu Z and Kopan R: Notch
signaling is required for the formation of mesangial cells from a
stromal mesenchyme precursor during kidney development.
Development. 141:345–354. 2014. View Article : Google Scholar
|