Introduction
Gastric cancer (GC) is one of the most common human
cancers and the second leading cause of cancer-related mortality
worldwide, as well as specifically in China (1–3).
Despite the fact that the therapeutic strategies against malignant
tumors have markedly improved in recent years, the metastasis of
malignant tumor cells is the cause of most cancer-related deaths
and remains one of the most enigmatic aspects of cancers, including
GC (2,4). Therefore, identifying
metastasis-associated gene expression and the molecular regulatory
mechanisms would provide a promising molecular target for the
treatment of cancer.
Long non-coding RNAs (lncRNAs) are transcripts
longer than 200 nucleotides, that lack protein coding capacity
(5). Accumulating evidence
indicates that lncRNAs are involved in cellular apoptosis, cell
proliferation, migration and invasion (6–8)
through a variety of mechanisms, including chromosome remodeling,
RNA processing, localization, mRNA stability, translation and even
as a competing endogenous RNA (9–11). The
dysregulation of lncRNAs in tumor formation and progression has
been investigated, with a particular focus on the mechanisms of
action of lncRNAs. To date, the majority of studies have analyzed
the functions of lncRNAs, the underlying mechanisms of dysregulated
lncRNAs in tumorigenesis and their potential roles as prognostic
markers or therapeutic targets for specific cancers (12). However, little is known about the
transcriptional regulatory mechanisms responsible for the aberrant
transcription of lncRNAs in cancers. In the present study, we
characterized the regulatory mechanisms through which the
transcription factor, activating enhancer-binding protein 4
(TFAP4), activates the expression of lncRNA TRERNA1, which plays an
important role in invasion and metastasis in GC, by binding to its
promoter. Thus, the present study may provide a novel potential
therapeutic strategy and target for GC. Our findings may also
contribute to the improvement of individualized treatment decisions
for GC patients.
Materials and methods
Tissue collection and ethics
statement
A total of 48 paired fresh gastric cancer tissues
and adjacent non-tumorous gastric tissues were analyzed in the
present study (clinicopathological characteristics shown in
Table I). The patients had
undergone surgical resection between May, 2010 and December, 2014
at the Affiliated Jiangyin Hospital of Medical School (Jiangyin,
Jiangsu, China) of Southeast University, China. The study was
approved by the Research Ethics Committee of Southeast University,
and written informed consent was obtained from all patients.
 | Table I.Association of TFAP4 expression with
clinicopathological characteristics of patients with GC. |
Table I.
Association of TFAP4 expression with
clinicopathological characteristics of patients with GC.
|
| Expression of
TFAP4 |
|
|---|
|
|
|
|
|---|
| Characteristic | T>N | T≤N | P-value |
|---|
| Age (years) |
|
| 0.868 |
|
<60 | 8 | 2 |
|
|
≥60 | 27 | 11 |
|
| Sex |
|
| 1.000 |
|
Female | 10 | 5 |
|
|
Male | 25 | 8 |
|
| Lauren
classification |
|
| 0.033 |
|
Intestinal type | 12 | 9 |
|
| Diffuse
type | 23 | 4 |
|
| Histological
grade |
|
| 0.506 |
|
High | 12 | 5 |
|
|
Moderate | 7 | 4 |
|
|
Poor | 16 | 4 |
|
| TNM staging |
|
| 0.675 |
| Stage
I/II | 23 | 7 |
|
| Stage
III/IV | 12 | 6 |
|
| Lymph node
metastasis |
|
| 0.021a |
| No | 11 | 9 |
|
|
Yes | 24 | 4 |
|
Cell culture
The GC cell lines, AGS, SGC-7901 and BGC-823, were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences (Shanghai, China). The MKN-74 cell
line was purchased from Cellcook Biotech Co., Ltd (Guangzhou,
China). Cells were cultured in RPMI-1640 medium supplemented with
10% fetal bovine serum (FBS; Wisent Inc., St. Bruno, QC, Canada),
100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen,
Carlsbad, CA, USA) in an incubator humidified air at 37°C with 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
Total RNA was first extracted from the cultured
cells or tissues using TRIzol reagent (Invitrogen). Subsequently,
RNA was reverse transcribed into cDNA using the PrimeScript™ RT
reagent kit with gDNA Eraser (Takara, Dalian, China). Quantitative
(real-time) PCR expression analyses were performed using the
SYBR® Premix Ex Taq™ II kit (Takara). The results were
normalized to the expression level of β-actin and all RT-qPCR data
were evaluated using the 2−ΔΔCq method (13), and their gene-specific primers as
follows. TFAP4 forward, 5′-GCAGGCAATCCAGCACAT-3′ and reverse,
5′-GGAGGCGGTGTCAGAGGT-3′; β-actin forward,
5′-AAAGACCTGTACGCCAACAC-3′ and reverse,
5′-GTCATACTCCTGCTTGCTGAT-3′. The RT-qPCR thermocycling conditions
for TFAP4 and β-actin began with an initial hold for 2 min at 95°C,
followed by 40 cycles of denaturation at 95°C, annealing at 60°C
and extension at 72°C all for 30 sec. Each experiment was performed
in triplicate.
Plasmid construction and cell
transfection
The cDNA TRERNA1 was synthesized by Genewiz (Suzhou,
China) and then cloned into the pcDNA3.1 plasmid (Invitrogen,
Carlsbad, CA, USA). The cDNA of TFAP4 was produced by
reverse-transcription PCR (RT-PCR) and then cloned into the
HindIII/EcoRI sites of the pcDNA3.1 plasmid. The
primer sequences for TFAP4 PCR amplification were as follows:
Forward, 5′-CCCAAGCTTATGGAGTATTTCATGGTGCC-3′ and reverse,
5′-GGTGGAATTCGGGGGGTAGTCAGGGAA-3′. Short hairpin RNA (shRNA)
targeting TFAP4 was synthesized by Genewiz, and ligated into the
BglII/HindIII sites of the pSUPER-EGFP vector
(OligoEngine, Seattle, WA, USA) after annealing. The sequences were
as follows:
5′-GATCCCCGTGATAGGAGGGCTCTGTAGTTCAAGAGACTACAGAGCCCTCCTATCACTTTTTGGAAA-3′
and
5′-AGCTTTTCCAAAAAGTGATAGGAGGGCTCTGTAGTCTCTTGAACTACAGAGCCCTCCTATCACGGG-3′.
Transfection was performed using Lipofectamine 2000 according to
the manufacturer's instructions (Invitrogen). For RT-qPCR assays,
at 24 h post-transfection, the cells were lysed and total cellular
RNA was extracted using TRIzol reagent (Invitrogen), and the
transfection efficiency was determined by RT-qPCR.
Cell migration and invasion
assays
Wound healing, cell migration and invasion assays
were performed as previously described (14). Briefly, a scratch wound was
generated using a 10 µl blunt pipette tip, and cells were cultured
in serum-free medium, after being washed with phosphate-buffered
saline (PBS) to remove floating cells. Cell migration and invasion
assays were performed with a Transwell chamber with 8-µm pore size
(EMD Millipore, Billerica, MA, USA). Cells were seeded into the
upper chamber, and the cells migrating through the pores or
invading through the Matrigel were fixed and stained with 0.5%
crystal violet (Beyotime Institute of Biotechnology, Nantong,
China) following 24–36 h of incubation.
Luciferase reporter assay
The TRERNA1 promoter fragments containing the
predicted TFAP4 binding site were amplified by PCR (primers are
listed in Table II), and then
subcloned into the downstream of the luciferase reporter gene in
the pGL3 Luciferase Reporter Vectors (Promega Corporation, Madison,
WI, USA). The wild-type sequence with wild-type E2 site
(5′-CAGCTG-3′) was
5′-CTAGAGGCACTGACTTTCCGCTTCCCAGCTGTGTGAGGTCTTGCTCAATTTC-3′, and the
mutant-type sequence with mutant-type E2 site (5′-TGATCA-3′) was
5′-CTAGAGGCACTGACTTTCCGCTTCCTGATCATGTGAGGTCTTGCTCAATTTC-3′. These
sequences were synthesized by Genewiz and subcloned into the pGL3
plasmid, separately. The luciferase activities were determined
using a dual luciferase assay kit (Promega Corporation) according
to the manufacturer's protocol. The relative luciferase activity
was normalized to the Renilla luciferase activity.
 | Table II.Primer sets designed to construct
luciferase reporter vectors containing the predicted E-box sequence
in the promoter region of the TRERNA1. |
Table II.
Primer sets designed to construct
luciferase reporter vectors containing the predicted E-box sequence
in the promoter region of the TRERNA1.
| Loci | Primer | 5′→3′ |
|---|
| E1 E3 | Sense |
ATATGCTAGCTAAGCACACACCACCTTGCC |
|
| Antisense |
ATATAAGCTTGTTCCCACCCCACTCACAAA |
| E2 | Sense |
ATATGCTAGCCACTGACTTTCCGCTTCCC |
|
| Antisense |
ATATAAGCTTTTAGCACTGGTTCCCTTCT |
| E3 | Sense |
ATATGCTAGCCAGGCGTCAGAAGGGAACCA |
|
| Antisense |
ATATAAGCTTCAACCAGCCAAGGCGGAGTA |
Chromatin immunoprecipitation (ChIP)
assay
ChIP assays were performed using the ChIP kit (EMD
Millipore) according to the manufacturer's instructions in MKN-74
cells. Antibodies against TFAP4 [AP-4(C-18)X, dilution 1:500] and
normal mouse IgG were obtained from Santa Cruz Biotechnology (Santa
Cruz, CA, USA). Primers designed to amplify the conserved E-box in
the promoter of the TRERNA1 are listed in Table III.
 | Table III.Primer sets designed to amplify the
conserved E-box sequence in the promoter region of the TRERNA1. |
Table III.
Primer sets designed to amplify the
conserved E-box sequence in the promoter region of the TRERNA1.
| Loci | Primer | 5′→3′ |
|---|
| E1 | Sense (P1) |
ATTATAAGCACACACCACC |
|
| Antisense (P1) |
ATAGCAAAAGAAAAACACC |
| E2 | Sense (P2) |
CACTGACTTTCCGCTTCCC |
|
| Antisense (P2) |
TTAGCACTGGTTCCCTTCT |
| E3 | Sense (P3) |
CAGGCGTCAGAAGGGAACCA |
|
| Antisense (P3) |
CAACCAGCCAAGGCGGAGTA |
Statistical analysis
Statistical analyses were performed using SPSS 20.0
software (IBM Corp., Armonk, NY, USA) or GraphPad Prism 5 software
(GraphPad Software, Inc., La Jolla, CA, USA). The significance of
differences in the TFAP4-dependent regulation of TRERNA1 expression
and transcriptional activity of E-box (E1-E3, E2 and E3) of the
TRERNA1 promoter region between TFAP4 and control groups were
estimated using a Student's t-test. The association between TFAP4
expression and pathological features of GC was analyzed with the
Chi-square (χ2) test. The correlation between the TFAP4
and TRERNA1 expression levels was analyzed using Pearson's
correlation coefficient test. All P-values presented were two-sided
and P<0.05 was considered to indicate a statistically
significant difference.
Results
TFAP4 upregulates the expression level
of lncRNA TRERNA1
Transcription factors are the most important
regulators of gene transcription by promoting or blocking specific
genes (15). In order to clarify
the transcription factors that may contribute to the regulation of
TRERNA1, in this study, we first screened the potential
transcription factors and their possible binding sites by scanning
the TRERNA1 gene promoter from upstream −4 kb to the downstream +1
kb region through the online analytical website (http://gene-regulation.com/pub/databases.html). The
predicted results indicated that a variety of transcription
factors, such as TFAP4, CCAAT enhancer binding protein beta
(CEBPB), heat shock factor 1 (HSF1), Jun proto-oncogene (JUN),
nuclear factor (NFE2L2) and cAMP response element binding protein 1
(CREB1) could bind to the promoter region of TRERNA1 (data not
shown). TFAP4 was considered as a priority candidate, since there
were multiple binding sites in the vicinity to the transcription
start site (TSS) of TRERNA1 (Fig.
1A).
In order to clarify the effect of TFAP4 on the
expression of TRERNA1, we constructed TFAP4 overexpression and
silencing vectors, and transfected these constructs into GC cells.
As displayed in Fig. 1B, the
enhanced expression of TFAP4 markedly increased the expression
level of TRERNA1 in SGC-7901 and AGS cells. In addition, we
observed a significant decrease in the TRERNA1 level following the
silencing of TFAP4 in MKN-74 and BGC-823 cells (Fig. 1C). These results indicated that the
expression of TRERNA1 was regulated by TFAP4 in GC cell lines.
TFAP4-regulated transcriptional
activity of TRERNA1 depends on the E-box of its promoter
To determine whether the TFAP4-dependent regulation
of the expression of TRERNA1 is attributed to activated
transcription, a series of constructs containing different
fragments of E-box (E1-E3, E2 and E3) of the TRERNA1 promoter
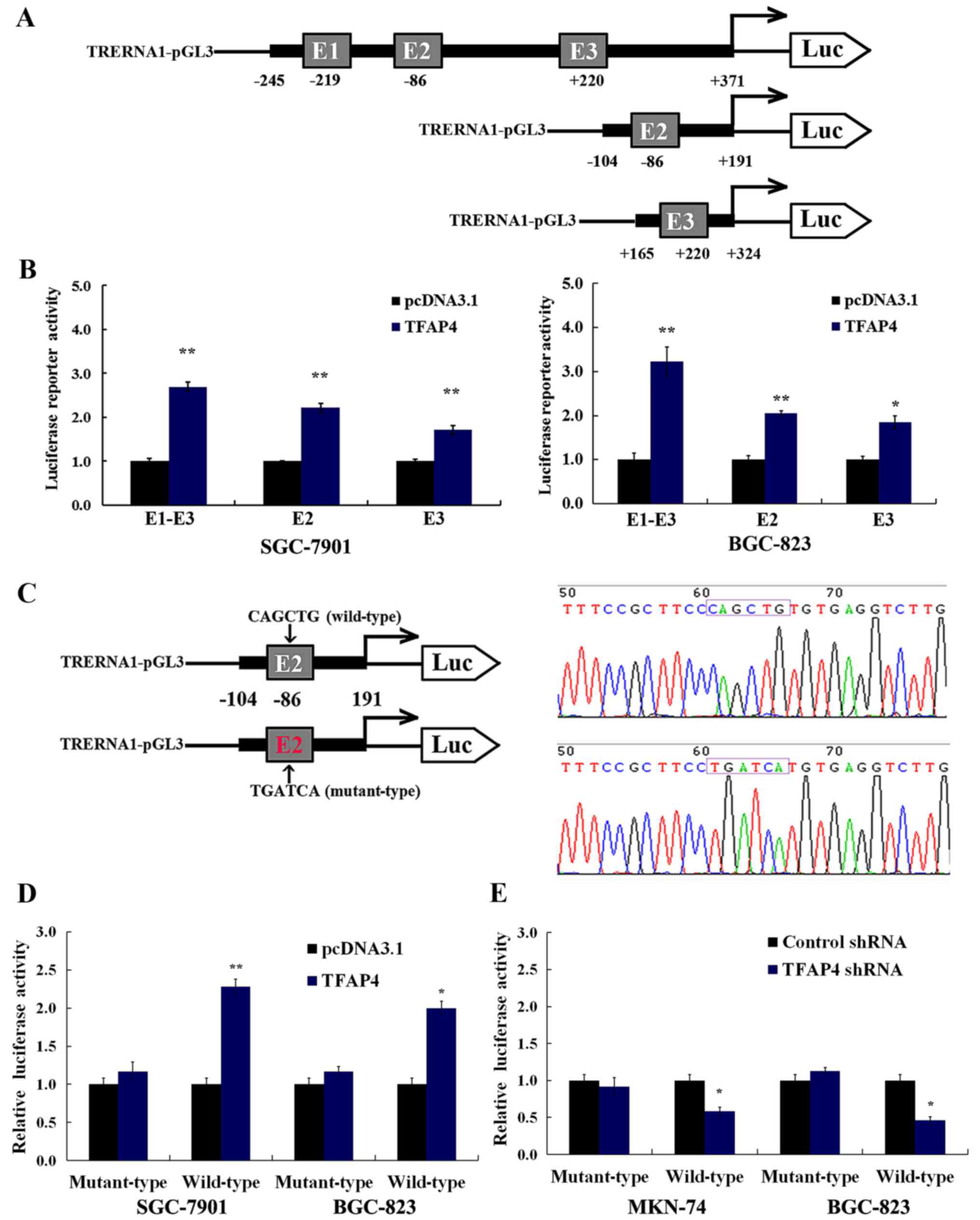
region were generated (Fig. 2A). As
displayed in Fig. 2B, the
overexpression of TFAP4 significantly enhanced TRERNA1 gene
promoter activity in the SGC-7901 and BGC-823 cells, particularly
in the E1-E3 fragment containing three TFAP4 binding sites in the
TRERNA1 promoter (Fig. 2B).
A previous study revealed that promoters activated
by TFAP4 are located closer to the TSS than those present at
TFAP4-suppressed promoters (16).
In this study, to further characterize the role of the E-box motif
situated on TRERNA1 as a transcriptional activator, we selected the
E2 site to construct the dual luciferase reporter vector containing
the motif of both the wild- (CAGCTG) and mutant-type (TGATCA) at
the E2 sites (Fig. 2C).
Dual-luciferase assays revealed that the overexpression or
silencing of TFAP4 significantly affected the transcriptional
activity of the wild-type E2 sites in SGC-7901, BGC-823 and MKN-74
cells (Fig. 2D and E). In contrast
to the wild-type, TFAP4 had no significant impact on the activity
of the E2-mutant TRERNA1 promoter in GC cells (Fig. 2D and E). These results also
demonstrated that mutations abolished TFAP4 binding to E2.
Collectively, our results revealed that TFAP4 regulation of the
transcriptional activity of TRERNA1 was dependent on the CACGTG
motifs in the TRERNA1 promoter.
TFAP4 specifically binds to the E-box
motif of TRERNA1 promoter in vitro
Previous studies have revealed that TFAP4 is
effective for both the suppression and/or activation of target
genes. TFAP4 downregulates the expression level of p16 and p21 to
suppress cell senescence by directly binding the putative E-box
site (CAGCTG) (16,17). In this study, in order to determine
whether TFAP4 promotes TRERNA1 transcriptional activity by directly
binding to its promoter, we designed PCR primers (Table III) specific for the amplification
of the three E-box sites (E1, E2 and E3) in the TRERNA1 promoter
region (Fig. 3A). As displayed in
Fig. 3B, the results indicated that
TFAP4 bound to the promoters of TRERNA1 in the vicinity of their
E-box binding motifs E1, E2 and E3, which reflected specificity of
the TFAP4 binding to the E-box motifs located in the promoter of
TRERNA1 (Fig. 3B). Notably, ChIP
different enrichment observed among E1, E2 and E3 (Fig. 3B), indicated that the maximal
binding capacity of TFAP4 to the TRERNA1 promoter presented at E2.
These results demonstrated that TFAP4 directly regulated lncRNA
TRERNA1 transcription by targeting the E-box of its promoter
region, particularly to the E2 in GC cells.
Elevated TFAP4 is positively related
to metastasis in GC cases
Numerous studies have revealed that TFAP4 plays a
vital role in carcinogenesis and tumor development (16,18,19).
In this study, in order to understand the expression pattern of
TFAP4 in GC tissues and the association between the expression of
TFAP4 with the clinicopathological features of GC patients, we
analyzed the expression of TFAP4 in 48 pairs of GC tissues and
non-tumorous tissues. As revealed by RT-qPCR analysis, the
expression levels of TFAP4 were markedly higher in the tumor
tissues compared with the adjacent non-tumor tissues (Fig. 4), and a significant association was
observed between the expression level of TFAP4, lymph node
metastasis and Lauren classification in GC specimens, although no
other significant associations were observed concerning the TFAP4
level and GC patients (Table I).
These results indicated the potential role of TFAP4 during GC
development and dissemination through the regulation of
TRERNA1.
Increased expression of TRERNA1 is
positively related to the elevated TFAP4 level and promotes the
migration and invasion ability of GC cells
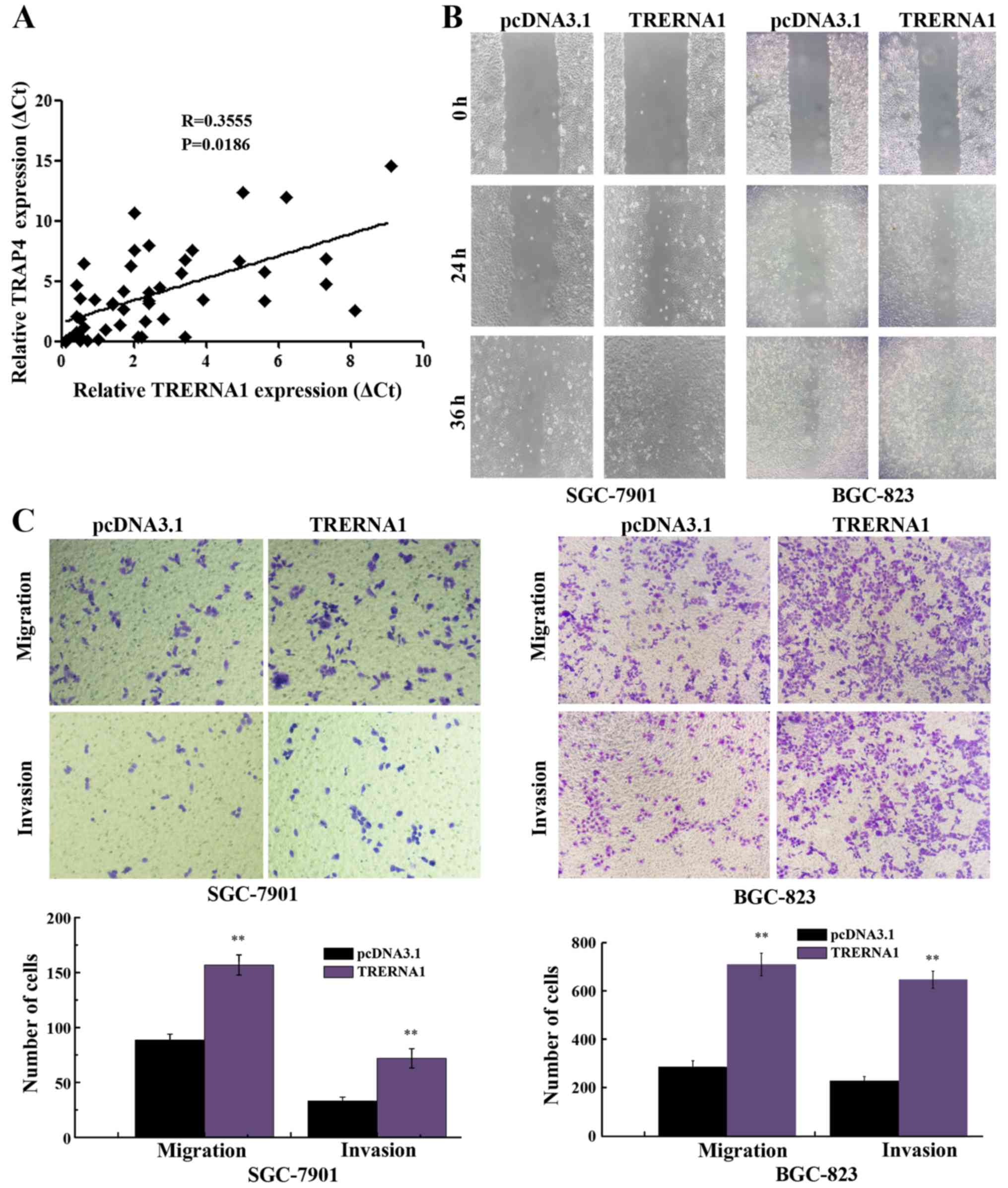
Since TRERNA1 was revealed to be a direct
transcriptional target of TFAP4 in GC cells, we investigated
whether TFAP4 influenced the expression level of TRERNA1 in GC
tissue specimens. As expected, the increased expression of TRERNA1
was observed to have a strong positive correlation with increased
TFAP4 expression in GC cases (Fig.
5A). Subsequently, to assess the potential role of TRERNA1 in
GC cell migration and invasion, we transfected TRERNA1
overexpression construct into BGC-823 and SGC-7901 cells, in which
the expression level of TRERNA1 is relatively low among human GC
cell lines (20). Wound healing
assay revealed that the enforced expression of TRERNA1 in GC cells
markedly enhanced the ability of the BGC-823 and SGC-7901 cells
compared with the control (Fig.
5B). Similarly, the enforced expression of TRERNA1 increased
the migration and invasion ability of the BGC-823 and SGC-7901
cells (Fig. 5C). These data further
indicated that the increased expression level of TRERNA1
upregulated by TFAP4 promoted the migration and invasion of GC
cells.
Discussion
GC is the fifth most common malignancy and the
second leading cause of cancer-related mortality worldwide
(21). Tumor metastasis is
responsible for ~90% of cancer-associated deaths, yet this process
remains poorly understood (4,5,22–26).
Therefore, it is urgent to uncover the pivotal genes and underlying
molecular mechanisms in cancer metastasis. With the rapid
development of sequencing technology, researchers found that only
~2% of the human genome DNA includes protein-coding sequences
(27). Non-coding RNAs, including
lncRNAs have gained widespread attention as a potentially crucial
molecule in disease onset and development in recent years (28), and hundreds of lncRNAs have been
identified to be dysregulated in human cancers (29–31).
Accumulating evidence has demonstrated that dysregulated lncRNAs,
such as H19HOXA11-AS and UCA1 have already been confirmed to
contribute to cell invasion and metastasis in GC (32–35).
TRERNA1 has been reported to act as a
metastasis-promoting oncogene in lung and breast cancer (36,37).
In the present study, we observed that TRERNA1 was significantly
upregulated in GC tissues compared with paired normal tissues. Data
from in vitro and in vivo experiments have also
indicated that TRERNA1 acts as an onco-lncRNA, which promotes the
metastasis and invasion of GC cells (20). A recent study demonstrated that
miR-190a inhibited epithelial-mesenchymal transition (EMT) in
hepatocarcinoma cells via targeting the lncRNA TRERNA1 (38). By bioinformatics analysis, in this
study, it was suggested that TFAP4 could be considered as a
potential regulator of TRERNA1. In addition, a loss-of-function and
gain-of-function study revealed that TFAP4 regulated the expression
of TRERNA1, and double luciferase activity assay revealed that
TFAP4 regulated the transcriptional activity of TRERNA1.
Furthermore, ChIP-PCR revealed that TFAP4 bound directly to the
E-box site of the TRERNA1 promoter to regulate its expression. We
also observed that the expression level of TFAP4 was significantly
related to the expression level of TRERNA1 in GC cases. These
results indicated that TFAP4 regulated the expression of TRERNA1 in
gastric carcinogenesis.
TFAP4 belongs to the helix-loop-helix family and
plays an important regulatory role by binding to the conserved
E-box (CAGCTG) sequence in transcriptional networks, thus affecting
cell proliferation, differentiation, cell lineage determination and
other essential biological processes (39). Previous studies have revealed that
TFAP4 regulates the expression of genes such as p21 and p16
(17,40), caspase-9 (41) and HDM2 (42). According to recent research, the
overexpression of TFAP4 has been reported in colorectal, breast,
lung and prostate cancer. TFAP4 has been shown to induce EMT and
enhance the migration and invasion of colorectal cancer cells
(16–18,40,43).
In GC, research has revealed that the expression of TFAP4 is
significantly higher in tumor tissue than in non-neoplastic tissue,
and an elevated TFAP4 expression significantly and positively
correlates with the degree of tumor differentiation, depth of tumor
invasion, lymph node metastasis and TNM stage (19). The downregulation of TFAP4 has been
shown to inhibit proliferation, induce cell-cycle arrest and
promote the apoptosis of GC cells (44). TFAP4 performs critical functions in
GC by participating in signaling pathways such as the
ECM-receptor-interaction and spliceosome (45). Therefore, the function and molecular
mechanisms of TFAP4 in gastric carcinogenesis warrant further
investigation.
The genome-wide characterization of TFAP4 DNA
binding sites and genes directly regulated by TFAP4 were identified
using ChIP, next-generation sequencing and bioinformatics analysis
(16). These studies on
TFAP4-mediated transcriptional regulation have mainly focused on
protein-coding genes; however the regulation of TFAP4 to lncRNAs
has not yet been investigated. In the present study, our findings
indicated that upregulated lncRNA TRERNA1 expression, which could
contribute to gastric cell invasion and metastasis was induced by
TFAP4 by directly binding to the CACGTG motifs in the promoter of
TRERNA1. The present study clarified the mechanism of upregulated
TRERNA1, which was involved in the invasion and metastasis of GC,
and also revealed a new field of TFAP4 regulation of the expression
of lncRNA in tumorigenesis. Furthermore, our study indicated that
the TFAP4-TRERNA1 axis may be a promising novel therapeutic target
for the intervention therapy of GC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81672414 and 81472548),
the Jiangsu Provincial Natural Science Foundation-Youth Foundation
(BK20160667), the Foundational Research Funds for the Central
University (2242016k41034) and the Foundation for Young Talents and
Natural Science in Higher Education of Anhui Province (KJ2017A213,
gxyq2018035).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HF conceived and designed the study. HW performed
the experiments and wrote the manuscript. XL, PG and WS provided
assistance for clinical sample collection, preservation, data
analysis and statistical analysis. MZ, YL and ZZ were involved in
interpreting the results, manuscript editing and manuscript review.
All authors have read and approved the content of the
manuscript.
Ethics approval and consent to
participate
The study was approved by the Research Ethics
Committee of Southeast University, and written informed consent was
obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heo JB and Sung S: Vernalization-mediated
epigenetic silencing by a long intronic noncoding RNA. Science.
331:76–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jariwala N and Sarkar D: Emerging role of
lncRNA in cancer: A potential avenue in molecular medicine. Ann
Transl Med. 4:2862016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui H, Wang L, Gong P, Zhao C, Zhang S,
Zhang K, Zhou R, Zhao Z and Fan H: Deregulation between miR-29b/c
and DNMT3A is associated with epigenetic silencing of the CDH1
gene, affecting cell migration and invasion in gastric cancer. PLoS
One. 10:e01239262015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karin M: Too many transcription factors:
Positive and negative interactions. New Biol. 2:126–131.
1990.PubMed/NCBI
|
|
16
|
Jackstadt R, Röh S, Neumann J, Jung P,
Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A,
et al: AP4 is a mediator of epithelial-mesenchymal transition and
metastasis in colorectal cancer. J Exp Med. 210:1331–1350. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jackstadt R, Jung P and Hermeking H: AP4
directly downregulates p16 and p21 to suppress senescence and
mediate transformation. Cell Death Dis. 4:e7752013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gong H, Han S, Yao H, Zhao H and Wang Y:
AP4 predicts poor prognosis in nonsmall cell lung cancer. Mol Med
Rep. 10:336–340. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xinghua L, Bo Z, Yan G, Lei W, Changyao W,
Qi L, Lin Y, Kaixiong T, Guobin W and Jianying C: The
overexpression of AP-4 as a prognostic indicator for gastric
carcinoma. Med Oncol. 29:871–877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu H, Hu Y, Liu X, Song W, Gong P, Zhang
K, Chen Z, Zhou M, Shen X, Qian Y and Fan H: LncRNA TRERNA1
function as an enhancer of SNAI1 promotes gastric cancer
metastasis by regulating epithelial-mesenchymal transition. Mol
Ther Nucleic Acids. 8:291–299. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang SY, Miah A, Pabari A and Winslet M:
Growth factors and their receptors in cancer metastases. Front
Biosci. 16:531–538. 2011. View
Article : Google Scholar
|
|
26
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Heery R, Finn SP, Cuffe S and Gray SG:
Long non-coding RNAs: Key regulators of epithelial-mesenchymal
transition, tumour drug resistance and cancer stem cells. Cancers.
9:E382017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ning S, Zhang J, Wang P, Zhi H, Wang J,
Liu Y, Gao Y, Guo M, Yue M, Wang L and Li X: Lnc2Cancer: A manually
curated database of experimentally supported lncRNAs associated
with various human cancers. Nucleic Acids Res. 44:D980–D985. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang ZQ, He CY, Hu L, Shi HP, Li JF, Gu
QL, Su LP, Liu BY, Li C and Zhu Z: Long noncoding RNA UCA1 promotes
tumour metastasis by inducing GRK2 degradation in gastric cancer.
Cancer Lett. 408:10–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: LncRNA HOXA11-as promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gumireddy K, Li AP, Yan JC, Setoyama T,
Johannes GJ, Orom UA, Tchou J, Liu Q, Zhang L, Speicher DW, et al:
Identification of a long non-coding RNA-associated RNP complex
regulating metastasis at the translational step. EMBO J.
32:2672–2684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Ren Y, Yang X, Xiong X, Han S, Ge
Y, Pan W, Zhou L, Yuan Q and Yang M: miR-190a inhibits
epithelial-mesenchymal transition of hepatoma cells via targeting
the long non-coding RNA treRNA. FEBS Lett. 589:4079–4087. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Atchley WR and Fitch WM: A natural
classification of the basic helix-loop-helix class of transcription
factors. Proc Natl Acad Sci USA. 94:5172–5176. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jung P, Menssen A, Mayr D and Hermeking H:
AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl
Acad Sci USA. 105:15046–15051. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsujimoto K, Ono T, Sato M, Nishida T,
Oguma T and Tadakuma T: Regulation of the expression of caspase-9
by the transcription factor activator protein-4 in
glucocorticoid-induced apoptosis. J Biol Chem. 280:27638–27644.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ku WC, Chiu SK and Chen YJ, Huang HH, Wu
WG and Chen YJ: Complementary quantitative proteomics reveals that
transcription factor AP-4 mediates E-box-dependent complex
formation for transcriptional repression of HDM2. Mol Cell
Proteomics. 8:2034–2050. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cao J, Tang M, Li WL, Xie J, Du H, Tang
WB, Wang H, Chen XW, Xiao H and Li Y: Upregulation of activator
protein-4 in human colorectal cancer with metastasis. Int J Surg
Pathol. 17:16–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu X, Zhang B, Guo Y, Liang Q, Wu C, Wu
L, Tao K, Wang G and Chen J: Down-regulation of AP-4 inhibits
proliferation, induces cell cycle arrest and promotes apoptosis in
human gastric cancer cells. PLoS One. 7:e370962012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Y: Transcriptional regulatory network
analysis for gastric cancer based on mRNA microarray. Pathol Oncol
Res. 23:785–791. 2017. View Article : Google Scholar : PubMed/NCBI
|