Introduction
Head and neck cancer is still a significant health
problem in the world that seriously affects the quality of life of
patients. Histologically, the most common form of head and neck
cancer is squamous cell carcinoma (HNSCC). HNSCC occurs in the oral
and nasal cavity, paranasal sinuses, salivary glands, pharynx and
larynx, and HNSCC risk factors include chewing or smoking tobacco,
heavy alcohol consumption, and infection of Epstein-Barr virus
(EBV) or human papilloma virus (HPV) (1,2). Head
and neck cancer is the sixth most commonly diagnosed malignancy in
the world with >500,000 new cancer cases annually (1), and the overall 5-year survival rate is
low due to tumor local recurrence and regional lymph node
metastasis (2). Early detection and
prediction of prognosis could help medical oncologists to
effectively manage this deadly disease.
MicroRNAs (miRNAs) are a class of non-coding small
RNAs of ~18–25 nucleotides in length. miRNAs function to
post-transcriptionally regulate gene expression by degradation
and/or inhibition of the target mRNA and its translation (3,4). Thus,
miRNAs play important roles in the human body, during events such
as embryo development, stem cell differentiation, and cell growth
and apoptosis, and altered expression of a given miRNA could result
in disease development and progression, including human cancers
(3,4). Early studies have revealed that
aberrant expression of different miRNAs was associated with HNSCC
development and progression (5–12).
Altered levels of particular miRNAs were assessed as diagnostic
markers for HNSCC metastasis (9),
while another study revealed that circulating miRNAs could serve as
biomarkers to assess the efficacy of therapy and the prognosis of
HNSCC (10). miRNAs were also
researched as molecular targets or biomarkers in HNSCC therapy
(13).
In this study, we focused on miR-20a-5p, which has
been reported to be associated with radioresistance of HNSCC cells
(14) and to promote proliferation
and migration of human hepatocellular and colorectal cancer cells
(15,16). We assessed the expression, function
and mechanism of action of miR-20a-5p in HNSCC and its role in the
regulation of HNSCC cell proliferation, migration and invasion
in vitro.
Materials and methods
Cell lines and culture
HNSCC cell lines PCI-37A and PCI-37B were generated
from one HNSCC patient (37A was derived from a primary tumor, and
37B was derived from lymph node metastasis of the same HNSCC
patient). These cell lines were characterized in the Cancer
Institute of Pittsburgh University (Pittsburgh, PA, USA) and
cultured in low glucose Dulbecco's modified Eagle's medium (DMEM;
HyClone Laboratories; Thermo Fisher Scientific, Inc., Logan, UT,
USA) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL;
Thermo Fisher Scientific, Inc., Gaithersburg, MD, USA), 100 U/ml
penicillin (Sigma-Aldrich Chemical Co., St. Louis, MO, USA) and 100
µg/ml streptomycin (Sigma-Aldrich) in a humidified incubator with
5% CO2 at 37°C.
Tissue specimens
Surgical tissue specimens were obtained from 15
HNSCC patients at the Department of Oromaxillofacial-Head and Neck
Surgery, School of Stomatology (Shenyang China Medical University
between August 2014 and September 2014. The present study was
approved by the Ethics Committee of China Medical University and
all the patients signed an informed consent form before being
enrolled in this study. None of the patients had received any
chemotherapy or radiotherapy prior to surgery, and all tissue
specimens were obtained from surgical resection of the primary
tumors, along with paired samples of adjacent normal tissues. The
tissues were snap-frozen in liquid nitrogen and stored at −80°C.
Histological diagnosis was performed and confirmed by the
Department of Pathology, School of Stomatology, China Medical
University, and the clinicopathological data are displayed in
Table I.
 | Table I.Clinicopathological data of
patients. |
Table I.
Clinicopathological data of
patients.
| Patient no. | Age (years) | Sex | Primary tumor
site | Tumor
differentiation | Lymph node
metastasis | TNM | Clinical stage |
|---|
| 1 | 59 | Male | Bucca | Moderate | N | T2N0M0 | II |
| 2 | 54 | Male | Soft palate | High | N | T2N0M0 | II |
| 3 | 50 | Male | Tongue | Moderate | Y | T2N1M0 | III |
| 4 | 51 | Male | Mouth floor | Poor | N | T2N0M0 | II |
| 5 | 47 | Male | Soft palate | Moderate | N | T1N0M0 | II |
| 6 | 68 | Male | Tongue | Moderate | N | T2N0M0 | II |
| 7 | 61 | Male | Gingiva | Moderate | Y | T1N2bM0 | IVA |
| 8 | 72 | Male | Tongue | Poor | N | T2N0M0 | II |
| 9 | 63 | Male | Tongue | Poor | Y | T2N2cM0 | IVA |
| 10 | 58 | Male | Tongue | Moderate | N | T2N0M0 | II |
| 11 | 50 | Male | Tongue | Moderate | N | T2N0M0 | II |
| 12 | 64 | Female | Bucca | Moderate | N | T2N0M0 | II |
| 13 | 66 | Female | Bucca | Moderate | Y | T4N1M0 | IVA |
| 14 | 54 | Female | Tongue | Moderate | N | T2N0M0 | II |
| 15 | 61 | Female | Tongue | Poor | N | T2N0M0 | II |
RNA isolation and real-time polymerase
chain reaction (RT-PCR)
Total RNA from cell lines and tissue samples was
isolated using TRIzol reagent (Takara Biotechnology, Co., Ltd.,
Dalian, China) and subsequently reverse-transcribed into cDNA using
the Super M-MLV reverse-transcription kit (BioTek, Beijing, China)
according to the manufacturers' instructions. qPCR amplification
was then conducted using the ABI PRISM 7500 Sequence Detection
System (ABI7500; Applied Biosystems; Thermo Fisher Scientific,
Inc., Foster City, CA, USA) with the following conditions: 95°C for
30 sec, then 95°C for 5 sec, and 60°C for 30–34 sec for 40 cycles.
The relative miR-20a-5p expression was normalized to U6 expression
levels. The qPCR primers for miR-20a-5p were:
5′-CGACAGTGACCGATTTCTCCT-3′ and 5′-GTGCAGGGTCCGAGGTATTC-3′; while
the U6 primers were: 5′-CTCGCTTCGGCAGCACA-3′ and
5′-AACGCTTCACGAATTTGCGT-3′. The relative expression level of
miR-20a-5p was determined using the 2−ΔΔCt analytical
method. In the analysis of the tissue specimens, a calibration
sample was used to ensure all reactions occurred in similar
conditions. All experiments were carried out in triplicate.
Transient transfection
miR-20a-5p mimics and inhibitor were obtained from
GenePharma (Shanghai, China), and transiently transfected into
HNSCC cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Carlsbad, CA, USA) according to the
manufacturer's instructions. The cells were subjected to different
assays thereafter.
CCK-8 assay
The Cell Counting Kit-8 (CCK-8) assay kit (Beyotime
Institute of Biotechnology, Shanghai, China) was used to evaluate
tumor cell viability. In brief, cells were seeded into 96-well
plates at a density of ~3,000 cells/well and cultured for 24 h
before transfection with miR-20a-5p mimics or the inhibitor. The
cells were then further cultured for 0, 24, 48, 72 and 96 h,
respectively. At the end of each experiment, the culture medium was
replaced with 100 µl fresh serum-free DMEM plus 10 µl CCK-8 per
well, and then the cells were incubated at 37°C for an additional 1
h. The optical density value at 450 nm (OD450) was measured using a
plate reader (model ELX-800; BioTek Instruments, Inc., Winooski,
VT, USA). All experiments were carried out in triplicate and
repeated at least thrice.
Wound healing assay
HNSCC cells were seeded at a density of
3×105 cells/well in 6-well culture plates, incubated for
24 h, and then transfected with miR-20a-5p mimics or the inhibitor.
After the cells had formed a monolayer, scratches were made on the
cell monolayer with a sterile 200-µl pipette tip. The cells were
washed twice with phosphate-buffered saline (PBS; HyClone; Thermo
Fisher Scientific, Inc.) and further incubated in serum-free
medium. The wound area was photographed at 0, 12 and 24 h under an
inverted phase-contrast microscope. The cell migration distance was
then estimated using software. All experiments were carried out in
triplicate and repeated at least once.
Transwell migration and invasion
assays
Cell migration and invasion capacity was assessed
using Transwell chambers (Corning Inc., Corning, NY, USA). In the
migration experiment, the Transwell chamber filters were not coated
with Matrigel (BD Biosciences, San Jose, CA, USA), however they
were coated with 500 µl of 30% Matrigel for the invasion assay.
Briefly, the Transwell chambers were placed into a 24-well plate,
and 800 µl DMEM containing 20% FBS was added to the lower chambers.
The corresponding upper chambers were seeded with 1×104
transfected cells in 200 µl DMEM without FBS. The plates were then
cultured for 24 h, and at the end of the experiments, the cells on
the surface of the upper chamber were removed using a cotton swab
and washed with PBS twice. The cells that had migrated to the
bottom side of the filters were fixed in 4% paraformaldehyde for 20
min at room temperature and then stained with 0.5% crystal violet
(Amresco, LLC, Solon, OH, USA) for 5 min. The filters were then
reviewed, and the number of cells was counted under an inverted
phase-contrast microscope (magnification, ×200) in five random
fields/Transwell chamber.
Western blot analysis
A C-C motif chemokine receptor 7 (CCR7)-specific
monoclonal antibody (mouse anti-human CCR7 antibody) and a rabbit
anti-TNFRSF21 antibody were purchased from Boyan Technology Co.,
Ltd. (Shanghai, China; cat. no. BYK-1305R) and Abnova (Walnut, CA,
USA; cat. no. PAB0156), respectively, while a monoclonal
anti-β-actin antibody (Wanleibio, Shenyang, China; cat. no.
WL01845;) was obtained from Sigma-Aldrich and used as an internal
control. Cells were transfected with miR-20a-5p mimics, inhibitor,
or miRNA control for 48 h, washed in ice-cold PBS, and lysed on a
plate with ice-cold cell lysis buffer (Wanleibio, Shenyang, China).
Cells were then centrifuged for 10,000 × g at 4°C for 10 min. The
protein concentration was assayed using the BCA assay (Wanleibio),
and 40-µg each of these protein samples were fractionated on a 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) gel and transferred onto polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA). For western
blotting, the membranes were blocked in 5% skim milk solution for 1
h and incubated with the TNFRSF21 and CCR7 primary antibody (1:100)
and the anti-β-actin antibody (1:1,000) overnight at 4°C. The next
day the membranes were washed with PBS-Tween-20 (PBST) thrice and
then further incubated with horseradish peroxidase-conjugated sheep
anti-rabbit IgG (cat. no. WLA023; Wanleibio) at a dilution of
1:5,000. The protein bands were subsequently visualized by enhanced
chemiluminescence (Wanleibio) and quantified using Gel-Pro-Analyzer
software (Media Cybernetics, Rockville, MD, USA).
Luciferase reporter assay
Luciferase reporter assays were carried out with a
Luciferase detection kit (Promega, Madison, WI, USA). The wild-type
(WT) and mutated type (MT) 3′-untranslational regions of TNFRSF21
cDNA were cloned into the pmirGLO reporter luciferase vector. For
the luciferase reporter assay, PCI-37B cells were co-transfected
with 25 pM miR-20a-5p mimic or negative control (NC) and 0.5 µg of
the pmirGLO-3′-UTR WT or MT plasmid using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity
was then assessed 48 h after transfection using the Dual-Luciferase
Reporter Assay system (Promega). The experiments were conducted in
triplicate and repeated three times.
Statistical analysis
All data analyses were performed using SPSS 13.0
statistical software (SPSS, Inc., Chicago, IL, USA), and the
χ2 test and Student's t-test were used to compare two
groups of data. A P-value of <0.05 was considered to indicate a
statistically significant result.
Results
Upregulated miR-20a-5p levels in HNSCC
cell lines and tissues
Our qRT-PCR data revealed that miR-20a-5p expression
was higher in 12 out of 15 paired tumor specimens (80%) than in the
adjacent normal tissues (P<0.05). miR-20a-5p expression was
lower in two tumor specimens (13%) than in the paired adjacent
normal tissues (P<0.05), and one specimen pair (7%) exhibited no
significant difference in miR-20a-5p expression (P>0.05;
Fig. 1A). In HNSCC cell lines,
miR-20a-5p expression was 2.5 times higher in PCI-37B cells than in
PCI-37A cells (Fig. 1B).
Expression of miR-20a-5p promotes
HNSCC cell proliferation
We then assessed the effects of miR-20a-5p mimics
and an inhibitor on HNSCC cell viability, migration and mechanism
of action. We transiently transfected PCI-37B cells with miR-20a-5p
plasmids and found that miR-20a-5p expression was increased by
2.3-fold in the mimics group, whereas miR-20a-5p expression was
reduced by 0.6-fold in the inhibitor group (Fig. 2A).
The tumor cell viability CCK-8 assay revealed that
the number of cells in the mimics group was greater than that in
the PCI-37B parental and mimics-NC groups, whereas the number of
cells in the inhibitor group was less than that in PCI-37B parental
and inhibitor-NC groups (Fig. 2B),
indicating that miR-20a-5p promoted PCI-37B cell proliferation.
Expression of miR-20a-5p promotes
HNSCC cell migration
We next performed tumor cell wound healing and
Transwell assays and found that the wound width was narrower in the
miR-20a-5p mimics group than in the PCI-37B parental and mimics-NC
groups after 12 and 24 h in culture (P<0.05; Fig. 3A). In contrast, the wound width was
wider in the miR-20a-5p inhibitor group than in the PCI-37B
parental and inhibitor-NC groups (P<0.01; Fig. 3A). Similarly, the Transwell
migration assay further confirmed this finding (P<0.01; Fig. 3B).
Expression of miR-20a-5p promotes
HNSCC cell invasion
The Transwell invasion assay revealed that compared
with that in the control group, upregulation of miR-20a-5p
expression using the mimics significantly increased the number of
cells that invaded through the Matrigel-coated membrane into the
lower chamber, whereas downregulation of miR-20a-5p expression led
to the opposite result (P<0.01; Fig.
4).
miR-20a-5p targets TNFRSF21 in HNSCC
cells
To explore the mechanism of miR-20a-5p action in
HNSCC, we used algorithms (TargetScan, PicTar and miRanda) to
predict potential downstream targets of miR-20a-5p and found that
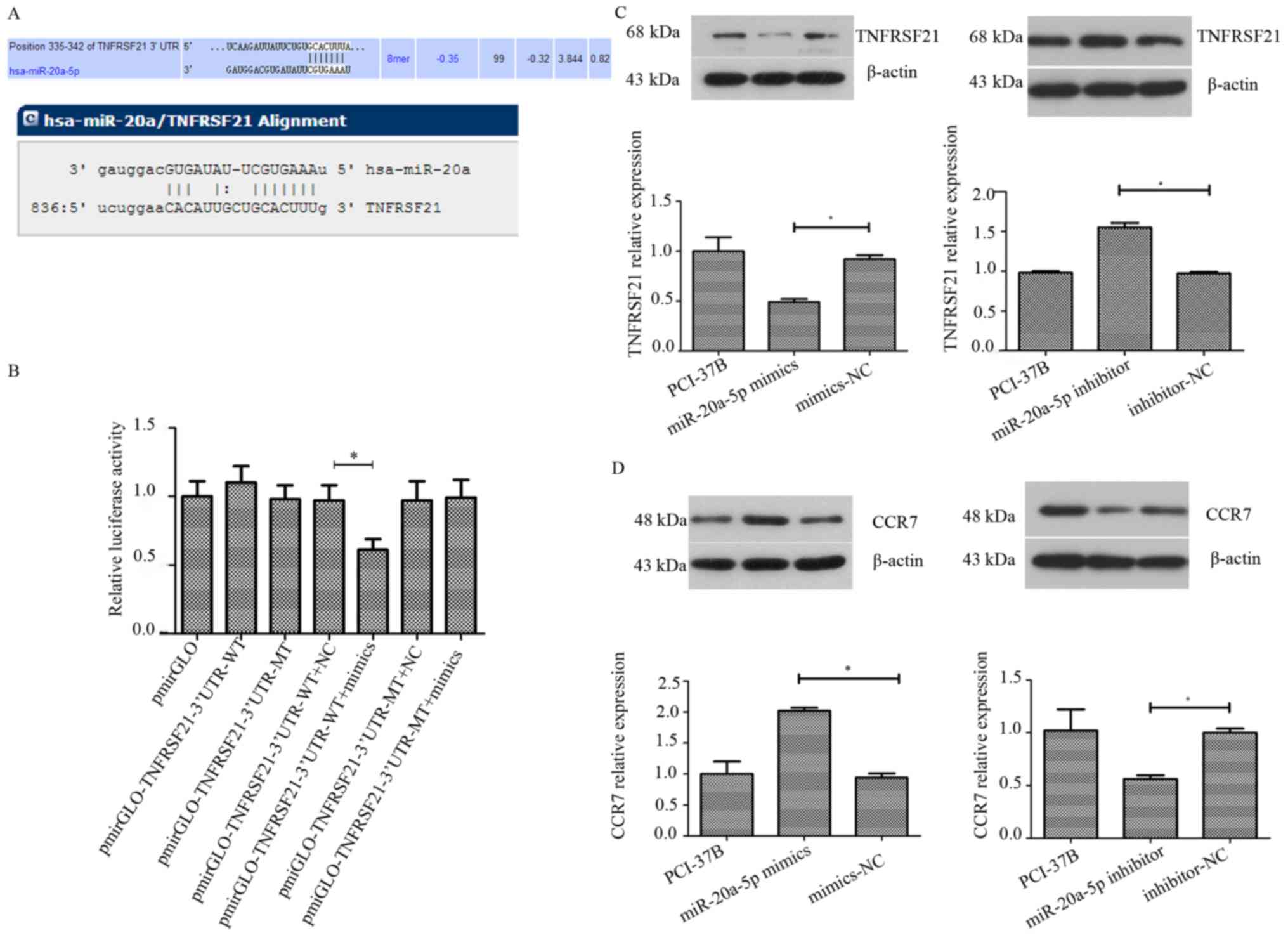
TNFRSF21 could be one of the miR-20a-5p targets (miR-20a-5p had
8-mer nucleotides that matched between the TNFRSF21 3′-UTR and
miR-20a-5p, which was predicted by TargetScan, miRDB and miRanda
programs; Fig. 5A). We therefore
conducted luciferase reporter and western blot assays. Our data
revealed that luciferase activity was downregulated by ~40% in the
WT group transfected with miR-20a-5p mimics compared with that in
the control group (P<0.05; Fig.
5B). However, there was no obvious alteration of luciferase
activity in the MT group (P>0.05; Fig. 5B). Moreover, the western blotting
results revealed that upregulation of miR-20a-5p expression reduced
the level of TNFRSF21 protein in PCI-37B cells by 0.53-fold
compared with expression in the control group (P<0.05), whereas
downregulation of miR-20a-5p expression increased the level of
TNFRSF21 protein in PCI-37B cells by 1.6-fold compared with that in
the control group (P<0.05; Fig. 5C
and D). These data revealed that TNFRSF21 is a direct
downstream target of miR-20a-5p.
miR-20a-5p upregulates CCR7 level in
HNSCC cells
Our previous data revealed the role of CCR7 in HNSCC
metastasis by demonstrating that CCR7 expression promoted HNSCC
cell migration and invasion through several signaling pathways
(17–21). In this study, we also found that
miR-20a-5p mimics promoted HNSCC cell migration and invasion
(Figs. 3 and 4). We hypothesized that miR-20a-5p may be
upstream of signaling in this CCR-7-related tumor cell migration
and invasion event. Although all three computer databases did not
support the notion that miR-20a-5p is able to directly target CCR7,
our western blot data revealed that upregulation of miR-20a-5p
expression did increase the level of CCR7 protein in PCI-37B cells
by 2.15-fold compared with that of the control group P<0.05);
however, downregulation of miR-20a-5p expression reduced the level
of CCR7 protein in PCI-37B cells by 0.56-fold compared with that of
the control group (P<0.05; Fig.
5C), indicating that an indirect effect of miR-20a-5p on CCR-7
expression in HNSCC cells in vitro.
Discussion
To date, the role of miR-20a-5p in HNSCC remains to
be defined, although a number of different studies has revealed
aberrant miR-20a-5p expression in a variety of diseases and
cancers, including ovarian (22),
cervical (23) and gastric cancers
(24). Altered miR-20a-5p
expression acts as an oncogene to promote tumor cell proliferation
and invasion in these cancers, but in skin squamous cell carcinoma
(25), thyroid cancer (26), and oral squamous cell carcinoma
(27,28), miR-20a-5p was revealed to function
as a tumor suppressor gene to inhibit tumor cancer cell
proliferation and invasion, indicating that there are organ sites-
or target-dependent effects of miR-20a-5p in human cancers. In the
present study, we demonstrated miR-20a-5p overexpression in both
HNSCC tissues and metastatic HNSCC cells, which let us speculate
that miR-20a-5p could be an oncogene in HNSCC. Our further
experiments demonstrated that upregulation of miR-20a-5p expression
promoted HNSCC cell proliferation, migration and invasion capacity
in vitro, whereas downregulation of miR-20a-5p expression
inhibited HNSCC cell proliferation, migration and invasion capacity
in vitro. A previous study reported that miR-20a-5p
expression induced resistance of nasopharyngeal cancer cells to
radiation therapy (14), while
another study revealed that miR-20a-5p increased the capacity of
colorectal cancer invasion and metastasis (16). Our present study further supported
these studies, although controversial data do exist; for example,
Chang et al (27) profiled
differentially expressed miRNAs in oral SCC cell lines and found
that the miR-17-92 cluster (miR-17, miR-19b, miR-20a and miR-92a)
was significantly downregulated in oral SCC cells (particularly,
miR-17 and miR-20a), loss of which was associated with advanced
oral squamous cancer tumor-node-metastasis (TNM) stage and lymph
node metastasis. However, Hu et al (28) linked miR-20a expression to HPV16 E7,
a risk factor for HNSCC, and revealed that miR-20a expression was
significantly higher in oral SCC tissues compared to adjacent
non-tumor tissues and that silencing of HPV-16 E7 expression
downregulated the level of miR-20a in tumor cells and, in turn,
reduced tumor cell proliferation, invasion and metastasis. Yu et
al (29) revealed that when
cytokine secretion in the cell microenvironment was changed, the
role of miR-20a-5p in breast cancer cells was also altered. A given
miRNA was able to play a different role in different cells by
targeting and inhibiting expression of different genes (30,31).
All of these data indicated that the role of miR-20a-5p in the
development of various human cancers remains to be defined and
thus, further studies with a larger sample size and more HNSCC cell
lines are warranted to confirm our data.
By targeting the expression of a particular gene,
miRNAs exert their biological effects on cells; thus, miRNA
involvement in different targeting genes could have different roles
in human cancers or diseases. In our present study, we investigated
the potential genes targeted by miR-20a-5p in HNSCC cells and our
bioinformatic analysis revealed that TNFRSF21 could be a potential
target of miR-20a-5p, and our luciferase reporter assay confirmed
this bioinformatic prediction, revealing that miR-20a-5p bound to
the TNFRSF21 3′-UTR, but not to the mutant one. Our western blot
results revealed that TNFRSF21 expression was inversely correlated
with miR-20a-5p expression, while CCR7 expression was positively
correlated with miR-20a-5p expression, which clearly suggest that
miR-20a-5p indirectly affected CCR7 expression. This is because a
given miRNA could inhibit the expression of the targeting gene, but
will not upregulate the expression of the targeting genes. However,
previous studies have revealed that miR-20a-5p could directly
target different genes in different human cancers, such as TNKS2,
EGR2 and LIMK1, to manipulate tumor cell phenotypes and gene
expression (23–25), which could help to explain why
miR-20a-5p possesses different roles in different human cancers as
an oncogene or tumor-suppressor gene. In our present study,
miR-20a-5p was able to directly target and regulate TNFRSF21
expression in HNSCC cells.
TNFRSF21, also called death receptor 6 (DR6), is a
member of the tumor necrosis factor receptor superfamily (TNFRSF)
(32,33). TNFRSF21 is widely expressed in all
types of tissues and various cultured cells (32) and after binding to the corresponding
ligands, TNFRSF21 is able to induce cell apoptosis (34,35).
However, a previous study revealed that negative lymphocytic
HLA-DR6 was associated with a favorable 5-year survival of HNSCC
patients (36). However, since we
did not find a qualified anti-TNFRSF21 antibody commercially
available for immunohistochemical analysis of TNFRF21 expression in
tumor tissues, we will leave such a project for our future study by
collecting fresh tissue samples for western blot analysis of
TNFRSF21 expression. Moreover, CCR7 is a member of the G
protein-coupled receptor family and was reported to be induced by
the Epstein-Barr virus (37) and
overexpressed in different human cancers, such as non-small cell
lung and (38) gastric cancer
(39), and esophageal squamous cell
carcinoma (40). In the present
study, we found that CCR7 expression was upregulated in cells
transfected with miR-20a-5p mimics and downregulated in cells
transfected with miR-20a-5p inhibitor; thus, we speculate that
reduced expression TNFRF21 by miR-20a-5p could be responsible for
CCR7 upregulation in HNSCC cells. Our research group has focused on
studying the impact of CCR7 on HNSCC, and a previous study revealed
that CCR7 was able to promote HNSCC cell proliferation, migration
and invasion by activating NF-κB (21). Other researchers have reported that
NF-κB can directly or indirectly regulate TNFRSF21 expression to
protect the survival signaling from TNFRSF21-induced cell apoptosis
(41). If this is true, our present
study may have linked these molecules in HNSCC cells, e.g., CCR7
can upregulate the expression of NF-κB and in turn activate
TNFRSF21. In our present study, we found that upregulated
miR-20a-5p expression inhibited TNFRSF21 expression, but
upregulated CCR7 expression. We also found that miR-20a-5p was able
to directly target TNFRSF21 to inhibit TNFRSF21 expression in
PCI-37B cells. However, we are not sure whether there is a negative
feedback loop among TNFRSF21, CCR7, and NF-κB and how TNFRSF21
downregulated CCR7 expression in PCI-37B cells, thus we will
undertake further investigation in our future study.
Our present study is just a proof-of-principle and
does have a number of limitations. For example, we only used a
single HNSCC cell line for gene knock-in and knockdown, which is
not an appropriate research design. However, PCI-37A possesses a
stronger adhesive property and has a poor gene transfection
efficiency. Our future studies will acquire additional HNSCC cells
for validation of our present data. Furthermore, we did not provide
mechanistic data for how TNFRSF21 regulates CCR-7 expression in
HNSCC cells. We should have performed a gene knockout experiment to
assess the effect of TNFRSF21 on the regulation of CCR7 expression
and altered HNSCC cell proliferation, migration and invasion in
vitro. In conclusion, our present study revealed the oncogenic
effect of miR-20a-5p on the promotion of HNSCC cell proliferation,
migration, and invasion by downregulation of TNFRSF21. Further
research is warranted to elucidate the precise mechanisms
underlying the role of miR-20a-5p in HNSCC; for example, we will
carefully select HNSCC cells with high or low expression of
miR20a-5p to assess the effects of miR20a-5p knockdown and
knock-in, respectively on the regulation of HNSCC phenotypes and
gene targets. We will also perform in vivo experiments to
assess the effects of miR-20a-5p on the regulation of tumor
formation and growth in a nude mouse xenograft model of HNSCC
cells. In addition, further research will reveal the underlying
mechanism of miR-20a-5p upregulation in HNSCC to provide a
molecular basis for a better understanding of HNSCC
development.
Acknowledgements
The authors would like to thank the staff in The
Central Laboratories, School of Stomatology, China Medical
University for their technical assistance.
Funding
The present study was supported in part by grants
from the National Natural Science Foundation of China (no.
81372877), the National Young Scholars Science Foundation of China
(no. 81102058), the Public Welfare Fund Project for Science of
Liaoning Province (no. 2011002001), the Excellent Talent Fund
Project of Higher Education of Liaoning Province (no. LJQ2014087)
and the Doctoral Scientific Research Foundation of Liaoning
Province (no. 201501002).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
CFS and FYL designed and conceived the study. HW,
PP, MDL, SW and SJ performed the experiments. HW completed most of
the experimental work and prepared the manuscript. CFS and FYL
reviewed and edited the manuscript. All authors read and approved
the final version of this manuscript and ensured the accuracy or
integrity of any part of this work.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethics Committee of China Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grégoire V, Lefebvre JL, Licitra L and
Felip E; EHNS-ESMO-ESTRO Guidelines Working Group: Squamous cell
carcinoma of the head and neck: EHNS-ESMO-ESTRO Clinical Practice
Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 21
(Suppl 5):v184–v186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bernstein E, Caudy AA, Hammond SM and
Hannon GJ: Role for a bidentate ribonuclease in the initiation step
of RNA interference. Nature. 409:363–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeng Y and Cullen BR: Sequence
requirements for micro RNA processing and function in human cells.
RNA. 9:112–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arantes LM, Laus AC, Melendez ME, de
Carvalho AC, Sorroche BP, De Marchi PR, Evangelista AF,
Scapulatempo-Neto C, de Souza Viana L and Carvalho AL: MiR-21 as
prognostic biomarker in head and neck squamous cell carcinoma
patients undergoing an organ preservation protocol. Oncotarget.
8:9911–9921. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sousa LO, Sobral LM, Matsumoto CS,
Saggioro FP, López RV, Panepucci RA, Curti C, Silva WA Jr, Greene
LJ and Leopoldino AM: Lymph node or perineural invasion is
associated with low miR-15a, miR-34c and miR-199b levels in head
and neck squamous cell carcinoma. BBA Clin. 6:159–164. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hudcova K, Raudenska M, Gumulec J, Binkova
H, Horakova Z, Kostrica R, Babula P, Adam V and Masarik M:
Expression profiles of miR-29c, miR-200b and miR-375 in tumour and
tumour-adjacent tissues of head and neck cancers. Tumour Biol.
37:12627–12633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Koshizuka K, Hanazawa T, Fukumoto I,
Kikkawa N, Okamoto Y and Seki N: The microRNA signatures:
Aberrantly expressed microRNAs in head and neck squamous cell
carcinoma. J Hum Genet. 62:3–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Carvalho AC, Scapulatempo-Neto C, Maia
DC, Evangelista AF, Morini MA, Carvalho AL and Vettore AL: Accuracy
of microRNAs as markers for the detection of neck lymph node
metastases in patients with head and neck squamous cell carcinoma.
BMC Med. 13:1082015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hou B, Ishinaga H, Midorikawa K, Shah SA,
Nakamura S, Hiraku Y, Oikawa S, Murata M and Takeuchi K:
Circulating microRNAs as novel prognosis biomarkers for head and
neck squamous cell carcinoma. Cancer Biol Ther. 16:1042–1046. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B,
Waldron J, et al: Comprehensive MicroRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramdas L, Giri U, Ashorn CL, Coombes KR,
El-Naggar A, Ang KK and Story MD: miRNA expression profiles in head
and neck squamous cell carcinoma and adjacent normal tissue. Head
Neck. 31:642–654. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rahimy E, Kuo SZ and Ongkeko WM:
Evaluation of non-coding RNAs as potential targets in head and neck
squamous cell carcinoma cancer stem cells. Curr Drug Targets.
15:1247–1260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang D, Bian G, Pan Y, Han X, Sun Y, Wang
Y, Shen G, Cheng M, Fang X and Hu S: MiR-20a-5p promotes
radio-resistance by targeting Rab27B in nasopharyngeal cancer
cells. Cancer Cell Int. 17:322017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Y, Wang X, Cheng J, Wang Z, Jiang T,
Hou N, Liu N, Song T and Huang C: MicroRNA-20a-5p targets RUNX3 to
regulate proliferation and migration of human hepatocellular cancer
cells. Oncol Rep. 36:3379–3386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng D, Zhao S, Tang H, Zhang D, Sun H,
Yu F, Jiang W, Yue B, Wang J, Zhang M, et al: MicroRNA-20a-5p
promotes colorectal cancer invasion and metastasis by
downregulating Smad4. Oncotarget. 7:45199–45213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang L, Liu F, Xu Z, Guo N, Zheng X and
Sun C: Chemokine receptor 7 via proline-rich tyrosine kinase-2
upregulates the chemotaxis and migration ability of squamous cell
carcinoma of the head and neck. Oncol Rep. 28:1659–1664. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Zhang X, Thomas SM, Grandis JR,
Wells A, Chen ZG and Ferris RL: Chemokine receptor 7 activates
phosphoinositide-3 kinase-mediated invasive and prosurvival
pathways in head and neck cancer cells independent of EGFR.
Oncogene. 24:5897–5904. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li P, Liu F, Sun L, Zhao Z, Ding X, Shang
D, Xu Z and Sun C: Chemokine receptor 7 promotes cell migration and
adhesion in metastatic squamous cell carcinoma of the head and neck
by activating integrin alphavbeta3. Int J Mol Med. 27:679–687.
2011.PubMed/NCBI
|
|
20
|
Zhao ZJ, Liu FY, Li P, Ding X, Zong ZH and
Sun CF: CCL19-induced chemokine receptor 7 activates the
phosphoinositide-3 kinase-mediated invasive pathway through Cdc42
in metastatic squamous cell carcinoma of the head and neck. Oncol
Rep. 25:729–737. 2011.PubMed/NCBI
|
|
21
|
Liu FY, Zhao ZJ, Li P, Ding X, Guo N, Yang
LL, Zong ZH and Sun CF: NF-κB participates in chemokine receptor
7-mediated cell survival in metastatic squamous cell carcinoma of
the head and neck. Oncol Rep. 25:383–391. 2011.PubMed/NCBI
|
|
22
|
Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T,
Liu M, Li X and Tang H: miR-20a promotes proliferation and invasion
by targeting APP in human ovarian cancer cells. Acta Biochim
Biophys Sin. 42:318–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang HW, Wang F, Wei Q, Zhao YF, Liu M, Li
X and Tang H: miR-20a promotes migration and invasion by regulating
TNKS2 in human cervical cancer cells. FEBS Lett. 586:897–904. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Zhang Z, Yu M, Li L, Du G, Xiao W
and Yang H: Involvement of miR-20a in promoting gastric cancer
progression by targeting early growth response 2 (EGR2). Int J Mol
Sci. 14:16226–16239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou J, Liu R, Luo C, Zhou X, Xia K, Chen
X, Zhou M, Zou Q, Cao P and Cao K: MiR-20a inhibits cutaneous
squamous cell carcinoma metastasis and proliferation by directly
targeting LIMK1. Cancer Biol Ther. 15:1340–1349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiong Y, Zhang L and Kebebew E: MiR-20a is
upregulated in anaplastic thyroid cancer and targets LIMK1.
PLoS One. 9:e961032014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang CC, Yang YJ, Li YJ, Chen ST, Lin BR,
Wu TS, Lin SK, Kuo MY and Tan CT: MicroRNA-17/20a functions to
inhibit cell migration and can be used a prognostic marker in oral
squamous cell carcinoma. Oral Oncol. 49:923–931. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu J, Ge W and Xu J: HPV 16 E7 inhibits
OSCC cell proliferation, invasion, and metastasis by upregulating
the expression of miR-20a. Tumour Biol. 37:9433–9440. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu Z, Willmarth NE, Zhou J, Katiyar S,
Wang M, Liu Y, McCue PA, Quong AA, Lisanti MP and Pestell RG:
microRNA 17/20 inhibits cellular invasion and tumor metastasis in
breast cancer by heterotypic signaling. Proc Natl Acad Sci USA.
107:8231–8236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Singh B, Ronghe AM, Chatterjee A, Bhat NK
and Bhat HK: MicroRNA-93 regulates NRF2 expression and is
associated with breast carcinogenesis. Carcinogenesis.
34:1165–1172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu S, Patel SH, Ginestier C, Ibarra I,
Martin-Trevino R, Bai S, McDermott SP, Shang L, Ke J, Ou SJ, et al:
MicroRNA93 regulates proliferation and differentiation of normal
and malignant breast stem cells. PLoS Genet. 8:e10027512012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pan G, Bauer JH, Haridas V, Wang S, Liu D,
Yu G, Vincenz C, Aggarwal BB, Ni J and Dixit VM: Identification and
functional characterization of DR6, a novel death domain-containing
TNF receptor. FEBS Lett. 431:351–356. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sedý J, Bekiaris V and Ware CF: Tumor
necrosis factor superfamily in innate immunity and inflammation.
Cold Spring Harb Perspect Biol. 7:a0162792014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pfeffer K: Biological functions of tumor
necrosis factor cytokines and their receptors. Cytokine Growth
Factor Rev. 14:185–191. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thorburn A: Death receptor-induced cell
killing. Cell Signal. 16:139–144. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tisch M, Kyrberg H, Weidauer H, Mytilineos
J, Conradt C, Opelz G and Maier H: Human leukocyte antigens and
prognosis in patients with head and neck cancer: Results of a
prospective follow-up study. Laryngoscope. 112:651–657. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sharma N, Benechet AP, Lefrancois L and
Khanna KM: CD8 T cells enter the Splenic T cell zones independently
of CCR7, but the subsequent expansion and trafficking patterns of
effector T cells after infection are dysregulated in the absence of
CCR7 migratory cues. J Immunol. 195:5227–5236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Takanami I: Overexpression of CCR7 mRNA in
nonsmall cell lung cancer: Correlation with lymph node metastasis.
Int J Cancer. 105:186–189. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mashino K, Sadanaga N, Yamaguchi H, Tanaka
F, Ohta M, Shibuta K, Inoue H and Mori M: Expression of chemokine
receptor CCR7 is associated with lymph node metastasis of gastric
carcinoma. Cancer Res. 62:2937–2941. 2002.PubMed/NCBI
|
|
40
|
Ding Y, Shimada Y, Maeda M, Kawabe A,
Kaganoi J, Komoto I, Hashimoto Y, Miyake M, Hashida H and Imamura
M: Association of CC chemokine receptor 7 with lymph node
metastasis of esophageal squamous cell carcinoma. Clin Cancer Res.
9:3406–3412. 2003.PubMed/NCBI
|
|
41
|
Kasof GM, Lu JJ, Liu D, Speer B, Mongan
KN, Gomes BC and Lorenzi MV: Tumor necrosis factor-alpha induces
the expression of DR6, a member of the TNF receptor family, through
activation of NF-kappaB. Oncogene. 20:7965–7975. 2001. View Article : Google Scholar : PubMed/NCBI
|