Introduction
Cholangiocarcinoma, a highly aggressive tumor
derived from bile duct epithelial cells, is one of the most severe
forms of cancer with a 5-year survival rate <10% (1,2).
During the last decades, the incidence and mortality rate of
cholangiocarcinoma have been increasing globally (3,4). The
survival quality and prognosis of patients with cholangiocarcinoma
are poor as a result of early cancer cell invasion and metastasis
(2). Radical surgery is the only
curative treatment for cholangiocarcinoma, while patients gain
little benefit, as they are usually diagnosed at an advanced stage
(5,6). Thus, the exploration of powerful
markers which may provide prognostic value for cholangiocarcinoma
patients is of great significance. Currently, prognostic microRNA
(miRNA) expression signatures in various cancers, such as colon
cancer (7), clear cell renal cell
carcinoma (8) and cervical cancer
(9) have attracted the attention of
researchers. Therefore, the miRNA signature has been regarded as an
important change in cholangiocarcinoma progression and therapy
(10).
miRNAs, a class of small non-coding RNAs of ~22–23
nucleotides in length, are considered to play pivotal roles in
post-transcriptional gene regulation. It has been confirmed that
miRNAs regulate malignancies by binding to the partially
complementary recognition sequences in the 3-untranslated region of
mRNAs, which causes target mRNA translation inhibition or
degradation (11). A number of
miRNAs play vital roles in tumorigenesis, such as cell
proliferation, apoptosis, autophagy, migration, invasion and
metastasis (12,13). Accordingly, miRNAs have a large
potential to serve as markers in the diagnosis, prognosis and
targeted therapies of cancers.
Although a large number of miRNAs have been
identified in predicting the clinical outcome in
cholangiocarcinoma, there are some limitations in previous studies.
These may be due to molecular and clinical heterogeneity in
different studies, relatively limited numbers of miRNAs, along with
methodological differences in detection and analysis. The Cancer
Genome Atlas (TCGA) is a National Cancer Institute effort to
profile >20 different tumor types using genomic platforms and to
make raw and processed data available to researchers worldwide
(14). TCGA provides a collection
of clinical data, RNA sequence, DNA copy number variations, DNA
methylation and miRNA sequence profiles for cholangiocarcinoma. The
aim of the present study was to identify the differentially
expressed miRNAs between cholangiocarcinoma and normal tissues by
analyzing high-throughput data downloaded from TCGA database.
Furthermore, we evaluated the prognostic performance of the
differentially expressed miRNAs and established a novel three-miRNA
signature which could effectively predict survival in
cholangiocarcinoma patients.
Materials and methods
TCGA dataset of
cholangiocarcinoma
The miRNA sequencing data and corresponding clinical
information for cholangiocarcinoma patients (up to June 29, 2017)
were downloaded from TCGA data portal (https://portal.gdc.cancer.gov). The inclusion criteria
were set as follows: i) samples with both miRNA sequencing data and
clinical information; and ii) samples with detailed prognostic
information. Finally, a total of 45 samples were enrolled in this
study, including 36 cholangiocarcinoma tissues and 9 matched normal
tissues.
Exploration of the differentially
expressed miRNAs in cholangiocarcinoma
The RNA-Seq data of cholangiocarcinoma with 1,046
miRNAs were analyzed on the Illumina HiSeq miRNA Seq platform.
Subsequently, the R language package ‘edgeR’ was used for the
calculation of differentially expressed miRNAs (15). The expression difference of
individual miRNAs was characterized by log2FC and
adjusted P-value. LogFC indicates the fold change in expression of
each miRNA between cholangiocarcinoma tissues and normal tissues.
Upregulated and downregulated miRNAs were determined based on
log2FC >1 and log2FC <-1 respectively,
with adjusted P<0.01. The miRNAs which had expression mean value
<1 were excluded.
Selection of the cut-off point for the
Kaplan-Meier survival analysis
Cutoff Finder (http://molpath.charite.de/cutoff) was used to
determine a cut-off point for patient stratification into two
groups (16). Then, the differences
in patient overall survival (OS) between the high-level and the
low-level group were evaluated by Kaplan-Meier survival analysis
(log-rank method). The miRNAs with a P-value <0.01 were regarded
to display statistically significant differences between groups and
were considered for further analysis.
Association between miRNA signature
index and OS
A value of one or zero was assigned to patients
according to each miRNA value. Subsequently, each miRNA value was
scored in the signature. Thus, each patient would have a score,
defined as miRNA signature index. We set index as high-risk and
low-risk into two new groups according to the index value. Then,
Kaplan-Meier survival analysis (log-rank method) was performed to
evaluate the differences in patient OS between these two
groups.
Information collection of the
validation dataset
An independent cohort of cholangiocarcinoma patients
(GSE53870) (17) downloaded from
Gene Expression Omnibus (GEO) database was used for the prognostic
signature validation. There consisted of 63 cholangiocarcinoma
patients and corresponding prognostic information in the GSE53870
dataset.
Target gene prediction of three
prognostic miRNAs and functional analysis
The target genes of the three prognostic miRNAs were
predicted using TargetScan (http://www.targetscan.org/), miRDB (http://www.mirdb.org/miRDB/), and miRanda (http://www.microrna.org/) online analysis tools. To
further increase the bioinformatics analysis reliability, Venn
diagram was carried out to identify the overlapping target genes.
Furthermore, the Gene Ontology (GO) annotation and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis of target genes were performed using the Database for
Annotation, Visualization and Integrated Discovery (DAVID) online
tool (https://david.ncifcrf.gov/). The results
were then presented in a bubble diagram.
RNA isolation and qRT-qPCR
Total RNA was extracted from cells using an HP Total
RNA kit (Omega Biotech, Stamford, CT, USA) according to the
manufacturer's protocol. Synthesis of cDNA with reverse
transcriptase (RT) was performed with an M-MLV First Strand kit
(Life Technologies; Thermo Fisher Scientific, Inc., Gaithersburg,
MD, USA). Primer sequences for miR-10b, miR-22, miR-551b and U6
detection were obtained from RiboBio (Guangzhou, China). The RT
primers for mature miRNAs and U6 were designed according to the
concept of a stem-loop RT primer (18). RT-qPCR analysis was carried out
using Platinum SYBR-Green qPCR SuperMix-UDG kits (Life
Technologies) according to the manufacturer's protocol. Real-time
PCR was performed on an Applied Biosystems ABI PRISM 7500 Real-Time
PCR system (Thermo Fisher Scientific, Inc.). Ct values of miRNAs
were equilibrated to U6, which was used as an internal control.
Relative expression was calculated using the 2−ΔΔCq
method.
Cell culture and transfection
Human cholangiocarcinoma cell line HUCCT1 (cat. no.
JCRB0425) was purchased from the Japanese Collection of Research
Bioresources Cell Bank (JCRB; Osaka, Japan) and human intrahepatic
biliary epithelial cell line (HiBEC) (cat. no. 5100) was purchased
from the ScienCell Research Laboratories (San Diego, CA, USA) and
were cultured under standard conditions. When HUCCT1 cells reached
50–70% confluence, miR-551b mimics and negative control were
transfected using Invitrogen™ Lipofectamine 2000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. miR-551b mimics (sense, 5′-GCGACCCAUACUUGGUUUCAG-3′ and
antisense, 5′-GAAACCAAGUAUGGGUCGCUU-3′) and corresponding negative
control were purchased from GenePharma (Shanghai, China).
Proliferation assay
HUCCT1 cells were transfected with miR-551b mimics
or negative control for 48 h, and then were plated into 96-well
plates at a density of 5×103 cells/well and then 10 µl
of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT, 5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was
added and incubated for 4 h. The supernatant was then replaced with
100 µl of dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA) and
read at 490 nm using a multifunction microplate reader (POLARstar
OPTIMA; BMG, Offenburg, Germany). Concerning the colony formation
assay, HUCCT1 cells (1×103 cells/dish) were seeded on
35-mm petri dishes. Cells were further cultured for two weeks to
allow colonies to form. At the indicated time-point, colonies were
fixed with 4% paraformaldehyde, stained with 0.1% crystal violet
solution and rinsed. Then, images were captured using a Nikon
camera (Nikon Corp., Tokyo, Japan) and the colony number was
counted.
Apoptosis assay
HUCCT1 cells were transfected with miR-551b mimics
or negative control for 48 h, then were harvested by trypsinization
in a tube and were washed twice in ice-cold phosphate-buffered
saline (PBS). After staining using an Annexin V-FITC/7-AAD
apoptosis detection kit (Becton-Dickinson, Franklin Lakes, NJ, USA)
according to the manufacturer's protocol, the cell apoptosis rate
was assessed using a FACSCalibur flow cytometer (BD Biosciences,
San Diego, CA, USA).
Statistical analysis
Mann-Whitney U test was used to compare the
expression levels of miRNAs between two different groups of each
clinical characteristic. Univariate/multivariate Cox proportional
hazard regression analyses were performed to compare each clinical
parameter and prognostic miRNA signature (high-risk vs. low-risk).
All statistical analysis was performed by SPSS 20.0 (SPSS, Inc.,
Chicago, IL, USA). Statistical significance was defined as a
two-sided P-value <0.05, unless specifically indicated.
Results
Exploration of differentially
expressed miRNAs
A total of 45 samples were enrolled in our study,
including 36 cholangiocarcinoma tissues and 9 matched normal
tissues. The specific clinical characteristics included sex, age at
diagnosis, stage, T stage, lymph node status, metastasis and
histological type (Table I).
According to the cut-off criteria of P<0.01 and
|log2FC|>1.0, a total of 100 miRNAs were found to be
differentially expressed in cholangiocarcinoma vs. normal tissues,
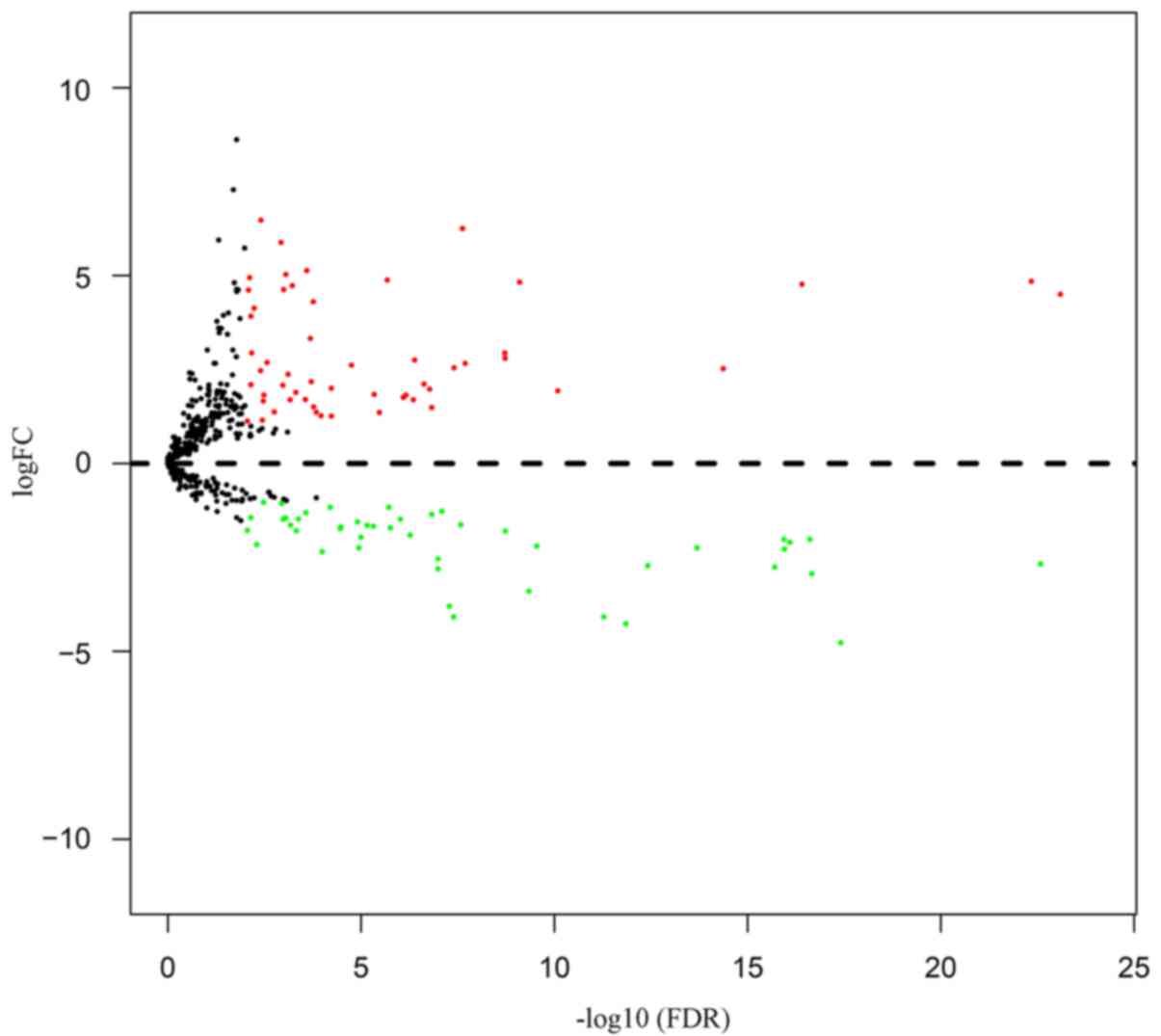
including 54 upregulated and 46 downregulated miRNAs (Table II). In order to show the above
differentially expressed miRNA more clearly, we present the results
as a Volcano plot (Fig. 1).
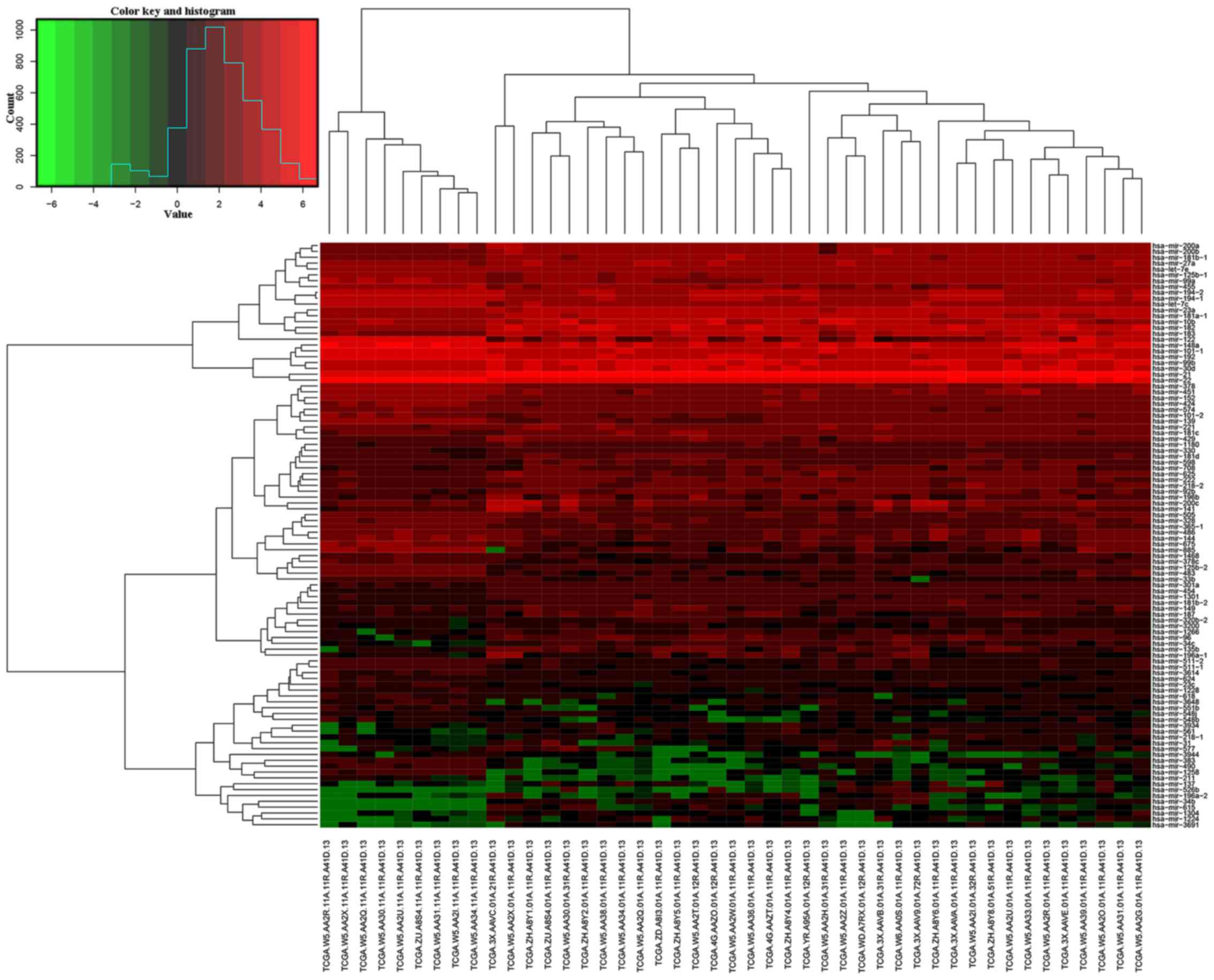
Furthermore, it was obvious that cholangiocarcinoma tissues could
be clearly discriminated from normal tissues in terms of
differentially expressed miRNA patterns using unsupervised
hierarchic cluster analysis (Fig.
2).
 | Table I.Clinical characteristics of the
cholangiocarcinoma patients (n=36). |
Table I.
Clinical characteristics of the
cholangiocarcinoma patients (n=36).
| Variables | Cases, n (%) |
|---|
| Sex |
|
Female | 20 (55.56) |
|
Male | 16 (44.44) |
| Age at diagnosis
(years) |
|
≤60 | 14 (38.89) |
|
>60 | 22 (61.11) |
| Stage |
|
I+II | 28 (77.78) |
|
III+IV | 8
(22.22) |
| T stage |
|
T1+T2 | 31 (86.11) |
|
T3+T4 | 5
(13.89) |
| Lymph node
status |
| N0 | 26 (72.22) |
| N1 | 5
(13.89) |
| NX | 5
(13.89) |
| Metastasis |
| M0 | 28 (77.78) |
| M1 | 5
(13.89) |
| MX | 3
(8.33) |
| Histological
type |
|
Intrahepatic | 30 (83.33) |
|
Hilar+distal | 6
(16.67) |
 | Table II.Differentially expressed miRNAs
between cholangiocarcinoma and normal tissues. |
Table II.
Differentially expressed miRNAs
between cholangiocarcinoma and normal tissues.
| Upregulated
miRNAs | Downregulated
miRNAs |
|---|
| miRNAs | logFC | P-value | FDR | miRNAs | logFC | P-value | FDR |
|---|
| hsa-mir-182 | 4.50 | 1.72E-26 | 8.03E-24 | hsa-mir-148a | −2.67 | 1.12E-25 | 2.61E-23 |
| hsa-mir-183 | 4.85 | 2.93E-25 | 4.56E-23 | hsa-mir-1258 | −4.77 | 3.35E-20 | 3.91E-18 |
| hsa-mir-96 | 4.77 | 5.87E-19 | 3.92E-17 | hsa-mir-378 | −2.93 | 2.32E-19 | 2.17E-17 |
| hsa-mir-21 | 2.53 | 1.09E-16 | 4.26E-15 | hsa-mir-101-1 | −2.02 | 3.13E-19 | 2.44E-17 |
| hsa-mir-27a | 1.93 | 2.97E-12 | 8.15E-11 | hsa-let-7c | −2.10 | 1.39E-18 | 8.10E-17 |
| hsa-mir-34c | 4.83 | 3.39E-11 | 7.92E-10 | hsa-mir-99a | −2.28 | 2.15E-18 | 1.11E-16 |
| hsa-mir-200b | 2.80 | 8.91E-11 | 1.89E-09 | hsa-mir-505 | −2.02 | 2.44E-18 | 1.14E-16 |
| hsa-mir-92b | 2.94 | 9.35E-11 | 1.90E-09 | hsa-mir-139 | −2.76 | 4.65E-18 | 1.97E-16 |
| hsa-mir-200a | 2.67 | 1.05E-09 | 2.04E-08 | hsa-mir-125b-2 | −2.25 | 5.65E-16 | 2.03E-14 |
| hsa-mir-34b | 6.26 | 1.28E-09 | 2.39E-08 | hsa-mir-378c | −2.72 | 1.15E-14 | 3.83E-13 |
| hsa-mir-181d | 2.54 | 2.29E-09 | 3.97E-08 | hsa-mir-490 | −4.27 | 4.53E-14 | 1.41E-12 |
| hsa-mir-23a | 1.49 | 1.07E-08 | 1.49E-07 | hsa-mir-675 | −4.08 | 1.80E-13 | 5.25E-12 |
| hsa-mir-222 | 1.98 | 1.26E-08 | 1.69E-07 | hsa-mir-1468 | −2.20 | 1.10E-11 | 2.84E-10 |
| hsa-mir-181b-1 | 2.11 | 1.83E-08 | 2.37E-07 | hsa-mir-483 | −3.40 | 1.85E-11 | 4.55E-10 |
| hsa-mir-429 | 2.75 | 3.28E-08 | 4.14E-07 | hsa-mir-101-2 | −1.80 | 8.32E-11 | 1.85E-09 |
| hsa-mir-454 | 1.70 | 3.62E-08 | 4.45E-07 | hsa-mir-424 | −1.63 | 1.46E-09 | 2.62E-08 |
| hsa-mir-221 | 1.82 | 5.94E-08 | 6.93E-07 | hsa-mir-122 | −4.09 | 2.42E-09 | 4.04E-08 |
| hsa-mir-330 | 1.76 | 7.15E-08 | 8.14E-07 | hsa-mir-885 | −3.80 | 3.21E-09 | 5.18E-08 |
| hsa-mir-135b | 4.88 | 2.04E-07 | 2.11E-06 | hsa-mir-22 | −1.27 | 5.24E-09 | 8.15E-08 |
| hsa-let-7e | 1.36 | 3.31E-07 | 3.36E-06 | hsa-mir-383 | −2.81 | 6.81E-09 | 1.02E-07 |
| hsa-mir-181c | 1.83 | 4.60E-07 | 4.57E-06 | hsa-mir-551b | −2.54 | 7.01E-09 | 1.02E-07 |
| hsa-mir-708 | 2.62 | 2.01E-06 | 1.77E-05 | hsa-mir-574 | −1.36 | 1.09E-08 | 1.49E-07 |
| hsa-mir-99b | 1.26 | 7.07E-06 | 5.88E-05 | hsa-mir-192 | −1.91 | 4.51E-08 | 5.40E-07 |
| hsa-mir-181b-2 | 2.00 | 7.17E-06 | 5.88E-05 | hsa-mir-624 | −1.48 | 8.71E-08 | 9.69E-07 |
| hsa-mir-301a | 1.27 | 1.39E-05 | 0.000108 | hsa-mir-455 | −1.71 | 1.62E-07 | 1.76E-06 |
| hsa-mir-181a-1 | 1.36 | 1.91E-05 | 0.000144 | hsa-mir-152 | −1.17 | 1.82E-07 | 1.93E-06 |
| hsa-mir-1301 | 1.50 | 2.29E-05 | 0.00017 | hsa-mir-194-2 | −1.67 | 4.97E-07 | 4.84E-06 |
| hsa-mir-196b | 4.30 | 2.38E-05 | 0.000174 | hsa-mir-194-1 | −1.65 | 7.29E-07 | 6.95E-06 |
| hsa-mir-1266 | 2.18 | 2.70E-05 | 0.000194 | hsa-mir-618 | −1.96 | 1.08E-06 | 1.01E-05 |
| hsa-mir-561 | 3.32 | 2.91E-05 | 0.000206 | hsa-mir-144 | −2.25 | 1.25E-06 | 1.14E-05 |
| hsa-mir-200c | 5.13 | 3.64E-05 | 0.000254 | hsa-mir-1228 | −1.55 | 1.38E-06 | 1.24E-05 |
| hsa-mir-218-2 | 1.70 | 4.09E-05 | 0.000277 | hsa-mir-511-2 | −1.69 | 3.88E-06 | 3.35E-05 |
| hsa-mir-149 | 1.89 | 7.53E-05 | 0.000488 | hsa-mir-511-1 | −1.73 | 4.10E-06 | 3.48E-05 |
| hsa-mir-141 | 4.73 | 9.43E-05 | 0.000603 | hsa-mir-125b-1 | −1.17 | 7.87E-06 | 6.34E-05 |
| hsa-mir-625 | 1.69 | 0.00011 | 0.000683 | hsa-mir-548b | −2.35 | 1.28E-05 | 0.000102 |
| hsa-mir-10b | 2.38 | 0.000126 | 0.000774 | hsa-mir-3614 | −1.31 | 3.91E-05 | 0.000269 |
| hsa-mir-196a-1 | 5.03 | 0.000151 | 0.000883 | hsa-mir-33b | −1.48 | 6.25E-05 | 0.000417 |
| hsa-mir-1224 | 4.63 | 0.000177 | 0.001009 | hsa-mir-3648 | −1.79 | 7.18E-05 | 0.000472 |
| hsa-mir-3934 | 2.08 | 0.000189 | 0.00105 | hsa-mir-486 | −1.64 | 0.000107 | 0.000672 |
| hsa-mir-615 | 5.88 | 0.000218 | 0.001184 | hsa-mir-23c | −1.46 | 0.000151 | 0.000883 |
| hsa-mir-598 | 1.37 | 0.000332 | 0.00176 | hsa-mir-548j | −1.48 | 0.000184 | 0.001033 |
| hsa-mir-187 | 2.69 | 0.000544 | 0.002704 | hsa-mir-328 | −1.06 | 0.000206 | 0.001135 |
| hsa-mir-3200 | 1.82 | 0.000662 | 0.003252 | hsa-mir-365-1 | −1.03 | 0.000687 | 0.003343 |
| hsa-mir-320b-2 | 1.66 | 0.00071 | 0.00342 | hsa-mir-211 | −2.16 | 0.001105 | 0.005009 |
| hsa-mir-1180 | 1.15 | 0.000753 | 0.003587 | hsa-mir-451 | −1.44 | 0.001677 | 0.007107 |
| hsa-mir-526b | 6.47 | 0.000834 | 0.003895 | hsa-mir-3944 | −1.78 | 0.002201 | 0.00871 |
| hsa-mir-1304 | 2.47 | 0.000859 | 0.003972 |
|
|
|
|
| hsa-mir-31 | 4.13 | 0.001327 | 0.005901 |
|
|
|
|
| hsa-mir-3691 | 2.94 | 0.001532 | 0.006748 |
|
|
|
|
| hsa-mir-218-1 | 2.09 | 0.001665 | 0.007107 |
|
|
|
|
| hsa-mir-577 | 3.92 | 0.001689 | 0.007107 |
|
|
|
|
| hsa-mir-137 | 4.94 | 0.001893 | 0.007687 |
|
|
|
|
| hsa-mir-196a-2 | 4.62 | 0.002038 | 0.008205 |
|
|
|
|
| hsa-mir-30d | 1.13 | 0.00219 | 0.00871 |
|
|
|
|
Association between differentially
expressed miRNAs and clinical features
The differentially expressed miRNAs were further
analyzed upon the expression level and clinical characteristics.
Notably, miR-490 and miR-141 were found to be related with sex,
whereas miR-615, miR-135b, miR-92b, miR-23c and miR-149 were found
to be related with age at diagnosis. Furthermore, miR-301a was
associated with stage, whereas miR-551b, miR-222 and miR-221 were
associated with T stage, miR-92b and miR-615 were associated with
lymph node status, miR-101-1 and miR-301a were associated with
metastasis. In addition, we also found other 8 miRNAs which were
linked to histological type (Table
III).
 | Table III.Differentially expressed miRNAs
associated with clinical features. |
Table III.
Differentially expressed miRNAs
associated with clinical features.
| Variables | Upregulated miRNAs
identified in TCGA | Downregulated
miRNAs identified in TCGA |
|---|
| Sex |
| (female
vs. male) | miR-141 | miR-490 |
| Age at
diagnosis |
| (≤60
vs. >60) | miR-615, miR-135b,
miR-92b, miR-149 | miR-23c |
| Stage |
| (I+II
vs. III+IV) | miR-301a |
|
| T stage |
| (T1+T2
vs. T3+T4) | miR-222,
miR-221 | miR-551b |
| Lymph node
status |
| (N0 vs.
N1) | miR-92b,
miR-615 |
|
| Metastasis |
| (M0 vs.
M1) | miR-301a | miR-101-1 |
| Histological
type | miR-23a,
miR-196a-1, miR-27a, | miR-365-1,
miR-383 |
|
(intrahepatic vs.
hilar+distal) | miR-598, miR-31,
mir-181c |
|
Identification of three miRNAs
associated with OS
Of the aforementioned 100 miRNAs, we used Cutoff
Finder to determine a cut-off point and classified patients into
two groups based on the miRNA expression level. Subsequently, the
Kaplan-Meier survival analysis was performed to explore the
association between miRNA expression and OS in cholangiocarcinoma
patients. According to the survival analysis, three miRNAs were
identified to be significantly associated with OS in
cholangiocarcinoma patients. These three miRNAs were miR-10b,
miR-22 and miR-551b. These results illustrated that high expression
levels of miR-10b and miR-551b were considered as better prognostic
markers vs. the low level group (Fig.
3A and C). In contrast, compared to the high-level group, low
expression level of miR-22 revealed a longer survival rate and time
(Fig. 3B).
Definition of three-miRNA signature
index for cholangiocarcinoma prognosis
In order to establish the miRNA signature index, we
assigned a score for each patient. To be specific, patients who
belonged to the shorter survival group received one score for each
miRNA, while those who belonged to the longer survival group
received a 0 score for each miRNA. Subsequently, we calculated the
total score for each patient. Subsequently, we calculated the score
for each patient. According to these criteria, the highest score
was 3 and the lowest score was 0. Subsequently, we ranked 36
cholangiocarcinoma patients based on their miRNA signature index
and divided them into two new groups (Table IV). The high-risk group miRNA
signature index score was 2–3, while the score in the low-risk
group was 0–1. Markedly, Kaplan-Meier survival analysis illustrated
that these two groups were significantly associated with patient OS
(Fig. 4). In the low-risk group,
~80% patients showed 5-year survival, while none of the patients
survived >5 years in the high-risk group. Furthermore, the
median survival of the low-risk group was markedly longer than that
of the high-risk group (63.75 vs. 14.63 months). As a result, our
findings demonstrated that the survival rate, as well as the
survival time of patients were obviously enhanced in relation to a
lower miRNA signature index.
 | Table IV.Three-miRNA signature index for
survival analysis. |
Table IV.
Three-miRNA signature index for
survival analysis.
| Patient ID | Survival
status | Overall survival
(months) | miR-10b | miR-22 | miR-551b | miRNA index |
|---|
| TCGA-3X-AAVB | Alive | 13.22 | 0 | 0 | 0 | 0 |
| TCGA-3X-AAVE | Alive | 21.37 | 0 | 0 | 0 | 0 |
| TCGA-3X-AAVC | Alive | 23.31 | 0 | 0 | 0 | 0 |
| TCGA-W5-AA2H | Alive | 35.41 | 0 | 0 | 0 | 0 |
| TCGA-W5-AA33 | Alive | 47.64 | 0 | 0 | 0 | 0 |
| TCGA-W5-AA2R | Alive | 50.70 | 0 | 0 | 0 | 0 |
| TCGA-W5-AA2Q | Alive | 1.64 | 0 | 0 | 1 | 1 |
| TCGA-4G-AAZT | Alive | 13.81 | 0 | 0 | 1 | 1 |
| TCGA-ZH-A8Y8 | Alive | 19.79 | 0 | 0 | 1 | 1 |
| TCGA-ZH-A8Y4 | Dead | 24.36 | 0 | 0 | 1 | 1 |
| TCGA-W5-AA30 | Alive | 37.91 | 0 | 0 | 1 | 1 |
| TCGA-4G-AAZO | Alive | 38.70 | 0 | 0 | 1 | 1 |
| TCGA-ZH-A8Y5 | Alive | 40.41 | 0 | 0 | 1 | 1 |
| TCGA-W5-AA36 | Dead | 46.09 | 1 | 0 | 0 | 1 |
| TCGA-W5-AA38 | Alive | 48.36 | 0 | 0 | 1 | 1 |
| TCGA-W5-AA2Z | Alive | 53.06 | 0 | 0 | 1 | 1 |
| TCGA-W5-AA2I | Dead | 63.75 | 1 | 0 | 0 | 1 |
| TCGA-W5-AA2G | Alive | 64.96 | 1 | 0 | 0 | 1 |
| TCGA-W5-AA31 | Alive | 0.33 | 1 | 1 | 0 | 2 |
| TCGA-WD-A7RX | Dead | 0.69 | 1 | 0 | 1 | 2 |
| TCGA-ZU-A8S4 | Dead | 3.22 | 0 | 1 | 1 | 2 |
| TCGA-ZD-A8I3 | Dead | 5.56 | 0 | 1 | 1 | 2 |
| TCGA-W5-AA2X | Dead | 8.91 | 1 | 0 | 1 | 2 |
| TCGA-3X-AAV9 | Dead | 11.15 | 0 | 1 | 1 | 2 |
| TCGA-ZH-A8Y1 | Dead | 12.66 | 1 | 0 | 1 | 2 |
| TCGA-W5-AA34 | Dead | 18.25 | 1 | 0 | 1 | 2 |
| TCGA-W5-AA2U | Dead | 20.61 | 0 | 1 | 1 | 2 |
| TCGA-W5-AA2O | Dead | 21.04 | 1 | 0 | 1 | 2 |
| TCGA-ZH-A8Y2 | Dead | 23.05 | 1 | 0 | 1 | 2 |
| TCGA-W6-AA0S | Alive | 26.56 | 0 | 1 | 1 | 2 |
| TCGA-W5-AA2W | Dead | 30.38 | 1 | 0 | 1 | 2 |
| TCGA-W5-AA2T | Dead | 40.11 | 1 | 0 | 1 | 2 |
| TCGA-YR-A95A | Dead | 0.85 | 1 | 1 | 1 | 3 |
| TCGA-W5-AA39 | Dead | 5.59 | 1 | 1 | 1 | 3 |
| TCGA-3X-AAVA | Dead | 14.63 | 1 | 1 | 1 | 3 |
| TCGA-ZH-A8Y6 | Alive | 17.06 | 1 | 1 | 1 | 3 |
Taking into account the following clinical
parameters: Sex, age at diagnosis, stage, T stage, lymph node
status, metastasis and histological type, univariate and
multivariate Cox regression analysis was used to test the effect of
the three-miRNA signature (high-risk vs. low-risk) on OS. In
univariate analysis, stage [hazard ratio (HR)=3.104, P=0.040] and
three-miRNA signature (HR=6.013, P<0.0001) were associated with
OS in cholangiocarcinoma patients. In multivariate analysis,
three-miRNA signature (HR=6.124, P<0.0001) was revealed to be an
independent prognostic factor in cholangiocarcinoma patients
(Table V).
 | Table V.Univariate and multivariate analyses
of parameters associated with overall survival. |
Table V.
Univariate and multivariate analyses
of parameters associated with overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex |
| (male
vs. female) | 1.387
(0.544–3.534) | 0.494 |
|
|
| Age at
diagnosis |
| (>60
vs. ≤60) | 0.911
(0.350–2.375) | 0.849 |
|
|
| Stage |
| (III+IV
vs. I+II) | 3.104
(1.051–9.169) | 0.040 | 2.983
(0.981–9.072) | 0.054 |
| T stage |
| (T3+T4
vs. T1+T2) | 0.660
(0.150–2.896) | 0.582 |
|
|
| Lymph node
status |
| (N1 vs.
N0) | 2.289
(0.602–8.700) | 0.224 |
|
|
| Metastasis |
| (M1 vs.
M0) | 1.650
(0.462–5.891) | 0.440 |
|
|
| Histological
type |
|
(hilar+distal vs.
intrahepatic) | 1.197
(0.343–4.172) | 0.778 |
|
|
| Three-miRNA
signature |
| (high
risk vs. low risk) | 6.013
(2.621–13.796) | <0.0001 | 6.124
(2.582–14.525) | <0.0001 |
Three-miRNA signature verification in
the validation cohort
In order to validate the performance of the
prognostic miRNA signature, it was tested on the GSE53870 dataset
derived from GEO database. As displayed in Fig. 5, miR-22 and miR-551b were observed
to be markedly associated with OS in cholangiocarcinoma patients
(P<0.05), while miR-10b was found to be marginally significant
with patient OS (P=0.0511). Subsequently, we used the same
risk-score formula to calculate three-miRNA signature index for
each of the 63 patients and dichotomized them into low-risk and
high-risk group. Notably, these two groups were surprisingly
associated with patient OS. Based on the three-miRNA signature
index, >40% patients in the low-risk group showed 5-year
survival, while none of the patients survived >5 years in the
high-risk group. In addition, the median survival of the low-risk
group was significantly longer than that of high-risk group (34.20
vs. 12.80 months). These findings were similar to what was noted in
TCGA data. Collectively, these results proved the accuracy of the
prognostic miRNAs signature.
KEGG pathway enrichment and GO
annotation of three miRNA predicted genes
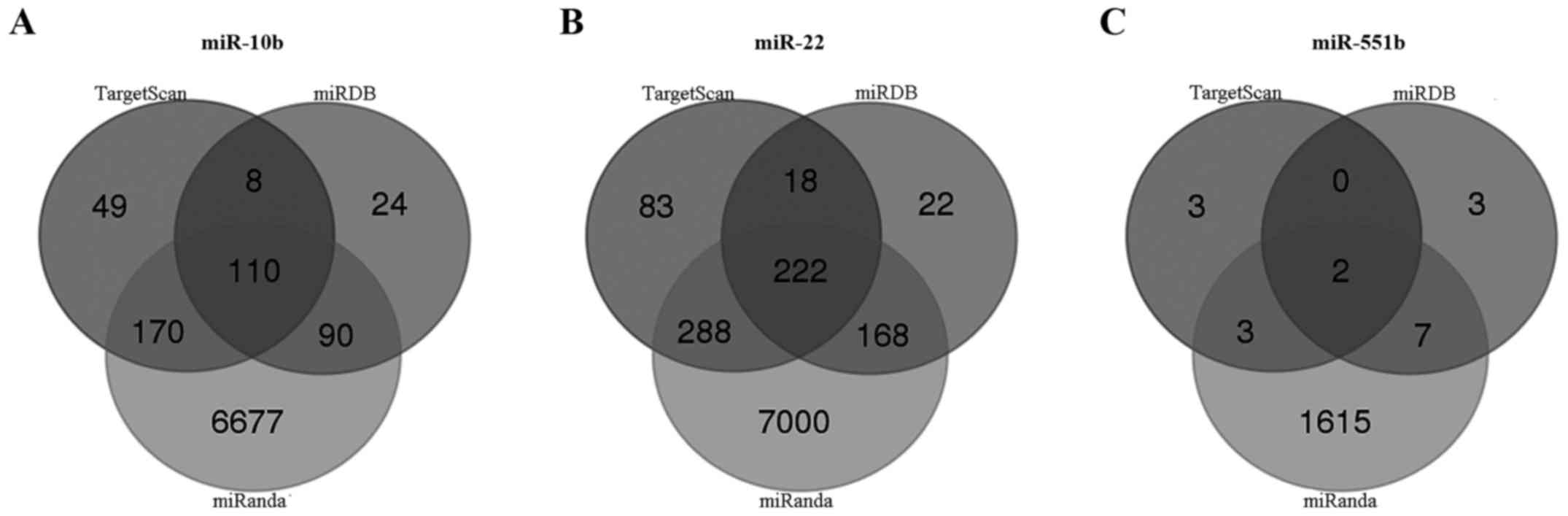
TargetScan, miRDB and miRanda online analysis tools
were used to predict the target genes of these three miRNAs
(miR-10b, miR-22 and miR-551b). A total of 110 overlapping genes of
miR-10b, 222 overlapping genes of miR-22, and 2 overlapping genes
of miR-551b were identified (Fig.
6). Then, enrichment analysis was performed to elucidate the
biological function of consensus target genes. Notably,
cancer-related pathways were found to be intensely activated
according to KEGG results, including small cell lung cancer,
chronic myeloid leukemia, prostate cancer, glioma, miRNAs in cancer
and proteoglycans in cancer. We hypothesized that these target
genes play pivotal roles in various types of cancers. Furthermore,
target genes were significantly enriched in the mTOR, FoxO and
HIF-1 signaling pathways (Fig. 7A).
The GO biological process terms were mainly enriched in the
regulation of metabolism, modification and transcription (Fig. 7B), which indicated that these three
miRNAs may be closely associated with biological function and gene
expression.
Effect of miR-551b overexpression on
the proliferation and apoptosis in HUCCT1 cells
To further validate this conclusion, we first
detected the expression of three miRNAs by RT-qPCR. According to
the data in Fig. 8, two of the
three tested miRNAs (miR-22 and miR-551b) yielded results quite
similar to those of the TCGA data. Although the performance of
miR-10b was not well-repeated, the direction of change was similar
to that noted in the TCGA data. These results indicated that the
differentially expressed miRNAs we identified by RNA-Seq data were
reliable. As miR-551b is the least studied member among these three
miRNAs in cancer, we here focused on its biological function in
cholangiocarcinoma. miR-551b mimics were transfected into HUCCT1
cells to upregulate the expression of miR-551b. Successful
overexpression of miR-551b in HUCCT1 cells was confirmed by RT-qPCR
(Fig. 9A). MTT and colony formation
assays revealed that overexpression of miR-551b significantly
inhibited proliferation (Fig. 9B)
and colony formation in HUCCT1 cells (Fig. 9C). In addition, flow cytometry
analysis showed that overexpression of miR-551b significantly
induced apoptosis in the HUCCT1 cells (Fig. 9D).
Discussion
In the last decade, miRNAs, as the master modulators
of multiple biological and pathological processes, are a ‘hot’
research topic in the field of cancer development. miRNAs are
regarded as a novel group of disease biomarkers for the stability
and universality in human tissues (19). Currently, growing investigations
have demonstrated specific miRNA profiles in multiple cancers,
emphasizing the pivotal roles of miRNAs in the initiation and
progression of cancer, including cholangiocarcinoma. Previous
studies have demonstrated that many miRNAs are crucial for the
initiation, progression, and metastasis of cholangiocarcinoma by
regulating various processes, including cancer cell proliferation,
apoptosis, adhesion, cell cycle arrest, migration and invasion
(20–23). To date, several studies have
identified a number of miRNAs with prognostic value in
cholangiocarcinoma, such as miR-126 (24), miR-192 (25), miR-26a (26), miR-203 (27), miR-106a (28) and miR-29a (29). Unfortunately, due to molecular and
clinical heterogeneity in different studies, the relatively limited
numbers of miRNAs to investigate, as well as the methodological
differences in detection and analysis, there still exist some
restrictions for applying the above specific miRNAs for
prognosis.
TCGA was constructed to contain a wide assortment of
high-throughput experimental data, which are available and will be
valuable to researchers worldwide. In our study, we analyzed
high-throughput data downloaded from TCGA database, and eventually
obtained 100 differentially expressed miRNAs between
cholangiocarcinoma and normal tissues, of which 54 were upregulated
and 46 were downregulated. Subsequently, we evaluated the
prognostic value of each differentially expressed miRNA. According
to previous research, the performance of a single biomarker in
predicting survival across the datasets is unstable, while the
combination of biomarkers increases the performance (30). Therefore, we established a novel
three-miRNA signature (high-risk vs. low-risk) with excellent
prognostic performance for cholangiocarcinoma patients. Although no
statistically significant associations were observed between our
three-miRNA signature and other clinical parameters (data not
shown), it was then identified to be an independent prognostic
factor and was successfully validated in an independent cohort from
the GEO database.
Emerging evidence has demonstrated that numerous
miRNAs are aberrantly expressed (upregulated or downregulated) in
various cancers (31). Markedly,
miRNAs show differential effects in multiple cancers, that is, they
serve not only as tumor suppressors, but also as oncogenic
promoters to hinder or aggravate cancer formation and malignant
transformation. miR-10b, first reported as an oncogene in breast
cancer, was found to induce the invasion and metastasis of breast
cancer cells (32). Notably, a
previous study also suggested that miR-10b may be a tumor
suppressor in patients with gastric cancer and the lower level of
miR-10b was detected in advanced stage small-cell carcinoma of the
cervix patients compared to the early ones (33). Furthermore, miR-22, located on
chromosome 17p13.3 (34), was
reported to retard cellular growth, invasion and metastasis in
cervical and breast cancer through inducing p53 expression and
concurrently targeting SIRT1, CDK6 as well as Sp1 to activate pRb
signaling pathway (35). However,
Budd et al (36) reported
that miR-22 in prostate cancer was overexpressed and promoted
prostate cancer tumorigenesis by directly targeting PTEN. As for
miR-551b, Lin et al (37)
reported that it was upregulated in lung adenocarcinoma tissues
compared to normal tissues and high level of miR-551b predicted a
longer survival. Conversely, Song et al (38) found that miR-551b was downregulated
in gastric cancer and suppressed EMT and metastasis in gastric
cancer by inhibiting ERBB4. Furthermore, the functions of these
three miRNAs in cholangiocarcinoma were poorly investigated. The
expression and clinical information have provided us some clues to
investigate the roles of these miRNAs in cholangiocarcinoma. Hence,
we explored the biological functions of miR-551b in HUCCT1 cells
and found that miR-551b overexpression inhibited proliferation and
induced apoptosis, which was similar to that noted in Yuan et
al recent study of gastric cancer (39).
To gain a deep insight into the molecular mechanisms
of these three miRNAs, we predicted their target genes and analyzed
the related pathways and GO annotations. Abnormal signaling
pathways play crucial roles in the pathogenesis and progression of
cholangiocarcinoma. We found that three miRNAs regulated several
key signaling pathways, including mTOR, FoxO and HIF-1 signaling
pathway. It has been well acknowledged that the PI3K/Akt/mTOR
signaling pathway plays an important role in cholangiocarcinoma,
and inhibition of mTOR kinase activity may be a viable approach for
future application in patients with cholangiocarcinoma (40). FoxO transcription factors have been
reported to play vital roles in tumorigenesis and drug resistance.
Guan et al (41) reported
that FoxO3 inactivation promoted human cholangiocarcinoma
tumorigenesis and chemoresistance through Keap1-Nrf2 signaling. As
for hypoxia inducible factor-1 (HIF-1), a family of heterodimeric
proteins which includes HIF-1α and HIF-1β subunits, has been
identified to play an important role in initiation and progression
of multiple cancers. Thongchot et al (42) reported that positive expression of
HIF-1α enhanced metastasis and predicted a poor prognosis of
cholangiocarcinoma. Therefore, further molecular investigations are
needed to confirm these predictions, and may provide new
therapeutic interventions in cholangiocarcinoma.
However, there are some limitations in interpreting
the above results. Firstly, a larger sample size was required to
validate our findings. Secondly, the miRNA expression profiles were
detected from bile duct tissues, which may not accurately reflect
the levels of miRNAs in saliva, serum, urine or stool. Hence, we
may need to explore the miRNAs signature in the above-mentioned
samples since they are conveniently available for monitoring.
In conclusion, through performing an integrative
analysis for differentially expressed miRNAs accompanied with
relevant clinical data, we established a novel three-miRNA
signature as an alternative prognostic predictor for
cholangiocarcinoma patients. Further investigations are required to
validate our findings and further functional studies are also
needed to explore the potential molecular mechanisms of these
miRNAs in cholangiocarcinoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Scientific Foundation of China (nos. 81402971,
81472248 and 81672434).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JC, QM and WD designed the study. JC, LS and JL
collected the data. JC performed the bioinformatical analysis. CZ
and LC performed the statistical analysis. JC wrote the main
manuscript and prepared all figures. KC, BY and WQ contributed to
the revision of the manuscript and were also involved in the
conception of the study. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Squadroni M, Tondulli L, Gatta G, Mosconi
S, Beretta G and Labianca R: Cholangiocarcinoma. Crit Rev Oncol
Hematol. 116:11–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blechacz B: Cholangiocarcinoma: Current
knowledge and new developments. Gut Liver. 11:13–26. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saha SK, Zhu AX, Fuchs CS and Brooks GA:
Forty-year trends in cholangiocarcinoma Incidence in the U.S:
Intrahepatic disease on the rise. Oncologist. 21:594–599. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fairweather M, Balachandran VP and
D'Angelica MI: Surgical management of biliary tract cancers. Chin
Clin Oncol. 5:632016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghouri YA, Mian I and Blechacz B: Cancer
review: Cholangiocarcinoma. J Carcinog. 14:12015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu J, Zhao J and Zhang R: Four microRNAs
signature for survival prognosis in colon cancer using TCGA Data.
Sci Rep. 6:383062016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ge YZ, Wu R, Xin H, Zhu M, Lu TZ, Liu H,
Xu Z, Yu P, Zhao YC, Li MH, et al: A tumor-specific microRNA
signature predicts survival in clear cell renal cell carcinoma. J
Cancer Res Clin Oncol. 141:1291–1299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang B, Li Y and Wang T: A three miRNAs
signature predicts survival in cervical cancer using bioinformatics
analysis. Sci Rep. 7:56242017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Puik JR, Meijer LL, Le Large TY, Prado MM,
Frampton AE, Kazemier G and Giovannetti E: miRNA profiling for
diagnosis, prognosis and stratification of cancer treatment in
cholangiocarcinoma. Pharmacogenomics. 18:1343–1358. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu HT and Gao P: The roles of microRNAs
related with progression and metastasis in human cancers. Tumour
Biol. 2016.(Epub ahead of print). View Article : Google Scholar
|
|
13
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chandran UR, Medvedeva OP, Barmada MM,
Blood PD, Chakka A, Luthra S, Ferreira A, Wong KF, Lee AV, Zhang Z,
et al: TCGA Expedition: A data acquisition and management system
for TCGA Data. PLoS One. 11:e01653952016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff finder: A
comprehensive and straightforward Web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang MY, Li SH, Huang GL, Lin GH, Shuang
ZY, Lao XM, Xu L, Lin XJ, Wang HY and Li SP: Identification of a
novel microRNA signature associated with intrahepatic
cholangiocarcinoma (ICC) patient prognosis. BMC Cancer. 15:642015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Varkonyi-Gasic E, Wu R, Wood M, Walton EF
and Hellens RP: Protocol: A highly sensitive RT-PCR method for
detection and quantification of microRNAs. Plant Methods. 3:122007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Tang H, Yin S and Dong C:
Downregulation of microRNA-138 enhances the proliferation,
migration and invasion of cholangiocarcinoma cells through the
upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep. 29:2046–2052.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu L, Huang F, Deng G, Nie W, Huang W, Xu
H, Zheng S, Yi Z and Wan T: MicroRNA-212 targets FOXA1 and
suppresses the proliferation and invasion of intrahepatic
cholangiocarcinoma cells. Exp Ther Med. 13:21092017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamanaka S, Campbell NR, An F, Kuo SC,
Potter JJ, Mezey E, Maitra A and Selaru FM: Coordinated effects of
microRNA-494 induce G2/M arrest in human cholangiocarcinoma. Cell
Cycle. 11:2729–2738. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu N, Jiang F, He TL, Zhang JK, Zhao J,
Wang C, Jiang GX, Cao LP, Kang PC, Zhong XY, et al: The Roles of
MicroRNA-122 overexpression in inhibiting proliferation and
invasion and stimulating apoptosis of human cholangiocarcinoma
cells. Sci Rep. 5:165662015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McNally ME, Collins A, Wojcik SE, Liu J,
Henry JC, Jiang J, Schmittgen T and Bloomston M: Concomitant
dysregulation of microRNAs miR-151-3p and miR-126 correlates with
improved survival in resected cholangiocarcinoma. HPB (Oxford).
15:260–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Silakit R, Loilome W, Yongvanit P, Chusorn
P, Techasen A, Boonmars T, Khuntikeo N, Chamadol N, Pairojkul C and
Namwat N: Circulating miR-192 in liver fluke-associated
cholangiocarcinoma patients: A prospective prognostic indicator. J
Hepatobiliary Pancreat Sci. 21:864–872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang LJ, Zhang KL, Zhang N, Ma XW, Yan SW,
Cao DH and Shi SJ: Serum miR-26a as a diagnostic and prognostic
biomarker in cholangiocarcinoma. Oncotarget. 6:18631–18640.
2015.PubMed/NCBI
|
|
27
|
Li J, Gao B, Huang Z, Duan T, Li D, Zhang
S, Zhao Y, Liu L, Wang Q, Chen Z and Cheng K: Prognostic
significance of microRNA-203 in cholangiocarcinoma. Int J Clin Exp
Pathol. 8:9512–9516. 2015.PubMed/NCBI
|
|
28
|
Cheng Q, Feng F, Zhu L, Zheng Y, Luo X,
Liu C, Yi B and Jiang X: Circulating miR-106a is a novel prognostic
and lymph node metastasis indicator for cholangiocarcinoma. Sci
Rep. 5:161032015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Deng Y and Chen Y: Increased expression of
miR-29a and its prognostic significance in patients with
cholangiocarcinoma. Oncol Res Treat. 40:128–132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du F, Yuan P, Zhao ZT, Yang Z, Wang T,
Zhao JD, Luo Y, Ma F, Wang JY, Fan Y, et al: A miRNA-based
signature predicts development of disease recurrence in HER2
positive breast cancer after adjuvant trastuzumab-based treatment.
Sci Rep. 6:338252016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fkih M, 'hamed I, Privat M, Ponelle F,
Penault-Llorca F, Kenani A and Bignon YJ: Identification of
miR-10b, miR-26a, miR-146a and miR-153 as potential triple-negative
breast cancer biomarkers. Cell Oncol. 38:433–442. 2015. View Article : Google Scholar
|
|
33
|
Huang L, Lin JX, Yu YH, Zhang MY, Wang HY
and Zheng M: Downregulation of six microRNAs is associated with
advanced stage, lymph node metastasis and poor prognosis in small
cell carcinoma of the cervix. PLoS One. 7:e337622012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Li Y, Ding M, Zhang H, Xu X and
Tang J: Molecular mechanisms and clinical applications of miR-22 in
regulating malignant progression in human cancer (Review). Int J
Oncol. 50:345–355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu D, Takeshita F, Hino Y, Fukunaga S,
Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A,
et al: miR-22 represses cancer progression by inducing cellular
senescence. J Cell Biol. 193:409–424. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Budd WT, Seashols-Williams SJ, Clark GC,
Weaver D, Calvert V, Petricoin E, Dragoescu EA, O'Hanlon K and
Zehner ZE: Dual action of miR-125b as a tumor suppressor and
OncomiR-22 promotes prostate cancer tumorigenesis. PLoS One.
10:e01423732015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin K, Xu T, He BS, Pan YQ, Sun HL, Peng
HX, Hu XX and Wang SK: MicroRNA expression profiles predict
progression and clinical outcome in lung adenocarcinoma. Onco
Targets Ther. 9:5679–5692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Song G, Zhang H, Chen C, Gong L, Chen B,
Zhao S, Shi J, Xu J and Ye Z: miR-551b regulates
epithelial-mesenchymal transition and metastasis of gastric cancer
by inhibiting ERBB4 expression. Oncotarget. 8:45725–45735.
2017.PubMed/NCBI
|
|
39
|
Yuan H, Chen Z, Bai S, Wei H, Wang Y, Ji
R, Guo Q, Li Q, Ye Y, Wu J, et al: Molecular mechanisms of lncRNA
SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett.
418:84–96. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ewald F, Norz D, Grottke A, Hofmann BT,
Nashan B and Jucker M: Dual inhibition of PI3K-AKT-mTOR- and
RAF-MEK-ERK-signaling is synergistic in cholangiocarcinoma and
reverses acquired resistance to MEK-inhibitors. Invest New Drugs.
32:1144–1154. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guan L, Zhang L, Gong Z, Hou X, Xu Y, Feng
X, Wang H and You H: FoxO3 inactivation promotes human
cholangiocarcinoma tumorigenesis and chemoresistance through
Keap1-Nrf2 signaling. Hepatology. 63:1914–1927. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thongchot S, Yongvanit P, Loilome W,
Seubwai W, Phunicom K, Tassaneeyakul W, Pairojkul C, Promkotra W,
Techasen A and Namwat N: High expression of HIF-1alpha, BNIP3 and
PI3KC3: Hypoxia-induced autophagy predicts cholangiocarcinoma
survival and metastasis. Asian Pac J Cancer Prev. 15:5873–5878.
2014. View Article : Google Scholar : PubMed/NCBI
|