Introduction
Nasopharyngeal carcinoma (NPC) is a type of cancer
arising from the nasopharynx epithelium and has a high prevalence
in south Asia and Africa. Its etiological agent includes
Epstein-Barr virus (EBV) infection, environmental and diet factors
and genetic susceptibility (1). Due
to its insidious location and lack of early symptoms, NPC patients
tend to present with a late-stage diagnosis. Although NPC is
radiosensitive, the patient 5-year survival rate remains low,
largely due to regional lymph node and distant metastasis and
locoregional recurrence. The outcomes of advanced-stage NPC are
markedly worse compared with those in earlier stages (2). Thus, identifying specific biomarkers
with diagnostic value and understanding the molecular mechanisms
that regulate the invasion and metastasis of NPC is urgently
needed.
MicroRNAs (miRNAs) are small non-coding
single-stranded RNAs of 19–25 nucleotides in length, which regulate
~60% of gene expression by binding to the 3′ untranslated region
(UTR) of the mRNA targets through the seed sequence (3). A growing body of literature indicates
that miRNAs play important roles in most biological processes, such
as development, proliferation, differentiation, cell cycle control
and cell death, tumorigenesis and adaptation to stress (4). Thus, miRNAs have emerged as potential
biomarkers or therapeutic targets for human diseases including
cancer.
To date, aberrant miRNA expression has been reported
in the development and progression of NPC. For example, miR-124
(5), miR-125a-5p (6) and miR-101 (7) acted as tumor suppressors by inhibiting
NPC cell growth, or by promoting cell apoptosis. miR-200a and
miR-200b suppressed NPC cell migration and invasion by targeting
ZEB2, CTNNB1 and Notch1 (8,9). miR-139-5p, miR-3188 and miR-29c were
reported to enhance the sensitivity of NPC to chemotherapy and
radiotherapy (10–12). Conversely, some miRNAs can act as
oncogenes in NPC. Upregulation of miR-19a and miR-222 promoted NPC
cell proliferation by directly targeting transforming growth factor
β receptor 2 or PTEN (13,14).
In our previous study, whole-genome small RNA deep
sequencing was performed on NPC cell lines C666-1, CNE2 and the
non-neoplastic cell line NP69, to characterize miRNA expression
profiles in NPC cells (unpublished data). The aberrantly expressed
miRNAs were subsequently validated by real-time quantitative PCR in
NPC and non-cancerous nasopharyngitis biopsies. Of these
differentially expressive miRNAs, miR-342-3p was identified to be
significantly downregulated in NPC cell lines and tissues. However,
thus far, the expression profile of miR-342-3p and its role in NPC
have not been characterized.
In the present study, we investigated the
tumor-suppressive roles of miR-342-3p in NPC cells. We revealed
that overexpression of miR-342-3p inhibited NPC cell proliferation
and invasion by directly targeting Cdc42.
Materials and methods
Patients and samples
Twenty-two NPC tissue samples and 15 nasopharyngitis
tissues were collected and total RNA was extracted for further
RT-qPCR assay. Formalin-fixed, paraffin-embedded tissues (FFPTs) of
10 NPC tissues were acquired from NPC patients who were firstly
diagnosed with NPC at the Sun Yat-sen University Cancer Center
(SYSUCC; Guangzhou, China) from January 2007 to December 2007. The
patients were histologically and clinically diagnosed with NPC and
assessed according to the TNM staging of The International Union
against Cancer. None of the patients were subjected to radiotherapy
or chemotherapy prior to biopsy sampling. The present study was
approved by the Research Ethics Committee of SYSUCC, and written
informed consent was obtained from all patients.
Immunohistochemical staining
Paraffin sections were prepared from NPC patients
and immunohistochemistry (IHC) assays were performed according to
the manufacturer's instructions to detect the protein expression.
The antibody rabbit anti-Cdc42 (1:200; cat. no. 10155-AP-1) was
purchased from Proteintech Group Inc., (Wuhan, China). In brief,
slides with paraffin sections underwent deparaffinage and aquation.
Then 3% hydrogen peroxide was used to block endogenous peroxidase
activity and antigen retrieval was performed by boiling slides in
ethylene diamine tetraacetic acid (EDTA) (1 mmol/l, pH 8.0). The
slides were incubated with the primary antibody anti-Cdc42
overnight at 4°C. The staining score was assessed by two
pathologists independently. The intensities were graded as 0
(negative), 1 (weakly positive), 2 (moderately positive) and 3
(strongly positive). The abundance of positive cells was graded
from 0 to 100%. The staining score was determined using the
following formula: Overall scores = percentage score × intensity
score × 100.
NPC cell culture and transfection
The human NPC cell lines C666-1, SUNE1, 6-10B and
5-8F used in the present study, were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10%
fetal bovine serum (FBS; Invitrogen Life Technologies; Thermo
Fisher Scientific, Inc.). The human immortalized nasopharyngeal
epithelial cell line, NP69, was grown in defined-keratinocyte
serum-free medium (KSFM) supplemented with EGF (Invitrogen Life
Technologies; Thermo Fisher Scientific, Inc.). The aforementioned
cells were cultured in a humified atmosphere with 5% CO2
at 37°C. All NPC cell lines and NP69 cell were provided generously
by Professor Musheng Zeng (Sun Yat-sen University Cancer Center,
Guangzhou, China). All cells were negatively tested for mycoplasma
contamination before use, and authenticated based on STR
fingerprinting before use at The Medicine Lab of Forensic Medicine
Department of Sun Yat-sen University. miR-342-3p mimics were
synthesized by Suzhou GenePharma Co., Ltd. (Suzhou, China) as
follows: miR-342-3p sense, 5′-UCUCACACAGAAAUCGCACCCGU-3′ and
antisense, 5′-GGGUGCGAUUUCUGUGUGAGAUU-3′). miR-342-3p mimics were
transfected into 6-10B cells at a final concentration of 15 nM
using Lipofectamine 3000 (Invitrogen Life Technologies; Thermo
Fisher Scientific, Inc.).
Reverse quantitative polymerase chain
reaction (RT-qPCR)
Total RNA was extracted from tissues or cells with
TRIzol reagent (Invitrogen Life Technologies; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. cDNA was
obtained using GoScript Reverse Transcription Mix (Promega Corp.,
Madison, WI, USA) and Bulge-Loop™ miRNA qPCR Primer set for
miR-342-3p (Guangzhou RiboBio Co., Ltd., Guangzhou, China). qPCR
was performed with GoTaq® qPCR Master Mix (Promega
Corp.). The standard cycling program was performed as follows:
hot-start activation: 95°C for 2 min, 1 cycle; denaturation: 95°C
for 15 sec, 40 cycles; annealing/extension: 60°C for 60 sec;
dissociation: 75°C, 1 cycle. The mRNA expression level was examined
by the 2−ΔΔCq method (15) and normalized by internal control U6.
The sequence of Cdc42 primer for RT-qPCR assay was as follows:
Forward, 5′-CCATCGGAATATGTACCGACTG-3′ and reverse,
5′-CTCAGCGGTCGTAATCTGTCA-3′.
Cell proliferation and colony
formation assay
Cell growth was determined by Cell Counting Kit-8
(CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). In
brief, 1,000 cells/well were suspended into a 96-well-plate and
pre-incubated for 24 h in a humidified incubator (5% CO2
at 37°C). Following the addition of 10 µl CCK-8 solution to each
well of the plate and incubation of the plate for 3 h, the
absorbance at 450 nm was determined using a microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments
were performed in triplicates.
Concerning the colony formation assay, 6-10B
miR-342-3p mimics, as well as their control cells were seeded into
a 6-well-plate (500 cells/well) with complete medium. The cells
were grown for 12 days at 37°C with 5% CO2. To visualize
the colony formation, the cells were fixed with methanol and
stained with 0.1% crystal violet (Beyotime Institute of
Biotechnology, Shanghai, China) and observed under an inverted
fluorescent microscope. The number of colonies >0.1 mm diameter
was counted. The colony formation of each cell line was performed
in triplicate.
Luciferase reporter assay
The wild-type (wt) or mutant (mt) Cdc42-3′UTR
luciferase reporter vector was designed on the basis of miR-342-3p
binding sites. In brief, 4×104 293T cells/well were
seeded into a 24-well plate. Following incubation for 24 h at 37°C
with 5% CO2, using Lipofectamine 3000 reagent
(Invitrogen Life Technologies; Thermo Fisher Scientific, Inc.),
1,000 ng firefly luciferase reporter plasmid was co-transfected
with 10 ng pRL-TK Renilla plasmid into cells, combined with
miR-342-3p mimics or negative control. In accordance with the
manufacturer's instructions, the luciferase and Renilla
signals were determined using the Dual-Luciferase Reporter Assay
kit (Promega Corp.). The experiments were repeated independently
three times.
Wound healing and invasion assay
A scratch wound healing assay was performed to
assess the cell migration rate. Cells transfected with miR-342-3p
mimics were cultured in a 6-well plate. Subsequently, as the cells
reached sub-confluence, with a sterile micropipette tip, a
scratching wound was generated and images were captured under an
inverted fluorescent microscope at 0 and 48 h. The culture medium
was replaced by serum-free RPMI-1640 medium. The distance between
either side of the scratch in images captured by the inverted
fluorescent microscope was assessed to evaluate the cell migration
rate.
Cell invasion assay was conducted using cell culture
inserts with 8-µm PET track-etched membranes (Falcon; BD
Biosciences, Franklin Lakes, NJ, USA). Tumor cells were placed into
the upper chambers (5×104 cells/well) with 200 µl
serum-free medium, whereas the lower chambers in the 24-well-plate
contained 650 µl complete medium with 10% FBS. After 16 h, the
cells were fixed with methanol and stained with 0.1% crystal
violet. Cells in the upper chambers were removed gently and the
cells which had invaded into the membranes were imaged under an
inverted fluorescent microscope. The number of invasive cells were
counted for statistical use. All the aforementioned assays were
performed three times.
Western blot analysis
Cells were harvested and lysed in RIPA lysis buffer
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) supplemented
with phenylmethylsulfonyl fluoride (PMSF). Total protein
concentration was quantified by BCA assay kit (Beyotime Institute
of Biotechnology) and Gen5 Software (CHS 2.06; BioTek Instruments,
Winooski, VT, USA). An equal volume of protein samples (30 µg) were
subjected to 10% SDS-PAGE and transferred onto polyvinylidene
difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA).
The membranes were then incubated with the primary antibodies,
including Snail, β-catenin, vimentin, cyclin D1, cyclin D3, CDK4,
Myc, Cdc42 and GAPDH at 4°C overnight and subsequently with the
secondary antibody (anti-rabbit IgG, HRP-linked antibody; 1:5,000;
cat. no. 7054; Cell Signaling Technology, Danvers, MA, USA) for 1 h
at room temperature. The signals from protein were detected by an
advanced ECL detection kit. The rabbit monoclonal antibodies used
in the present study were purchased from Cell Signaling Technology,
including those against human Snail (1:1,000; cat. no. 3879),
β-catenin (1:1,000; cat. no. 8480), vimentin (1:1000; cat. no.
5741), cyclin D1 (1:1,000; cat. no. 2978), cyclin D3 (1:1,000; cat.
no. 2936), CDK4 (1:1,000; cat. no. 12790), c-Myc (1:1,000; cat. no.
13987) and GAPDH (1:1000; cat. no. 5174); Cdc42 (1:800; cat. no.
10155-AP-1) was purchased from Proteintech Group Inc. (Wuhan,
China).
MicroRNA sequencing and DataSets
A whole genome microRNA sequencing was carried out
by Illumina Hiseq 2500 to detect the dysregulated miRNAs in NPC
cell C666-1 and immortalized nasopharyngeal epithelial cell NP69.
Data in the GEO DataSets (GSE32960 and GSE36682) was involved in
the study (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
starBase v2.0 (http://www.lncrnablog.com/starbase-v2-0-for-decoding-rna-interaction-networks/),
miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html)
and microRNA.org (http://microrna.org/) were applied to predict the
potential targets of miR-342-3p.
Statistical analysis
All statistical analyses were performed using SPSS
software version 20.0 (IBM Corp., Armonk, NY, USA). To assess the
differences among categorical variables, one-way analysis of
variance (ANOVA)/Student-Newman-Keuls (SNK) test and
independent-sample Student's t-test and Spearman rank correlation
were the four major methods used to analyze the results of the
present study. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-342-3p is downregulated in NPC
cell lines and tissues
To explore the deregulation of miRNAs in human NPC
oncogenesis, a whole genome microRNA sequencing using Illumina
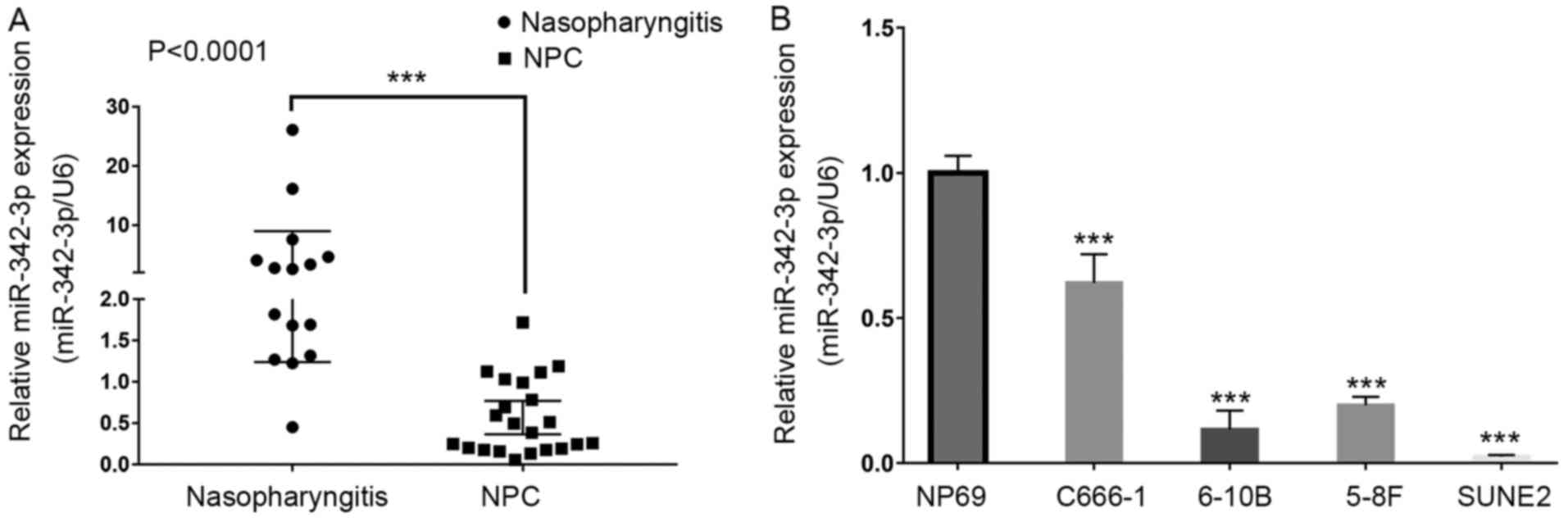
Hiseq 2500 was performed and its result revealed that the
expression level of miR-342-3p was clearly lower in NPC C666-1
cells than in non-tumor NP69 cells (Table I). To ascertain this finding, we
determined the expression level of miR-342-3p in four NPC cell
lines including C666-1, 6-10B, 5-8F and SUNE2 using RT-qPCR assays,
with the immortalized cell line NP69 used as the control. We
observed that the expression of miR-342-3p was markedly
downregulated in all four NPC cell lines compared with NP69
(Fig. 1B). In addition, the
expression of miR-342-3p was also detected in 22 NPC and 15
nasopharyngitis samples. Consistent with results in NPC cell lines,
it was observed that miR-342-3p was downregulated in NPC compared
with nasopharyngitises (P<0.0001, Fig. 1A). Furthermore, data in the GEO
DataSets (GSE32960 and GSE36682) revealed that the expression level
of miR-342-3p in NPC was significantly lower than that in normal
tissues using microarray, which was consistent with our results
(Table II).
 | Table I.miR-342-3p is downregulated in
nasopharyngeal carcinoma C666-1 cells compared with NP69 cells in
microRNA-Seq results. |
Table I.
miR-342-3p is downregulated in
nasopharyngeal carcinoma C666-1 cells compared with NP69 cells in
microRNA-Seq results.
|
| Mean expression
value |
|
|
|---|
|
|
|
|
|
|---|
| MicroRNA | NP69 | C666-1 | log2 (fold
change) | P-value |
|---|
| hsa-miR-342-3p | 327.16 | 26.36 | −3.64 (C666-1 vs.
NP69) | 0.006960605 |
 | Table II.The expression of miR-342-3p is
reduced in nasopharyngeal carcinoma compared with NP in MicroRNA
GEO datasets. |
Table II.
The expression of miR-342-3p is
reduced in nasopharyngeal carcinoma compared with NP in MicroRNA
GEO datasets.
|
|
| No. of cases |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| MicroRNA GEO
datasets accession no. | miRNA_ID | NP | NPC | log2 (fold
change) | P-value | Adjusted
P-value |
|---|
| GSE32960 | hsa-miR-342-3p | 312 | 18 | −1.479853 (NPC vs.
NP) | 7.95E-32 | 6.63E-30 |
| GSE36682 | hsa-miR-342-3p | 62 | 6 | −1.295291 (NPC vs.
NP) | 0.0004 | 0.0018 |
miR-342-3p inhibits NPC cell
proliferation in vitro
To investigate the biological function of miR-342-3p
in NPC cells, the miR-342-3p mimics were transfected into 6-10B
cells. The overexpression efficiency was validated by RT-qPCR assay
(Fig. 2A). miR-342-3p
overexpression significantly suppressed NPC cell viability as
assessed by CCK-8 assay (Fig. 2B).
In addition, compared with the scramble control, colony formation
ability of 6-10B cells was also decreased, which was induced by
miR-342-3p mimics (Fig. 2C). These
findings indicated that miR-342-3p may act as a suppressor in NPC.
In addition, cell cycle-related proteins cyclin D1, cyclin D3 and
CDK4 were downregulated following miR-342-3p overexpression
(Fig. 2D). Our results demonstrated
that miR-342-3p suppressed NPC cell proliferation in
vitro.
miR-342-3p inhibits NPC cell migration
and invasion in vitro
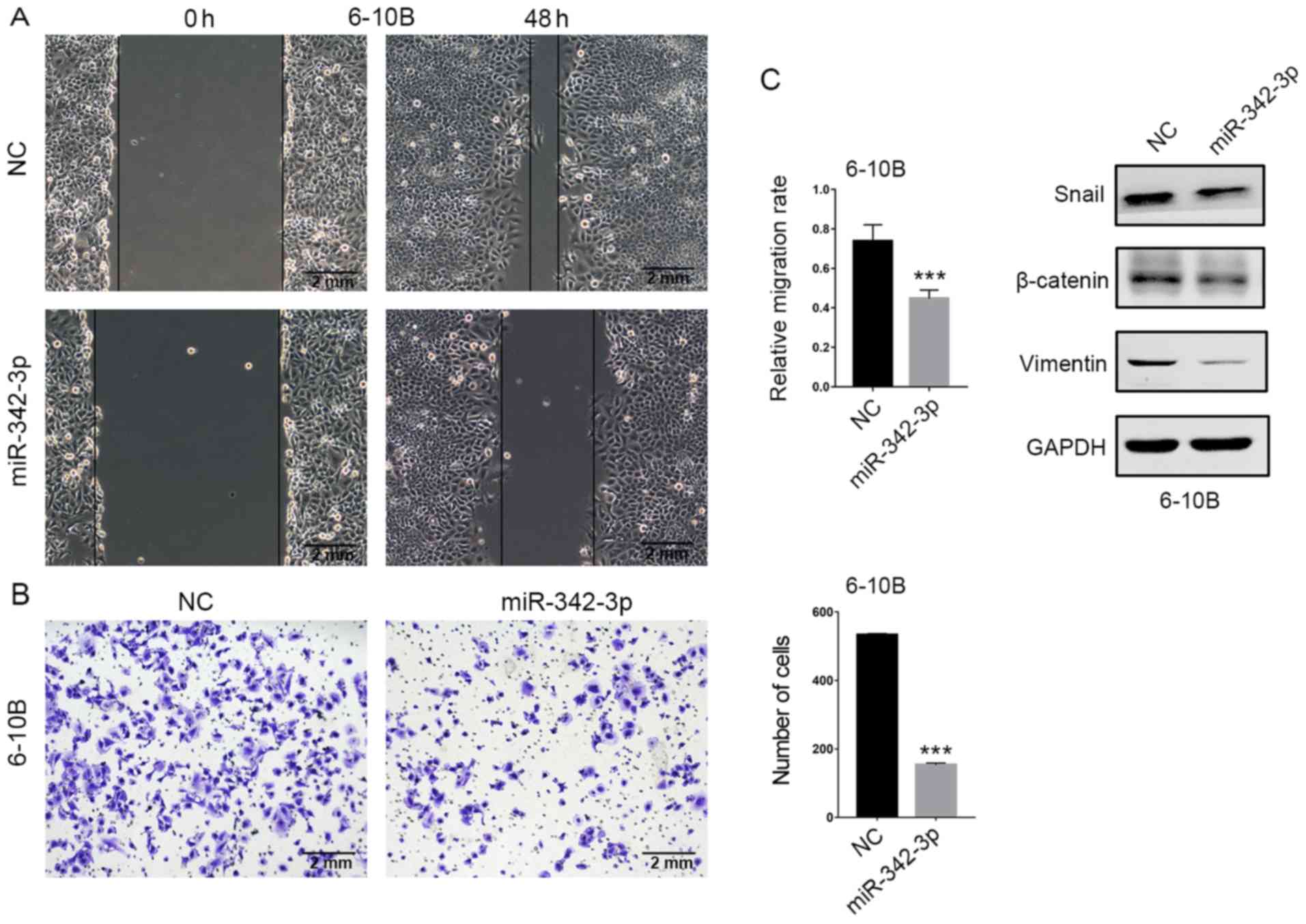
The effects of miR-342-3p overexpression on NPC cell
migration and invasion were determined by wound healing and
Transwell assays, respectively. In the wound healing assay, the gap
filling was markedly decreased in the miR-342-3p-overexpressing
6-10B cells compared with the control cells (Fig. 3A). In the Matrigel-coated Transwell
assay, the number of invading cells was markedly decreased in the
miR-342-3p-overexpressing 6-10B cells compared with control cells
(Fig. 3B). In addition, the
expression of transcription factor Snail and mesenchymal markers
β-catenin and vimentin were reduced in the
miR-342-3p-overexpressing 6-10B cells (Fig. 3C). Collectively, our results
indicated that miR-342-3p inhibited NPC cell migration and
invasion.
miR-342-3p directly targets Cdc42 in
NPC
To further explore how miR-342-3p regulated
proliferation and migration of NPC cells, we used multiple target
algorithms (starBase v2.0, miRWalk 2.0 and microRNA.org) to predict candidate targets of
miR-342-3p and observed that the 3′UTR of Cdc42 matched the ‘seed
sequence’ of miR-342-3p (Fig. 4A).
Luciferase reporter assay revealed that miR-342-3p overexpression
inhibited the luciferase activity of wild-type (wt) Cdc42 3′UTR but
not the mutant (mt) Cdc42 3′UTR (P<0.01, Fig. 4B). RT-qPCR and western blot assays
confirmed that both the mRNA and protein levels of Cdc42 were
downregulated when miR-342-3p was overexpressed in 6-10B cells
(Fig. 4C and D). Our results
determined that Cdc42 was one of the direct targets of miR-342-3p.
To further confirm the relationship between miR-342-3p and Cdc42,
we analyzed the miR-342-3p expression by RT-qPCR and Cdc42 protein
expression by immunohistochemistry assay simultaneously in 10 NPC
samples (Fig. 5A). As displayed in
Fig. 4E, miR-342-3p expression was
inversely correlated with Cdc42 expression (Fig. 4E Spearman's rank correlation
coefficient r=−0.749, P=0.013) in the same NPC specimens, which
indicated that miR-342-3p was actually involved in the in
vivo regulation of Cdc42 in NPC patients.
It has been reported that overexpression of Cdc42
was detected in different types of human cancer and correlated with
increased cancer progression and poorer outcome (16). However, the expression profile and
exact role of Cdc42 in NPC were not clear. In the present study, we
observed that the expression of Cdc42 in NPC cells was higher than
adjacent non-tumor epithelium as determined by immunohistochemistry
(Fig. 5A). To explore the role of
Cdc42 expression in NPC cells, we treated 6-10B cells with Cdc42
inhibitor ML141, and observed that ML141 treatment significantly
reduced NPC cell invasion ability (Fig.
5B and C). Our findings indicated that miR-342-3p may act as a
tumor suppressor in NPC by inhibiting Cdc42.
Discussion
miR-342 is encoded in an intron of the gene EVL,
whose protein product is an actin-associated protein involved in a
variety of processes related to cytoskeleton remodeling, cell
motility and polarity (17). A
previous study reported that both EVL and miR-342 genes were
coordinately downregulated in the majority of colorectal cancers
due to CpG island methylation upstream of the EVL/hsa-miR-342 locus
(18).
Reduced expression of miR-342 has been demonstrated
in colorectal cancer, hepatocellular carcinoma, cervical cancer,
osteosarcoma and extranodal natural killer (NK)/T-cell lymphoma,
nasal type (ENKTCL) tissues compared with normal tissues (19–22).
In addition, research has revealed that miR-342-3p may be used as a
potential therapeutic and prognostic biomarker. Breast cancer
patients with high miR-342 expression level have significantly
better survival compared to patients with low expression (9). Cittelly et al (23) reported that miR-342 was
downregulated in tamoxifen resistant breast tumor cell lines and
tamoxifen refractory human breast tumors. The expression level of
miR-342-3p was reduced in metastatic non-small cell lung cancer
samples compared to those in primary tumor samples. Functional
studies revealed that miR-342-3p exerted a tumor suppressive role
that was operated through regulation of cell proliferation,
apoptosis and cell cycle progression, invasion and migration. AGR2
(24), AEG-1 (21), Ikk-g, TAB2 and TAB3 (19), FOXM1 (20), TIAM1 (22) and DNMT1 (25) have been experimentally confirmed as
the direct targets for miR-342.
To date, the expression profile and biological
function of miR-342-3p in NPC have not been reported. Based on our
miRNA sequencing results and RT-qPCR validation, miR-342-3p was
identified to be downregulated in NPC tissues and cell lines
compared with normal controls. miR-342-3p overexpression inhibited
cell proliferation, epithelial-mesenchymal transition (EMT),
migration and invasion in NPC cells. These findings were generally
consistent with previous studies that suggested a possible tumor
suppressor function of miR-342-3p in other cancers.
In the present study, we revealed for the first time
that miR-342-3p may exert its function by specifically targeting
Cdc42 (cell division control protein) in NPC. Cdc42, a member of
the Rho GTPase family, acts as a molecular switch in multicellular
pathways. Activated Cdc42 mediates a signaling cascade leading to
the activation of more than twenty downstream effectors, and then
influences a variety of cellular responses such as cell polarity,
cytoskeleton remolding, proliferation, migration, cellular
transformation and gene expression. Cdc42 is overexpressed in a
number of human cancers, and in some instances, has been correlated
with poor prognosis. Despite the prominent association of Cdc42
activation with tumor development and progression, the regulation
and oncogenic role of Cdc42 in NPC cells remains to be elucidated.
Epstein-Barr Virus-encoded LMP1 was reported to interact with FGD4
to activate Cdc42 and thereby promote migration of NPC cells
(26). A traditional Chinese
medicine, berberine, may inhibit the anti-migration and
anti-invasion properties of NPC cells through effective
inactivation of Cdc42 (27). These
studies revealed that Cdc42 may be involved in the progression and
metastasis in NPC. In the present study, we provided evidence from
the luciferase activity assay and western blot analysis that Cdc42
was a direct target of miR-342-3p. In addition, the protein
expression levels of Cdc42 in most NPC cell lines were higher than
that in non-neoplastic cell line NP69 as determined by using
western blot analysis. Cdc42 expression in NPC biopsies was also
higher than adjacent non-tumor epithelium as revealed by
immunohistochemistry. These findings indicated that Cdc42
expression was activated in NPC and downregulation of miR-342-3p in
NPC may be the cause of Cdc42 activation. In addition, the
inhibition of Cdc42 activity by ML141 caused marked reduction of
NPC cell invasion ability, indicating that Cdc42 may play an
important role in NPC progression.
Our results revealed that reduced expression of
miR-342-3p in human NPC contributed to the enhanced proliferation
and invasion of NPC cells by direct targeting of the Cdc42 pathway.
Cdc42-selective small-molecule inhibitors have been used in a
variety of cancer models, including colon, breast and skin tumors
(28–30). Our study provided evidence that
miR-342-3p may be used as a potential biomarker and new therapeutic
target for Cdc42 regulation and NPC progression.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Nature Science Foundation of China (nos. 81772884,
81572466, 81772991 and 81629004).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
SM, HW and XFSZ conceived and designed the
experiments. SM, LS and MW wrote the study. RX and MZ collected and
prepared the tissues of NPC patients. LS and RX performed the
experiments. LS, NW, XZ and SM analyzed and interpreted the data.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Sun Yat-sen University Cancer Center (SYSUCC;
Guangzhou, China), and written informed consent was obtained from
all patients involved in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chua MLK, Wee JTS, Hui EP and Chan ATC:
Nasopharyngeal carcinoma. Lancet. 387:1012–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee AW, Ma BB, Ng WT and Chan AT:
Management of nasopharyngeal carcinoma: Current practice and future
perspective. J Clin Oncol. 33:3356–3364. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang J, Samuels DC, Zhao S, Xiang Y, Zhao
YY and Guo Y: Current research on non-coding ribonucleic acid
(RNA). Genes. 8:E3662017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Y, Li Z, Wu L, Wang Z, Wang X, Yu Y,
Zhao Q and Luo F: MiRNA-125a-5p: A regulator and predictor of
gefitinib's effect on nasopharyngeal carcinoma. Cancer Cell Int.
14:242014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu RS, Qiu EH, Zhu JJ, Wang JR and Lin HL:
MiR-101 promotes nasopharyngeal carcinoma cell apoptosis through
inhibiting Ras/Raf/MEK/ERK signaling pathway. Eur Rev Med Pharmacol
Sci. 22:150–157. 2018.PubMed/NCBI
|
|
8
|
Xia H, Ng SS, Jiang S, Cheung WK, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang X, Ni W and Lei K: miR-200b
suppresses cell growth, migration and invasion by targeting notch1
in nasopharyngeal carcinoma. Cell Physiol Biochem. 32:1288–1298.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shao Q, Zhang P, Ma Y, Lu Z, Meng J, Li H,
Wang X, Chen D, Zhang M, Han Y, et al: MicroRNA-139-5p affects
cisplatin sensitivity in human nasopharyngeal carcinoma cells by
regulating the epithelial-to-mesenchymal transition. Gene.
652:48–58. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q,
Luo X, Chen Y, Deng X, Liang Z, et al: miR-3188 regulates
nasopharyngeal carcinoma proliferation and chemosensitivity through
a FOXO1-modulated positive feedback loop with
mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 7:113092016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang JX, Qian D, Wang FW, Liao DZ, Wei
JH, Tong ZT, Fu J, Huang XX, Liao YJ, Deng HX, et al: MicroRNA-29c
enhances the sensitivities of human nasopharyngeal carcinoma to
cisplatin-based chemotherapy and radiotherapy. Cancer Lett.
329:91–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma F, Wang Z, Wang J, Liu X and Hu C:
MicroRNA-19a promotes nasopharyngeal carcinoma by targeting
transforming growth factor β receptor 2. Exp Ther Med.
14:1419–1426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu W, Chen X, Yu S, Wang R, Zhao R and Du
C: microRNA-222 promotes tumor growth and confers radioresistance
in nasopharyngeal carcinoma by targeting PTEN. Mol Med Rep.
17:1305–1310. 2018.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arias-Romero LE and Chernoff J: Targeting
Cdc42 in cancer. Expert Opin Ther Targets. 17:1263–1273. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krause M, Dent EW, Bear JE, Loureiro JJ
and Gertler FB: Ena/VASP proteins: Regulators of the actin
cytoskeleton and cell migration. Annu Rev Cell Dev Biol.
19:541–564. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Grady WM, Parkin RK, Mitchell PS, Lee JH,
Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz
AM, et al: Epigenetic silencing of the intronic microRNA
hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene.
27:3880–3888. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao L and Zhang Y: miR-342-3p affects
hepatocellular carcinoma cell proliferation via regulating NF-κB
pathway. Biochem Biophys Res Commun. 457:370–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li XR, Chu HJ, Lv T, Wang L, Kong SF and
Dai SZ: miR-342-3p suppresses proliferation, migration and invasion
by targeting FOXM1 in human cervical cancer. FEBS Lett.
588:3298–3307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang S, Liu L, Lv Z, Li Q, Gong W and Wu
H: MicroRNA-342-3p inhibits the proliferation, migration, and
invasion of osteosarcoma cells by targeting astrocyte-elevated
gene-1 (AEG-1). Oncol Res. 25:1505–1515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang H, Fan L, Zhan R, Wu S and Niu W:
Expression of microRNA-10a, microRNA-342-3p and their predicted
target gene TIAM1 in extranodal NK/T-cell lymphoma, nasal type.
Oncol Lett. 11:345–351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cittelly DM, Das PM, Spoelstra NS,
Edgerton SM, Richer JK, Thor AD and Jones FE: Downregulation of
miR-342 is associated with tamoxifen resistant breast tumors. Mol
Cancer. 9:3172010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xue X, Fei X, Hou W, Zhang Y, Liu L and Hu
R: miR-342-3p suppresses cell proliferation and migration by
targeting AGR2 in non-small cell lung cancer. Cancer Lett.
412:170–178. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu
R, Pan Z, Kang T and Huang W: MicroRNA-342 inhibits colorectal
cancer cell proliferation and invasion by directly targeting DNA
methyltransferase 1. Carcinogenesis. 32:1033–1042. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu HP, Chen CC, Wu CC, Huang YC, Liu SC,
Liang Y, Chang KP and Chang YS: Epstein-Barr virus-encoded LMP1
interacts with FGD4 to activate Cdc42 and thereby promote migration
of nasopharyngeal carcinoma cells. PLoS Pathog. 8:e10026902012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsang CM, Lau EP, Di K, Cheung PY, Hau PM,
Ching YP, Wong YC, Cheung AL, Wan TS, Tong Y, et al: Berberine
inhibits Rho GTPases and cell migration at low doses but induces G2
arrest and apoptosis at high doses in human cancer cells. Int J Mol
Med. 24:131–138. 2009.PubMed/NCBI
|
|
28
|
Bradshaw-Pierce EL, Pitts TM, Tan AC,
McPhillips K, West M, Gustafson DL, Halsey C, Nguyen L, Lee NV, Kan
JL, et al: Tumor P-glycoprotein correlates with efficacy of
PF-3758309 in in vitro and in vivo models of colorectal cancer.
Front Pharmacol. 4:222013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pelish HE, Peterson JR, Salvarezza SB,
Rodriguez-Boulan E, Chen JL, Stamnes M, Macia E, Feng Y, Shair MD
and Kirchhausen T: Secramine inhibits Cdc42-dependent functions in
cells and Cdc42 activation in vitro. Nat Chem Biol. 2:39–46. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Murray BW, Guo C, Piraino J, Westwick JK,
Zhang C, Lamerdin J, Dagostino E, Knighton D, Loi C, Zager M, et
al: Small-molecule p21-activated kinase inhibitor PF-3758309 is a
potent inhibitor of oncogenic signaling and tumor growth. Proc Natl
Acad Sci USA. 107:9446–9451. 2010. View Article : Google Scholar : PubMed/NCBI
|